Abstract
Functional interpretation of candidate gene lists is an essential task in modern biomedical research. Here, we present the 2011 update of g:Profiler (http://biit.cs.ut.ee/gprofiler/), a popular collection of web tools for functional analysis. g:GOSt and g:Cocoa combine comprehensive methods for interpreting gene lists, ordered lists and list collections in the context of biomedical ontologies, pathways, transcription factor and microRNA regulatory motifs and protein–protein interactions. Additional tools, namely the biomolecule ID mapping service (g:Convert), gene expression similarity searcher (g:Sorter) and gene homology searcher (g:Orth) provide numerous ways for further analysis and interpretation. In this update, we have implemented several features of interest to the community: (i) functional analysis of single nucleotide polymorphisms and other DNA polymorphisms is supported by chromosomal queries; (ii) network analysis identifies enriched protein–protein interaction modules in gene lists; (iii) functional analysis covers human disease genes; and (iv) improved statistics and filtering provide more concise results. g:Profiler is a regularly updated resource that is available for a wide range of species, including mammals, plants, fungi and insects.
INTRODUCTION
High-throughput technologies continue to transform biological and medical research. A decade after finalizing the human genome sequence (1), orchestrated efforts focus on exploring human variation and history (2,3), exhaustive identification of functional DNA elements (4), deciphering complex disease traits (5) and cancer (6), among others. Modern researchers routinely apply high-throughput tools like next-generation sequencing to unravel secrets of genomics, transcriptomics and proteomics and explain life, disease and death at the molecular level. As biotechnologies become exponentially more efficient and cost-effective, promising perspectives of personalized genomics and medicine emerge.
A common denominator of current high-throughput techniques is the abundance of data resulting from any single experiment. Thousands or millions of reads from genome or proteome-wide assays often highlight many hundreds of genes and proteins that are particularly informative of the experiment. Meaningful biological interpretation of these results is a crucial step. Functional profiling combines computational and statistical methods to link collections of biomolecules (genes, transcripts, proteins) with known, statistically significant functional features.
Functional profiling is an active area of research and numerous bioinformatics tools allow researchers to interpret their gene lists [e.g. (7–13), see Huang et al. (14) for a review]. Here, we are proud to present an update note of g:Profiler (15), a web server for functional interpretation and integration of gene lists in the context of versatile biological evidence. We combine rapid algorithms and rich visualization for interpreting gene lists, ranked lists and gene list collections. Our automated interpretative analysis takes advantage of systematized knowledge about gene and protein function, including Gene Ontology (GO) (16), Human Phenotype Ontology (HPO) (17) and biological pathway databases KEGG (18) and Reactome (19). We also cover experimental data like protein–protein interactions from BioGrid (20) and target sites of microRNAs [MicroCosm database (21)] and transcription factors [Transfac (22)]. Even more information can be retrieved from a large collection of microarrays from ArrayExpress (23) via gene coexpression searches, and from dozens of related organisms via gene homology searches. Frequent data updates from multispecies databases of Ensembl (24,25) and GOA (26) guarantee stable and consistent gene annotations. g:Profiler is available for 85 organisms, including common model organisms, mammals, plants, insects and fungi.
g:Profiler WEB SERVER
The g:Profiler web server comprises a collection of five closely integrated tools that perform functional profiling (g:GOSt, g:Cocoa), biomolecule integration tasks (g:Convert) and genomic data mining (g:Sorter, g:Orth). Each tool is briefly described below. A summary of existing and novel g:Profiler features can be found in Table 1.
Table 1.
