Abstract
RSAT (Regulatory Sequence Analysis Tools) comprises a wide collection of modular tools for the detection of cis-regulatory elements in genome sequences. Thirteen new programs have been added to the 30 described in the 2008 NAR Web Software Issue, including an automated sequence retrieval from EnsEMBL (retrieve-ensembl-seq), two novel motif discovery algorithms (oligo-diff and info-gibbs), a 100-times faster version of matrix-scan enabling the scanning of genome-scale sequence sets, and a series of facilities for random model generation and statistical evaluation (random-genome-fragments, random-motifs, random-sites, implant-sites, sequence-probability, permute-matrix). Our most recent work also focused on motif comparison (compare-matrices) and evaluation of motif quality (matrix-quality) by combining theoretical and empirical measures to assess the predictive capability of position-specific scoring matrices. To process large collections of peak sequences obtained from ChIP-seq or related technologies, RSAT provides a new program (peak-motifs) that combines several efficient motif discovery algorithms to predict transcription factor binding motifs, match them against motif databases and predict their binding sites. Availability (web site, stand-alone programs and SOAP/WSDL (Simple Object Access Protocol/Web Services Description Language) web services): http://rsat.ulb.ac.be/rsat/.
INTRODUCTION
This article presents an update of RSAT (Regulatory Sequence Analysis Tools), a software suite integrating a wide collection of modular tools for the detection of cis-regulatory elements in genome sequences. The web site has been running without interruption since 1998 (1–4). It includes various algorithms for sequence retrieval, motif discovery, sequence scanning with regular expressions or position-specific scoring matrices, random model generation, visualization and conversion utilities (sequences, matrices, background models and feature lists). As of December 2010, the web site supports 1794 genomes (including 1120 bacteria, 88 archaea, 98 fungi, 16 metazoa and 461 phages).
The web server offers an intuitive interface, where each program can be accessed either separately, or connected to the other tools via predefined analysis flows. Programs are documented at four levels: (i) manual pages give a systematic description of the functionalities and options; (ii) ‘demo’ buttons propose typical test cases; (iii) tutorial pages provide online practical courses, with a problem-based explanation of the biological questions and the bioinformatics approaches; (iv) a series of protocols have been published for the most popular tools (5–9), to provide step-by-step instructions about option choices and result interpretation. Furthermore, the web site hosts a forum enabling direct interactions between users and developers (announcements, bug reports, wish list, help and discussion).
The tools can also be used as stand-alone applications (Unix shell) and invoked remotely as web services (SOAP/WSDL (Simple Object Access Protocol/Web Services Description Language) interface), enabling diverse combinations in programmatic workflows.
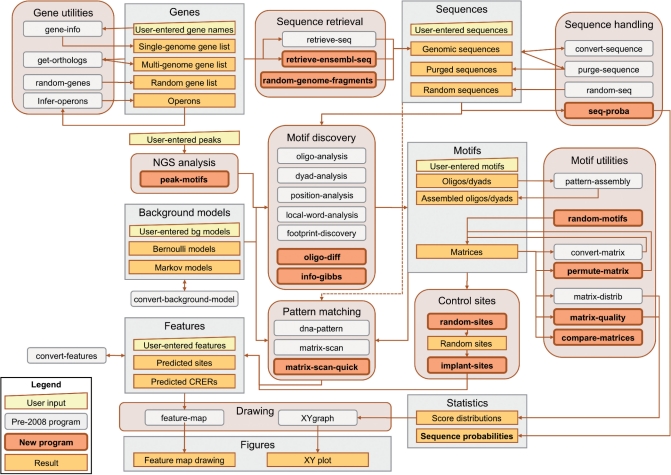
We describe hereafter 13 new programs (Table 1 and Figure 1) added to the 30 tools described in the 2008 NAR Web Software Issue (1).
Table 1.