Summary of g:Profiler features in 2007 and novel additions in 2011
| Feature | g:Profiler 2007 | Added in g:Profiler 2011 |
|---|---|---|
| Supported species |
|
|
| Biological evidence considered |
|
|
| Input for enrichment analysis |
|
|
| Methods |
|
|
| Tools |
|
|
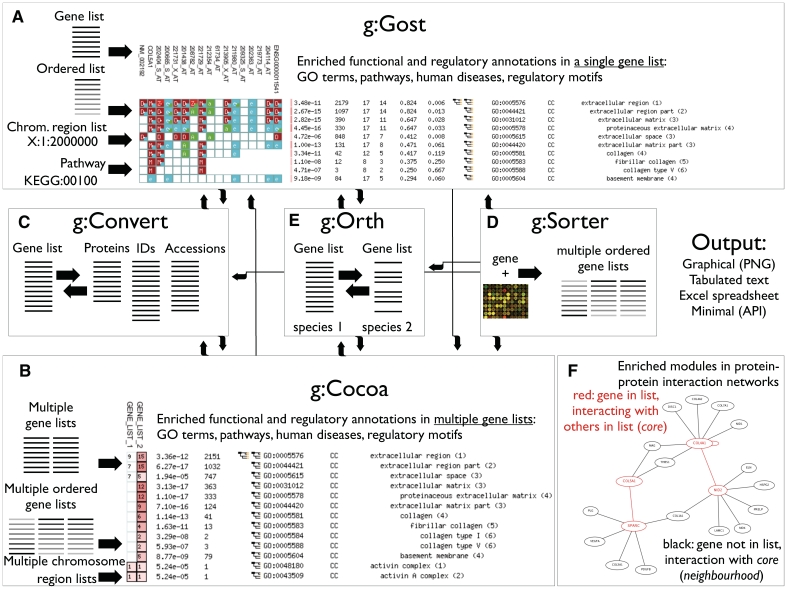
g:GOSt—gene group functional profiling
g:GOSt, the core of the g:Profiler, performs statistical enrichment analysis to provide interpretation to user-provided gene lists (Figure 1A). We study multiple sources of functional evidence, including Gene Ontology terms, biological pathways and regulatory motifs for transcription factors. In the 2011 update of g:Profiler, we added support for microRNA motifs, human disease annotations and protein–protein interactions (see below).
Figure 1.
g:Profiler tools (A–E) and data streams (arrows) for constructing analysis pipelines. (A) g:GOSt—functional profiling of gene lists, with sources of input shown on the left [also see (F)]. Output: genes are shown horizontally and annotations vertically, colours denote classes of functional evidence. (B) g:Cocoa—functional profiling of multiple gene lists. Output: gene lists are shown horizontally and annotations vertically, colours (intensity of red) denote strength of enrichment. (C) g:Convert—ID mapping service for kinds of molecules and databases. (D) g:Sorter—gene-based expression similarity search from microarrays. (E) g:Orth—mapping homologous genes of related species. (F) Identification of enriched protein–protein interaction modules in gene lists. A fragment of g:GOSt output includes a module of interacting query genes (core, red) and non-query genes interacting with the core module (neighbourhood, black).
Analysis of gene lists and significance estimation is carried out in our rapid C++ library. g:Profiler uses the widely applied hypergeometric distribution for significance estimation, and combines it with our custom multiple testing correction procedure g:SCS described earlier. This method is designed to eliminate false positives while considering the unevenly distributed structure of functional annotations, as confirmed in our simulations (15). The standard alternatives false discovery rate (FDR) and Bonferroni correction are also available.
The main input of the tool is a mixed list of genes or proteins, and the user may choose to treat the list as unordered or ordered. The user may also insert a GO or pathway ID to for annotated genes or specify chromosomal coordinates for genes in a region (see below). In case of an unordered gene list, a single enrichment analysis identifies all over-represented functional terms. The ordered option is useful when the genes are placed in some biologically meaningful order, for instance according to the differential expression in a given microarray experiment. g:Profiler then performs incremental enrichment analysis with increasingly larger numbers of genes from the top of the list (15). This optimization procedure identifies specific functional terms that associate to most dramatic changes in gene expression, as well as broader terms that characterize the gene set as a whole.
The main output of g:GOSt is a tabular graphical representation of detected functional enrichments in rows, input genes in columns and gene annotations in table cells. The visualization includes a broad spectrum of information for further consideration, including enrichment P-values, gene-to-term mappings, colour-coded evidence codes, hierarchy of enriched functional terms and peak enrichments in ordered gene lists. As a new feature, we visualize protein–protein interaction modules that are enriched in input gene lists (see below). Direct links with other g:Profiler tools, simple programmable interfaces and alternative output formats (textual, spreadsheet) allow smooth creation of analysis pipelines.
g:Cocoa—compact comparison of annotations (new in 2011)
In the 2011 g:Profiler update, we have added a new tool to the web server. g:Cocoa stands for ‘COmpact COmparison of gene Annotations’ and performs comparative analysis of multiple gene lists (Figure 1B). The input of g:Cocoa is presented in a multiline format that resembles FASTA, allowing users to construct and name multiqueries easily. Its output is a tabular graphic similar to g:GOSt, except that each column stands for a gene list rather than a single gene, and cell colours highlight significant enrichments. The compact output of g:Cocoa allows a condensed and minimal comparison of functional enrichments in dozens of gene lists.
In addition to enrichment analysis, g:Cocoa performs two further tasks for gene list comparison. First, one may use the tool to identify most meaningful gene lists in a multilist query. In that case, g:Cocoa evaluates the total statistical significance across all functional categories and orders the results accordingly. Second, one may use the tool to evaluate the functional similarity between gene lists. In that case, g:Cocoa compares the first list and all others, and orders the lists according to similarity. Total significance is quantified with log-sums of P-values, and the Euclidean distance over functional enrichments is used for similarity estimation. Such analyses have useful applications, for instance in comparing the performance of several bioinformatics algorithms in retrieving biologically meaningful results (27).
g:Convert—gene ID converter
g:Convert is a gene identifier conversion tool (Figure 1C). It uses information from Ensembl databases (24,25) to handle hundreds of types of IDs for genes, proteins, transcripts, microarray probesets for many species, experimental platforms and biological databases. The most important feature of g:Convert is flexibility—it accepts a mixed list of genes (identifiers, names, accessions) and recognizes their types automatically. Besides providing a front-end service for scientists through tabulated, textual and spreadsheet outputs, g:Convert is a unified interface for all g:Profiler tools and several others.
g:Convert accepts a gene list as input, and provides a table with converted identifiers, gene names and short descriptions as output. Our ID mapping strategy describes all biomolecules as triplets of Ensembl genes, transcripts and proteins, and stores these in a bidirectional hashtable-based index for fast performance. As a new feature in 2011, g:Convert and other g:Profiler tools unambiguously support several classes of numeric identifiers, notably EntrezGene accessions and Affymetrix Exon Array probesets.
The relevance of g:Convert to the community is illustrated by the fact that ∼30% of all web queries to g:Profiler involve g:Convert.
g:Sorter—expression similarity search
g:Sorter is a search tool for gene expression profiles (Figure 1D). It allows users to find similar gene expression profiles to their gene of interest in a large collection of public microarray datasets from ArrayExpress (23). g:Sorter is conceptually similar to our Multi Experiment Matrix (MEM) software that performs global coexpression search on large microarray collections (28). g:Sorter supports several commonly used distance measures and directions for expression similarity assessment on collections of microarrays from a single experiment. For instance, one may use positive Pearson correlation to find coexpressed genes or anticorrelation to discover genes with reverse regulatory modulation. g:Sorter analysis has been applied successfully in a number of occasions, for instance in uncovering the regulatory network of lysosomal biogenesis (29).
The main input of g:Sorter is a gene or a protein of interest (the query gene) and a microarray dataset ID from a large collection. g:Sorter links the query gene to microarray probesets and performs similarity searches. Its output features one or more gene lists that correspond to the identified coexpression or antiexpression patterns. In the 2011 g:Profiler update, we have added a convenient keyword-based search for finding relevant microarray datasets. Our search database now contains >4000 microarray experiments for 16 species.
g:Orth—orthology search
g:Orth is a tool for mapping homologous genes across related organisms (Figure 1E). The input of the tool is a list of genes of an organism. Given a selected target organism, g:Orth retrieves the genes of the target that are similar in sequence to the initial genes. g:Orth handles one-to-one and more complex orthology mappings using data from Ensembl and Ensembl Genomes. The orthology search in g:Orth has several applications in functional analysis. For instance, researchers of model organisms may retrieve human homologs of genes of interest, to study their hypotheses in the context of human health (30).
NEW DEVELOPMENTS IN g:Profiler IN 2011
Enriched protein–protein interaction modules in gene lists
g:Profiler now allows interpretation of gene lists in the context of protein–protein interaction networks (Figure 1F). We take advantage of a novel statistical strategy for identifying significant modules of interactions (subnetworks) in gene lists. Our strategy comprises a set of ‘core’ genes and a set of ‘neighbourhood’ genes. The core set includes all genes in the original input list that are connected by an interaction. The latter set includes all immediate interaction partners of the core set that do not belong to the original input. We use the hypergeometric test to decide if the input list contains significantly more interacting genes than expected, in contrast to the surrounding network neighbourhood. Intuitively, this strategy captures interaction modules (cores) in which a significant number of genes is present in the input gene list. g:Profiler also includes an extension of the above strategy for ordered gene lists. In this case, multiple cores and neighbourhoods are considered similarly to our incremental enrichment analysis, and the core with the lowest P-value is reported as the final result.
g:Profiler performs subnetwork enrichment analysis on single gene lists (g:GOSt) as well as multiple gene lists (g:Cocoa). We currently cover >300 000 protein–protein interactions from the Biogrid database (20), for seven species including human, mouse, fly and yeast. We provide a visual representation of the enriched subnetwork and its neighbourhood, and show related genes as text for further processing. Such analysis has several useful applications. For instance, we have studied yeast protein modules and physical complexes that are dysregulated in transcription factor knockout mutants (31). An example of such analysis is shown below (Case Study 2, Supplementary Figure S1).