Short description of the new programs supported on RSAT web site (since the publication in the 2008 web software issue of this journal)
| Task | Program name | Input | Output | Description |
|---|---|---|---|---|
| Sequences | retrieve-ensembl-seq | Gene names | Sequences | Retrieve upstream, downstream, intronic, exonic, UTR, transcript, mRNA, CDS or gene sequences for a list of genes from the EnsEMBL database. Multi-genome queries are supported, enabling automatic retrieval of sequences for all orthologs of query genes in selected taxa. |
| Motif discovery | oligo-diff | Two sequence sets | Differentially represented oligonucleotides | Compare oligonucleotide occurrences between two input sequence files, and return oligos that are significantly enriched in one of the files respective to the other one. |
| info-gibbs | Sequences | Over-represented motifs (matrices) | An enhanced gibbs sampler, based on a stochastic optimization of the information content of PSSMs. | |
| Pattern matching | matrix-scan-quick | Sequences+ motifs (PSSM) | Matching positions in input sequences | Scan a DNA sequence with a profile matrix. This implementation has restricted capabilities with respect to matrix-scan, but runs 100 times faster. |
| Motif comparisons | compare-matrices | Two sets of PSSM | Similarity scores + matrix alignments | Compare two collections of PSSMs, and return various similarity statistics + matrix alignments (pairwise, one-to-n). |
| Random model generation | random-genome-fragments | A genome supported in either RSAT or EnsEMBL | Randomly selected genome fragments | Select a set of fragments with random positions in a given genome, and return their coordinates and/or sequences. |
| random-motif | Randomly generated motifs (PSSM) | Generate random motifs with a given level of conservation in each column. | ||
| random-sites | Motif (PSSM) | Randomly generated sites (sequences) | Generate random sites given a motif (PSSM). | |
| implant-sites | Sequences + sites | Sequences with sites implanted | Implant given sites at random positions into given sequences. | |
| permute-matrix | 1 set of PSSM | Randomized PSSMs | Randomize a set of input matrices by permuting their columns. The resulting motifs have the same nucleotide composition and information content as the original ones. | |
| seq-proba | Sequences + background model | Sequence probability | Calculate the probability of a sequence, given a background model. Bernoulli or Markov models are supported. | |
| Work flows | matrix-quality | Motif (PSSM) + one or several sequence sets | Statistical analysis of score distributions | Evaluate the quality of a PSSM, by comparing score distributions obtained with this matrix in various sequence sets (positive set, negative set, etc.). Computes ROC curves indicating tradeoff between sensitivity and predictive value. |
| peak-motifs | Sequences | Discovered motifs + correspondences with motif databases + predicted binding sites + sequence composition | Pipeline for discovering motifs in massive ChIP-seq peak sequences. |
Note that additional programs are available as SOAP Web Services and/or with the stand-alone tools. PSSM: position-specific scoring matrix;
ROC: receiver operating characteristic.
Figure 1.
Flow chart of the Regulatory Sequence Analysis Tools (RSAT).
NEW PROGRAMS IN RSAT
Retrieving sequences from EnsEMBL on the fly
The tool retrieve-ensembl-seq (10) retrieves promoter (upstream), downstream, intronic, exonic, UTR, transcript, mRNA, Coding sequence (CDS) and gene sequences for all the organisms supported in the popular EnsEMBL database (11), and supports automated retrieval of sequences from orthologous or paralogous genes in a given taxon. Users can mask repeats, whenever these are annotated for the organism(s) of interest, as well as the coding part of retrieved sequences. Upstream and downstream sequences can be retrieved for any chosen size, relative to gene, transcript or CDS limits. By default, sequences of the chosen type are retrieved for each alternative transcript, but a specific option allows retrieval of non-redundant portions only for such sequence set.
Motif discovery
A strong focus of the RSAT suite is the development of algorithms for ab initio motif discovery in sequence sets. Three of the original algorithms have been recently enhanced in order to support the massive sets of sequences produced by next-generation sequencing: oligo-analysis (4) detects over- or underrepresented words; dyad-analysis (12) detects overrepresented spaced motifs, which are typically bound by dimeric transcription factors; position-analysis (13) detects oligonucleotides with heterogeneous positional distributions in a given sequence set.
Since 2008, two novel motif discovery algorithms have been added to the RSAT suite: oligo-diff (Defrance, M., unpublished data) detects oligonucleotides differentially represented between two input sequences and estimates their significance with the hypergeometric test; info-gibbs (14) discovers position-specific scoring matrices with high-information content using a Gibbs sampling optimization strategy.
Sequence scanning
The new tool matrix-scan-quick implements a subset of matrix-scan functionalities (9). This quick version, currently restricted to the detection of individual binding sites and their score distributions, has been optimized (with a 100-fold gain in execution time) to enable the scanning of genome-scale sequence sets. The program supports Bernoulli and higher order Markov background models, and can report the P-values of predicted sites. The additional functionalities of matrix-scan (enrichment analysis, prediction of cis-regulatory modules) are still supported by the original program, and will be optimized in the near future.
Assessing matrix quality
A common issue when working with position-specific matrices is to assess their quality, i.e. whether a matrix is able to separate correctly the true signal from the background. We have developed a workflow called matrix-quality (15) that computes theoretical and empirical score distributions to assess the reliability of position-specific matrices for predicting transcription factor binding sites. The underlying principle is to compare the score distributions obtained from various datasets in order to estimate their respective enrichment in binding sites, and this for all possible score threshold values. The theoretical distribution first provides an estimate of the false prediction rate. Empirical distributions then measure the enrichment of binding sites in various collections of sequences: known binding sites (positive control), all upstream regions of a genome, clusters of co-expressed genes, ChIP-seq peaks. As negative controls, empirical distributions are computed in the same sequence collections with column-permuted matrices. The comparison of those distributions permits the definition of score thresholds that optimize the tradeoff between sensitivity and positive predictive value. Typical applications of matrix-quality are (i) choice of the most accurate predictor among alternative matrices for the same transcription factor (e.g. coming from different databases, or built with different sets of sites); (ii) estimating the enrichment of ChIP-seq peaks for reference motifs (e.g. the pulled-down transcription factor) or for motifs discovered in the peak sequences themselves.
Motif comparison
The tool compare-matrices enables extensive comparisons between one or two collections of position-specific scoring matrices. A typical utilization is to compare a set of discovered motifs with databases of known transcription factor binding motifs. The web site includes collections from JASPAR (16), RegulonDB (17), UniPROBE (18) and DMMPMM (19). Users can also upload custom motifs, enabling the use of in-house collections or license-protected databases such as TRANSFAC (20). Another use of the custom motifs option is to compare motifs predicted by two different motif discovery algorithms.
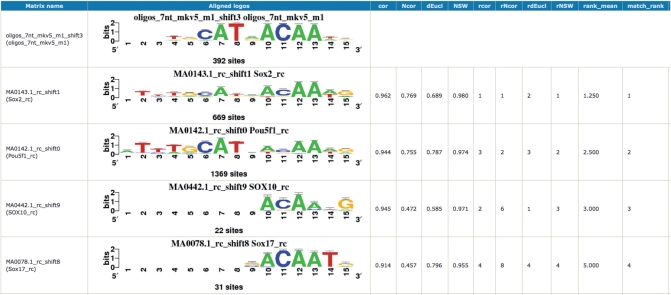
The tool integrates a wide variety of similarity/dissimilarity scoring metrics featured by other matrix comparison tools such as STAMP (21) or TOMTOM (22): sum of squared distances, Euclidian distance/similarity, Sandelin–Wasserman similarity (23), Kullback–Leibler distance as defined in (24), covariance, Pearson’s correlation. The program also computes length-normalized metrics, in order to avoid trivial alignments covering a small fraction of the motifs (e.g. the leftmost column of a query matrix aligned with the rightmost column of a reference matrix). Instead of having to choose between those metrics, the user can select several of them (or all) in order to compare their respective scores and compute a mean rank. Multiple thresholds can be specified, for instance a minimum of five aligned columns, a minimal correlation of 0.7 and a minimal normalized correlation of 0.4. Results are exported in various formats: tab-delimited file (one row per matrix comparison), motif similarity graph, HTML reports with pairwise or one-to-n aligned logos (Figure 2).