Enrichment analysis of disease genes in human and model organisms
The g:Profiler 2011 update provides means to interpret gene lists in the context of human disease. We have imported gene annotations from the HPO, a standardized vocabulary of phenotypic abnormalities encountered in human disease (17). HPO contains information from several important sources, notably including the Online Mendelian Inheritance in Man (OMIM) (32). Below we present an example analysis that takes advantage of these data for interpreting evolution in contemporary humans.
Due to ethical constraints, a significant portion of research on human disease is conducted in model organisms like mouse and rat. As HPO only describes human genes at the time, we have extended these annotations to model organisms using sequence homology information from Ensembl. Sequence similarity alone is not sufficient for reliable inference of translational disease models. However, we believe that the detailed resources of HPO, combined with stronger sources of evidence such as curated pathways, provide useful pointers for interpreting gene lists of many versatile organisms where detailed knowledge of disease and function is still lacking.
Functional analysis of single nucleotide polymorphisms and chromosomal regions
The 2011 update of g:Profiler allows direct functional analysis of single nucleotides and chromosomal regions. Users may submit entire collections of chromosomal coordinates or ranges, and g:Profiler automatically retrieves gene lists that are located in corresponding regions. Depending on the tool, the genes will be subjected to functional profiling (g:GOSt), orthology search (g:Orth) or gene ID mapping (g:Convert). Furthermore, g:Cocoa provides a special format to insert and study multiple collections of chromosomal regions.
Such an analysis has numerous useful applications. One may explore sequence variations like single nucleotide polymorphisms (SNP), copy number variations (CNVs) and chromosomal rearrangements that are responsible for human variation and history, complex disease traits and cancer, among others. These data are rapidly accumulated by genome sequencing efforts (2,3), genome-wide association studies (5) and cancer genomics projects (6). Below we present a case study to investigate the functional significance of genomic regions that have undergone positive selection in contemporary humans (Case Study 1, Figure 2).
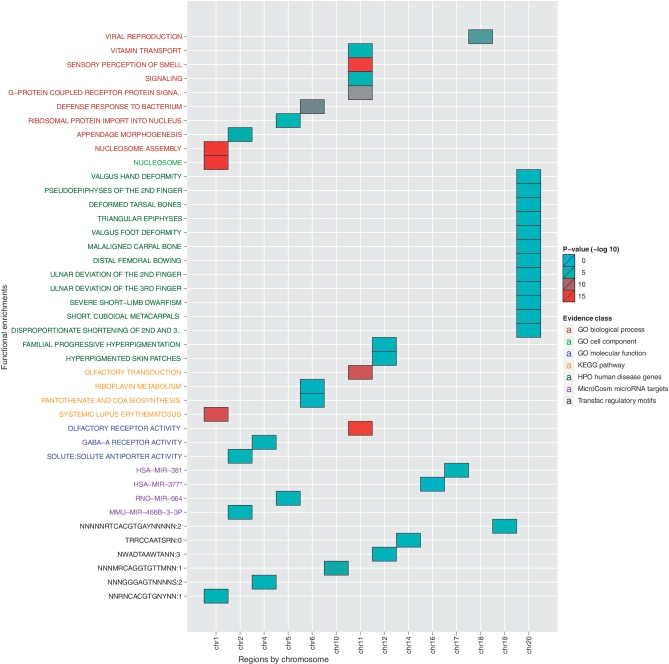
Figure 2.
Functional analysis of positively selected regions in modern human genome versus Neanderthal genome. Input genomic loci were grouped by chromosome and analysed with g:Cocoa. R was used for visualization. Enriched annotations are shown vertically and chromosomes horizontally. Coloured cells indicate statistically significant enrichments of corresponding functions (P < 0.05, red tones represent greater significance). Annotation axis labels are grouped and coloured according to data source. To avoid redundant annotations, only most significant function of any hierarchically related group is shown.
Custom statistical background for enrichment detection
In this g:Profiler update, we have improved our statistical methods and added a feature for customizing gene background in g:GOSt and g:Cocoa. Enrichment analysis involves a background set that determines the expected proportions of function-associated and non-associated genes in the input list. The default background usually corresponds to all known genes of a genome. However, such an approach may give misleading results when the user is initially focusing on a narrower set of genes. Relevant examples include microarrays with tissue-specific genes, or studies involving specific genes like transcription factors. While a default analysis of such genes would inevitably recover annotations like liver development or transcriptional regulation, customized background set of initial genes would help reduce global biases and uncover more specific annotations. An example illustrating customized backgrounds is shown below (Case Study 3, Figure 3).
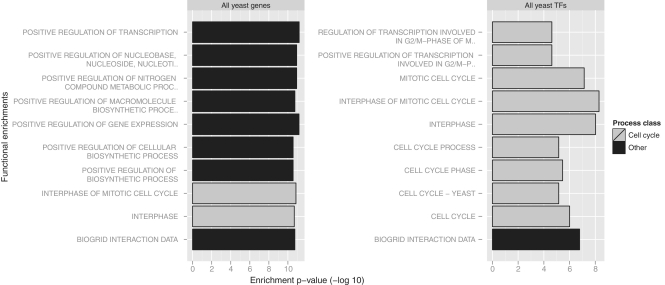
Figure 3.
Comparison of standard, global background gene list (left plot) and customized list (right plot) in functional enrichment analysis of nine core cell cycle transcription factors in yeast. Top 10 enriched categories are shown vertically and log-scale P-values horizontally. R was used for visualization. The global analysis reveals general functional enrichments of transcriptional regulation (black bars), while focused analysis with the custom background of all yeast TFs shows specific cell cycle-related terms (grey bars).
g:Profiler users may submit their backgrounds as mixed lists of gene IDs. Besides user-defined backgrounds, a few predefined backgrounds are available for precise analysis of most popular microarray platforms. Analysis with a custom background automatically discards unrelated genes and annotations to provide unbiased statistics.
Filtering of GO electronic annotations
Since the 2011 update of g:Profiler, we support filtering of GO annotations according to the quality of supporting evidence. A significant proportion of GO annotations result from automated analysis and are assigned the IEA evidence code (Inferred from Electronic Annotation). This concerns ∼36% of all functional annotations in current g:Profiler, and this proportion is even more dramatic in human data (77% of 2.7 million annotations). While IEA annotations are valuable in mapping gene functions, manual curation of experimental and computational data is generally of higher confidence. Therefore, it is sometimes advisable to exclude electronically inferred annotations from enrichment analysis and focus on stronger evidence. Excluding IEA annotations may also help reduce bias towards abundant and ubiquitous housekeeping genes like the ribosomes.
The IEA filter is enabled in g:GOSt and g:Cocoa via a single checkbox, and corresponding enrichment analyses account for altered structure of GO annotations.
g:Profiler for 85 species
The 2011 version of g:Profiler contains data for 85 species and this number is increasing with nearly every data reload. Besides mammalian and other species from Ensembl (24), we have expanded our repertoire to representatives of metazoa, plants and fungi from the Ensembl Genomes initiative (25). To our knowledge, g:Profiler is the only functional profiling software that supports such a broad collection of species. The current version of g:Profiler includes information for 50 million gene to function annotations, and provides functional profiling tools to different research communities, e.g. those of agricultural and plant genomics (33,34).
Advanced methods for automated analysis
In addition to improved experience of interactive analysis, 2011 g:Profiler update includes developments in the area of automated analysis and application programmable interfaces. All g:Profiler tools include text-based, tabulated and spreadsheet (Excel) output formats. Minimal output format with no HTML headers allows creation of software that interacts with g:Profiler. As an example of g:Profiler integration with programming languages, we have now implemented a simple package for GNU R that accesses the g:Profiler web server to mediate functionality of g:GOSt, g:Cocoa and g:Convert. The R package is available for download on our web site. We have also created an web service that is compatible with ENFIN Encore workflows (35). Since the 2011 update, we maintain archived versions of g:Profiler software and data to provide comparability to earlier studies.
CASE STUDIES
Functional analysis of positively selected regions in the human genome
To demonstrate the new features of g:Profiler, we present a case study with genomic regions that have undergone significant evolution during early human history. We retrieved 212 loci of positive selection from the recent analysis of the Neanderthal genome (3), grouped these into chromosome-specific lists and studied the functional enrichments of localized genes with the g:Cocoa chromosomal multiquery feature. These results were retrieved through the g:Profiler R interface, filtered for non-redundant terms and visualized using a custom script.