Figure 2.
Example of result from compare-matrices. Only the four best matches are displayed in the figure, the original Web page displayed five more matches. The second column displays a one-to-n alignment of matrix-logos. The next columns display multiple matching statistics, the corresponding ranks, and the mean rank. cor: Pearson’s correlation; Ncor: alignment width-normalized correlation; dEucl: Euclidian distance; NSW: width-normalized Sandelin–Wasserman similarity; rcor, rNcor, rdEucl, rNSW: ranks on the corresponding metrics; rank_mean: mean of these ranks; match_rank: rank of the alignments sorted by rank_mean.
Generating random data sets
Random data sets are highly useful to control the reliability of predictive programs. Since the early versions of RSAT, the programs random-seq and random-genes were used to build negative control sets, i.e. data sets supposed to contain no significant site (pattern matching) or motif (motif discovery). Several new tools have been added to these two programs in order to support other control types. We describe hereafter the ways to combine the previous and new tools in order to generate negative and positive control sets.
An essential parameter for building random sets is the choice of a suitable background model. The web site supports Markov models of any order between 0 and 7, calibrated with upstream non-coding sequences of all genes for each supported organism. The new program sequence-probability computes the probability of input sequences according to any of the supported background models, or yet to user-specified models.
The program random-seq generates random sequences according to Markov chains of any order. Such sequences are typically used to check the false positive rate of pattern matching algorithms (matrix-scan, matrix-scan-quick), and assess their capability to handle dependencies between adjacent nucleotides (higher order Markov models). The program random-genes enables another type of negative control, by selecting random gene sets from which natural genomic sequences (e.g. upstream non-coding) can be retrieved. Each of those genes may be regulated by some factors, but a random selection of sufficient size is unlikely to contain a significant proportion of co-regulated genes. Random gene selections thus provide a realistic framework for testing empirically the false positive rate of motif discovery algorithms. The new program random-genome-fragments selects sequences at random positions from a given genome, which can be used as negative controls for genome-wide location approaches such as ChIP-on-chip and ChIP-seq.
In addition to these negative controls, positive control sets can be built by inserting (artificial or natural) transcription factor binding sites at random positions in (artificial or natural) sequences: random-motifs generates random position-specific scoring matrices; random-sites generates binding sites on the basis of a matrix model; implant-sites inserts (real or fake) binding site sequences at random positions in (biological or randomly generated) sequences. The program permute-matrix performs random permutations among the columns of one or several input matrices. This method generates ‘realistic’ random models of motifs conserving the nucleotide composition, intra-column variability and information content of the original motifs.
A specialized workflow for analyzing motifs in ChIP-seq peak sets
peak-motifs combines several efficient motif discovery algorithms to extract transcription factor binding motifs and sites from large collection of peak sequences obtained from ChIP-seq or related technologies. Taking a full set of peak sequences as input (without size restriction), peak-motifs discovers exceptional motifs, compares them with motif databases, predicts binding site positions, and enables visualization in genome browsers (Thomas-Chollier, M., et al., submitted). In all studied cases, peak-motifs swiftly identified multiple relevant motifs. Like its constitutive modules, the whole workflow can be used as a stand-alone application, as well as SOAP/WSDL web services.
CONCLUSIONS
RSAT is one of the most comprehensive academic software suites for the analysis of cis-regulatory sequences to date. It integrates diverse, well-documented motif discovery and pattern matching modules and greatly facilitates their application to sequence sets belonging to numerous genomes, while offering particularly sophisticated means to statistically evaluate the returned motifs or sites, as well as to compare them with current knowledge (annotated genomes and motif collections). The modular conception of RSAT enables flexible and seamless module chaining to answer a variety of biological questions, problems and data types, and to address challenges coming from novel technologies. This point is particularly well illustrated by peak-motifs, which combines some of the very early tools of the suite (4,12,13) with some of the most recent ones (e.g. compare-matrices) to perform a comprehensive analysis of the huge sequence sets resulting from ChIP-seq experiments.