The results of this case study include several interesting functional features that may provide insights into the evolution of modern humans (Figure 2). For instance, GO functions like ‘defense response to bacterium’ (chr. 6, P = 10−8) and ‘viral reproduction’ (chr. 18, P = 10−7) as well as KEGG pathway for ‘systemic lupus erythematosus’ (P = 10−12) may indicate accumulated disease resistance. A number of enriched HPO disease gene annotations such as ‘malaligned carpal bone’ (chr. 20, P = 10−3) as well as the GO process ‘appendage morphogenesis’ (chr. 2, P = 10−6) may associate to the development of modern human hands. In addition, this analysis provides pointers to the evolution of gene regulation at multiple levels of organization, including epigenetic (GO cell component nucleosome, chr. 1, P = 10−15), transcriptional (six enrichments of transcription factor binding sites, P < 10−3) and post-transcriptional regulation (four enrichments of microRNA target sites, P < 0.02). In summary, these results provide interesting hypotheses for further study and show the usefulness of new g:Profiler features.
Identification of protein complexes in regulator perturbation data
We also provide an example of gene list interpretation in the context of protein–protein interactions. We previously studied differentially expressed genes in yeast transcription factor perturbations (ΔTF) and identified affected protein–protein interaction modules (31), using the method described above. Here, we investigated a collection of high-confidence protein complexes from a recent analysis (36) to interpret our ΔTF data, using the g:Profiler R interface and a custom script for filtering and visualization. In particular, we identified protein complexes that were fully listed in g:Profiler analysis of ΔTF data.
This case study highlights the presence of multiple important protein complexes among our enriched modules (Supplementary Figure S1). For instance, the global TFIIA complex is required for all transcription by RNA polymerase II and is therefore important in all processes involving transcription. In our data, all subunits of TFIIA are affected in multiple perturbation strains, including Δgal11, a subunit of the mediator complex that interacts with RNA Polymerase II. TFIIA is also dysregulated in the induced deletions of Rap1 and Gcr1. The latter are essential transcription factors with numerous global roles. In summary, the modules identified by our methods frequently overlap with actual protein complexes and may provide useful pointers for biologically meaningful gene list interpretation.
Application of background gene lists for improving specificity
The usefulness of custom background gene lists is shown in the following example. We studied the functional enrichments of nine core cell cycle transcription factors (TFs) in yeast (Mcm1, Mbp1, Swi4, Swi6, Fkh1, Fkh2, Ace2, Swi5, Ndd1), using the standard background (all yeast genes, ∼6000), as well as a user-defined background list (yeast transcription factors, ∼300). The former approach is reasonable when genes of interest are sampled from the genome-wide collection of genes, while the latter approach is more meaningful when focusing a priori on a narrow, predefined gene group such as transcription factors.
The comparison of top 10 results from the two queries highlights the differences of the two approaches (Figure 3, custom R graphic). The first, global analysis of the 9 TFs results in general functional annotations such as ‘positive regulation of transcription’ (P = 10−11) that would be expected from any such group of TFs. Annotations of this specificity are most informative given that the focus is the whole genome. When the analysis is restricted to the TF set beforehand, a customized background set allows one to bypass the a priori knowledge of general TF function and retrieve more specific annotations. In our example, the second analysis results in specific annotations like ‘interphase of mitotic cell cycle’ (P = 10−8). In summary, utilizing a biologically meaningful custom background can improve specificity in enrichment analysis.
DISCUSSION
With the ongoing development and maintenance of g:Profiler, we aim to provide a comprehensive functional profiling service that combines powerful visualization, sophisticated algorithms and up-to-date genomic and functional data. For this update, we implemented several improvements to address the needs of new and experienced g:Profiler users alike. Improved filtering and statistics produce results of greater confidence and more customization power. Multilist enrichment analysis in g:Cocoa delivers results and visualization in a condensed overview. Our special focus in this g:Profiler update relates to developments in biomedical and clinical research that are likely to lead to personalized genomics and medicine in the future. First, we include disease gene annotations in our enrichment tests to enable and promote discovery of associations to human health. Second, we incorporate algorithms for interpreting gene lists in the context of molecular interaction networks, as the concepts of biological systems and networks have become ever more important in the past decade. Last but not least, we provide direct means to study the functional enrichments of user-defined DNA regions and even single nucleotides through corresponding genes. Functional analysis of these data is likely to become increasingly important, as genome and transcriptome sequencing, genome-wide association studies and other biotechnologies reveal more DNA-based evidence to reason about human variation, history, complex disease traits and cancer.
Since the publication of g:Profiler in 2007, the tool has found extensive use in a number of projects in stem cell research (37–39), cancer genomics (40,41), machine learning and yeast genomics (42,43), microRNA regulatory networks (44), among others. g:Profiler is related to a whole family of bioinformatics tools that apply its functional profiling and biomolecule mapping methods for various tasks, like global coexpression analysis in large microarray collections [MEM (28)], module identification and interpretation in biological networks [GraphWeb (45)], transcriptome analysis in pathways [KEGGanim (46)] and optimal gene expression clustering with functional profiling [VisHiC (47)].
Future developments of g:Profiler will further elaborate on new and emerging sources of biomedical evidence. Functional interpretation of gene lists will greatly benefit from the comprehensive regulatory maps from ENCODE and modENCODE. Another important area of development involves deeper integration with network-based sources of evidence. Novel methods need to explore massive amounts of data from the next-generation sequencing of tumour catalogues, human variation studies and personalized genomics.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
EU FP6 COBRED; ENFIN; ETF7437 MEM; EITSA; ERDF through EXCS funding projects; M.Curie Biostar, U.Agur and A.Lind fellowships to (J.R.). Funding for open access charge: University of Tartu, Center of Excellence in Computer Science (EXCS).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Priit Adler, Raivo Kolde, Meelis Kull and Magali Michaut for valuable discussion and/or software contributions, and the anonymous reviewers for beneficial suggestions. University of Tartu HPC centre has provided computing resources.
REFERENCES
- 1.Lander ES. Initial impact of the sequencing of the human genome. Nature. 2011;470:187–197. doi: 10.1038/nature09792. [DOI] [PubMed] [Google Scholar]
- 2.The 1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MH, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–722. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The ENCODE Project Consortium. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.McCarthy MI, Abecasis GR, Cardon LR, Goldstein DB, Little J, Ioannidis JP, Hirschhorn JN. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 6.The International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang daW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 8.Doniger SW, Salomonis N, Dahlquist KD, Vranizan K, Lawlor SC, Conklin BR. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Al-Shahrour F, Minguez P, Tarraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khatri P, Voichita C, Kattan K, Ansari N, Khatri A, Georgescu C, Tarca AL, Draghici S. Onto-Tools: new additions and improvements in 2006. Nucleic Acids Res. 2007;35:W206–W211. doi: 10.1093/nar/gkm327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sealfon RS, Hibbs MA, Huttenhower C, Myers CL, Troyanskaya OG. GOLEM: an interactive graph-based gene-ontology navigation and analysis tool. BMC Bioinformatics. 2006;7:443. doi: 10.1186/1471-2105-7-443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Antonov AV, Dietmann S, Mewes HW. KEGG spider: interpretation of genomics data in the context of the global gene metabolic network. Genome Biol. 2008;9:R179. doi: 10.1186/gb-2008-9-12-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Huang daW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:193–200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Robinson PN, Mundlos S. The human phenotype ontology. Clin. Genet. 2010;77:525–534. doi: 10.1111/j.1399-0004.2010.01436.x. [DOI] [PubMed] [Google Scholar]
- 18.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:698–704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Parkinson H, Sarkans U, Kolesnikov N, Abeygunawardena N, Burdett T, Dylag M, Emam I, Farne A, Hastings E, Holloway E, et al. ArrayExpress update–an archive of microarray and high-throughput sequencing-based functional genomics experiments. Nucleic Acids Res. 2011;39:D1002–D1004. doi: 10.1093/nar/gkq1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Barrell D, Dimmer E, Huntley RP, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009–an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:396–403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kull M, Vilo J. Fast approximate hierarchical clustering using similarity heuristics. Biodata Min. 2008;1:9. doi: 10.1186/1756-0381-1-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adler P, Kolde R, Kull M, Tkachenko A, Peterson H, Reimand J, Vilo J. Mining for coexpression across hundreds of datasets using novel rank aggregation and visualization methods. Genome Biol. 2009;10:R139. doi: 10.1186/gb-2009-10-12-r139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sardiello M, Palmieri M, di Ronza A, Medina DL, Valenza M, Gennarino VA, Di Malta C, Donaudy F, Embrione V, Polishchuk RS, et al. A gene network regulating lysosomal biogenesis and function. Science. 2009;325:473–477. doi: 10.1126/science.1174447. [DOI] [PubMed] [Google Scholar]