AVAILABILITY
The main server is located in Belgium (http://rsat.bigre.ulb.ac.be/rsat/). Mirror servers are available in Mexico (http://embnet.ccg.unam.mx/rsa-tools/), Sweden (http://liv.bmc.uu.se/rsa-tools/), France (http://tagc.univ-mrs.fr/rsa-tools/; http://rsat01.biologie.ens.fr/rsa-tools/), and South Africa (http://anjie.bi.up.ac.za/rsa-tools/) These RSAT Web servers can be freely accessed by all users without login requirement.
FUNDING
M.T-C is supported by the Alexander von Humboldt foundation. A.M-R. was supported during her Ph.D. studies (Programa de Doctorado en Ciencias Biomédicas, Universidad Nacional Autónoma de México) by a fellowship from the Consejo Nacional de Ciencia y Tecnología (Mexico). The BiGRe laboratory is funded by the European Commission through the FP7 MICROME Collaborative Project (thematic area ‘BIO-INFORMATICS-Microbial genomics and bio-informatics’, contract number 222886-2), while the collaboration between BiGRe and TAGC laboratory is supported by the Belgian Program on Interuniversity Attraction Poles, initiated by the Belgian Federal Science Policy Office, project P6/25 (BioMaGNet). The collaboration between BiGRe and ENS has been stimulated by a 2-months invitation of JvH as visiting Professor at ENS. Funding for open access charge: Publication costs were covered by the European Commission through the FP7 MICROME Collaborative Project (thematic area ‘BIO-INFORMATICS-Microbial genomics and bio-informatics’, contract number 222886-2).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We ought particular thanks to Raphaël Leplae, Sylvain Brohée and Didier Croes for their invaluable help and willingness in installing and maintaining the RSAT server and the computer environment of the BiGRe laboratory. Sylvain Brohée has also been in charge of the installation and maintenance of fungal genomes. We are also thankful to the colleagues who help us to install and maintain the RSAT mirrors: Victor Moral Chavez and Romualdo Zayas-Lagunas (Centro de Ciencias Genomicas, Cuernavaca, Mexico), Erik Bongcam-Rudloff (BMC, Uppsala, Sweden), Fourie Joubert (University of Pretoria, South Africa), François-Xavier Théodule (Université Marseille-Méditerrannée, France) and Pierre Vincens (Ecole Normale Supérieure, Paris, France).
REFERENCES
- 1.Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Helden J. Regulatory sequence analysis tools. Nucleic Acids Res. 2003;31:3593–3596. doi: 10.1093/nar/gkg567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Helden J, Andre B, Collado-Vides J. A web site for the computational analysis of yeast regulatory sequences. Yeast. 2000;16:177–187. doi: 10.1002/(SICI)1097-0061(20000130)16:2<177::AID-YEA516>3.0.CO;2-9. [DOI] [PubMed] [Google Scholar]
- 4.van Helden J, Andre B, Collado-Vides J. Extracting regulatory sites from the upstream region of yeast genes by computational analysis of oligonucleotide frequencies. J. Mol. Biol. 1998;281:827–842. doi: 10.1006/jmbi.1998.1947. [DOI] [PubMed] [Google Scholar]
- 5.Janky R, van Helden J. Discovery of conserved motifs in promoters of orthologous genes in prokaryotes. Methods Mol. Biol. 2007;395:293–308. doi: 10.1007/978-1-59745-514-5_18. [DOI] [PubMed] [Google Scholar]
- 6.Sand O, van Helden J. Discovery of motifs in promoters of coregulated genes. Methods Mol. Biol. 2007;395:329–348. doi: 10.1007/978-1-59745-514-5_21. [DOI] [PubMed] [Google Scholar]
- 7.Defrance M, Janky R, Sand O, van Helden J. Using RSAT oligo-analysis and dyad-analysis tools to discover regulatory signals in nucleic sequences. Nat. Protoc. 2008;3:1589–1603. doi: 10.1038/nprot.2008.98. [DOI] [PubMed] [Google Scholar]