- 30.Yagi S, Hirabayashi K, Sato S, Li W, Takahashi Y, Hirakawa T, Wu G, Hattori N, Hattori N, Ohgane J, et al. DNA methylation profile of tissue-dependent and differentially methylated regions (T-DMRs) in mouse promoter regions demonstrating tissue-specific gene expression. Genome Res. 2008;18:1969–1978. doi: 10.1101/gr.074070.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reimand J, Vaquerizas JM, Todd AE, Vilo J, Luscombe NM. Comprehensive reanalysis of transcription factor knockout expression data in Saccharomyces cerevisiae reveals many new targets. Nucleic Acids Res. 2010;38:4768–4777. doi: 10.1093/nar/gkq232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Narsai R, Howell KA, Millar AH, O’Toole N, Small I, Whelan J. Genome-wide analysis of mRNA decay rates and their determinants in Arabidopsis thaliana. Plant Cell. 2007;19:3418–3436. doi: 10.1105/tpc.107.055046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McCarthy FM, Gresham CR, Buza TJ, Chouvarine P, Pillai LR, Kumar R, Ozkan S, Wang H, Manda P, Arick T, et al. AgBase: supporting functional modeling in agricultural organisms. Nucleic Acids Res. 2011;39:497–506. doi: 10.1093/nar/gkq1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kahlem P, Clegg A, Reisinger F, Xenarios I, Hermjakob H, Orengo C, Birney E. ENFIN–A European network for integrative systems biology. C. R. Biol. 2009;332:1050–1058. doi: 10.1016/j.crvi.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 36.Benschop JJ, Brabers N, van Leenen D, Bakker LV, van Deutekom HW, van Berkum NL, Apweiler E, Lijnzaad P, Holstege FC, Kemmeren P. A consensus of core protein complex compositions for Saccharomyces cerevisiae. Mol. Cell. 2010;38:916–28. doi: 10.1016/j.molcel.2010.06.002. [DOI] [PubMed] [Google Scholar]
- 37.Schulz H, Kolde R, Adler P, Aksoy I, Anastassiadis K, Bader M, Billon N, Boeuf H, Bourillot PY, Buchholz F, et al. The FunGenES database: a genomics resource for mouse embryonic stem cell differentiation. PLoS ONE. 2009;4:e6804. doi: 10.1371/journal.pone.0006804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jung M, Peterson H, Chavez L, Kahlem P, Lehrach H, Vilo J, Adjaye J. A data integration approach to mapping OCT4 gene regulatory networks operative in embryonic stem cells and embryonal carcinoma cells. PLoS ONE. 2010;5:e10709. doi: 10.1371/journal.pone.0010709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Billon N, Kolde R, Reimand J, Monteiro MC, Kull M, Peterson H, Tretyakov K, Adler P, Wdziekonski B, Vilo J, et al. Comprehensive transcriptome analysis of mouse embryonic stem cell adipogenesis unravels new processes of adipocyte development. Genome Biol. 2010;11:R80. doi: 10.1186/gb-2010-11-8-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hatzis P, van der Flier LG, van Driel MA, Guryev V, Nielsen F, Denissov S, Nijman IJ, Koster J, Santo EE, et al. Genome-wide pattern of TCF7L2/TCF4 chromatin occupancy in colorectal cancer cells. Mol. Cell. Biol. 2008;28:2732–2744. doi: 10.1128/MCB.02175-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vooder T, Valk K, Kolde R, Roosipuu R, Vilo J, Metspalu A. Gene expression-based approaches in differentiation of metastases and second primary tumour. Case Rep. Oncol. 2010;3:255–261. doi: 10.1159/000318010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stegle O, Parts L, Durbin R, Winn J. A Bayesian framework to account for complex non-genetic factors in gene expression levels greatly increases power in eQTL studies. PLoS Comput. Biol. 2010;6:e1000770. doi: 10.1371/journal.pcbi.1000770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tretyakov K, Laur S, Vilo J. G=MAT: linking transcription factor expression and DNA binding data. PLoS ONE. 2011;6:e14559. doi: 10.1371/journal.pone.0014559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gennarino VA, Sardiello M, Avellino R, Meola N, Maselli V, Anand S, Cutillo L, Ballabio A, Banfi S. MicroRNA target prediction by expression analysis of host genes. Genome Res. 2009;19:481–490. doi: 10.1101/gr.084129.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Reimand J, Tooming L, Peterson H, Adler P, Vilo J. GraphWeb: mining heterogeneous biological networks for gene modules with functional significance. Nucleic Acids Res. 2008;36:W452–W459. doi: 10.1093/nar/gkn230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adler P, Reimand J, Janes J, Kolde R, Peterson H, Vilo J. KEGGanim: pathway animations for high-throughput data. Bioinformatics. 2008;24:588–590. doi: 10.1093/bioinformatics/btm581. [DOI] [PubMed] [Google Scholar]
- 47.Krushevskaya D, Peterson H, Reimand J, Kull M, Vilo J. VisHiC–hierarchical functional enrichment analysis of microarray data. Nucleic Acids Res. 2009;37:W587–W592. doi: 10.1093/nar/gkp435. [DOI] [PMC free article] [PubMed] [Google Scholar]