- 8.Sand O, Thomas-Chollier M, Vervisch E, van Helden J. Analyzing multiple data sets by interconnecting RSAT programs via SOAP Web services-an example with ChIP-chip data. Nat. Protoc. 2008;3:1604–1615. doi: 10.1038/nprot.2008.99. [DOI] [PubMed] [Google Scholar]
- 9.Turatsinze JV, Thomas-Chollier M, Defrance M, van Helden J. Using RSAT to scan genome sequences for transcription factor binding sites and cis-regulatory modules. Nat. Protoc. 2008;3:1578–1588. doi: 10.1038/nprot.2008.97. [DOI] [PubMed] [Google Scholar]
- 10.Sand O, Thomas-Chollier M, van Helden J. Retrieve-ensembl-seq: user-friendly and large-scale retrieval of single or multi-genome sequences from Ensembl. Bioinformatics. 2009;25:2739–2740. doi: 10.1093/bioinformatics/btp519. [DOI] [PubMed] [Google Scholar]
- 11.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.van Helden J, Rios AF, Collado-Vides J. Discovering regulatory elements in non-coding sequences by analysis of spaced dyads. Nucleic Acids Res. 2000;28:1808–1818. doi: 10.1093/nar/28.8.1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.van Helden J, del Olmo M, Perez-Ortin JE. Statistical analysis of yeast genomic downstream sequences reveals putative polyadenylation signals. Nucleic Acids Res. 2000;28:1000–1010. doi: 10.1093/nar/28.4.1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Defrance M, van Helden J. info-gibbs: a motif discovery algorithm that directly optimizes information content during sampling. Bioinformatics. 2009;25:2715–2722. doi: 10.1093/bioinformatics/btp490. [DOI] [PubMed] [Google Scholar]
- 15.Medina-Rivera A, Abreu-Goodger C, Thomas-Chollier M, Salgado H, Collado-Vides J, van Helden J. Theoretical and empirical quality assessment of transcription factor-binding motifs. Nucleic Acids Res. 2011;39:808–824. doi: 10.1093/nar/gkq710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gama-Castro S, Salgado H, Peralta-Gil M, Santos-Zavaleta A, Muniz-Rascado L, Solano-Lira H, Jimenez-Jacinto V, Weiss V, Garcia-Sotelo JS, Lopez-Fuentes A, et al. RegulonDB version 7.0: transcriptional regulation of Escherichia coli K-12 integrated within genetic sensory response units (Gensor Units) Nucleic Acids Res. 2011;39:D98–D105. doi: 10.1093/nar/gkq1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robasky K, Bulyk ML. UniPROBE, update 2011: expanded content and search tools in the online database of protein-binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2011;39:D124–D128. doi: 10.1093/nar/gkq992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kulakovskiy IV, Favorov AV, Makeev VJ. Motif discovery and motif finding from genome-mapped DNase footprint data. Bioinformatics. 2009;25:2318–2325. doi: 10.1093/bioinformatics/btp434. [DOI] [PubMed] [Google Scholar]
- 20.Wingender E, Chen X, Fricke E, Geffers R, Hehl R, Liebich I, Krull M, Matys V, Michael H, Ohnhauser R, et al. The TRANSFAC system on gene expression regulation. Nucleic Acids Res. 2001;29:281–283. doi: 10.1093/nar/29.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mahony S, Benos PV. STAMP: a web tool for exploring DNA-binding motif similarities. Nucleic Acids Res. 2007;35:W253–W258. doi: 10.1093/nar/gkm272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandelin A, Wasserman WW. Constrained binding site diversity within families of transcription factors enhances pattern discovery bioinformatics. J. Mol. Biol. 2004;338:207–215. doi: 10.1016/j.jmb.2004.02.048. [DOI] [PubMed] [Google Scholar]
- 24.Aerts S, Van Loo P, Thijs G, Moreau Y, De Moor B. Computational detection of cis -regulatory modules. Bioinformatics. 2003;19(Suppl. 2):II5–II14. doi: 10.1093/bioinformatics/btg1052. [DOI] [PubMed] [Google Scholar]