Abstract
Short, linear motifs (SLiMs) play a critical role in many biological processes. The SLiMSearch 2.0 (Short, Linear Motif Search) web server allows researchers to identify occurrences of a user-defined SLiM in a proteome, using conservation and protein disorder context statistics to rank occurrences. User-friendly output and visualizations of motif context allow the user to quickly gain insight into the validity of a putatively functional motif occurrence. For each motif occurrence, overlapping UniProt features and annotated SLiMs are displayed. Visualization also includes annotated multiple sequence alignments surrounding each occurrence, showing conservation and protein disorder statistics in addition to known and predicted SLiMs, protein domains and known post-translational modifications. In addition, enrichment of Gene Ontology terms and protein interaction partners are provided as indicators of possible motif function. All web server results are available for download. Users can search motifs against the human proteome or a subset thereof defined by Uniprot accession numbers or GO term. The SLiMSearch server is available at: http://bioware.ucd.ie/slimsearch2.html.
INTRODUCTION
The purpose of the SLiMSearch (Short, Linear Motif Search) web server is to allow researchers to identify novel occurrences of user-defined Short Linear Motifs (SLiMs) in a set of sequences. SLiMs, also referred to as linear motifs or minimotifs, are functional microdomains that play a central role in many diverse biological pathways (1) through post-translational modification (including cleavage), subcellular localization and ligand binding (2). Once a SLiM has been defined, finding matches in a given set of protein sequences is a fairly trivial task. Several web-based methods to discover novel instances of known SLiMs are available, including ELM (2), MnM (3), SIRW (4) ScanProsite (5) and QuasiMotifFinder (6), which generally utilize databases of known motif patterns to search query protein sequences supplied by the user.
While finding matches is trivial, however, interpreting their biological significance is far from easy. Stochastic occurrences of small, degenerate motifs are common; distinguishing real occurrences from the background of random motif hits remains the greatest challenge in a priori motif discovery. One approach is to simply filter out motifs that are likely to occur numerous times by chance—ScanProsite (5), for example, has an option to ‘Exclude motifs with a high probability of occurrence’, while QuasiMotifFinder (6) uses the background occurrence of motifs in PfamA families (7) to assess the significance of hits. These strategies work well for longer, family descriptor motifs [such as are found in the Prosite database (8) used by both ScanProsite and QuasiMotifFinder] but are not so useful for SLiMs because of their tendency to occur by chance. Instead, additional contextual information such as sequence conservation (3,6,9,10), structural context (3,11) or even biological keywords (4) can be used to assess the likelihood of true functional significance for putatively functional sites.
Most motif search tools rely on pre-existing motif libraries, such as ELM (2), MnM (3) or Prosite (8). Those that permit users to define their own motifs, such as ScanProsite (5), are generally lacking the contextual information required to aid functional inference. Recent developments in de novo motif discovery has given rise to a number of tools that are capable of predicting entirely novel SLiMs from sets of protein sequences [e.g. PRATT (12), MEME (13), Dilimot (14), SLiMDisc (15), SLiMFinder (16) and FIRE-pro (17)]. Although SLiMFinder (16) estimates the statistical significance of returned motif predictions, correcting for biases introduced by evolutionary relationships within the data, assessing the ‘biological’ significance of predicted SLiMs remains challenging. One approach is to compare candidate SLiMs to existing motif libraries to identify similarities to previously known motifs (18).When a genuinely novel motif is predicted, however, knowledge of existing motifs is of limited use. Instead, it is useful to be able to establish the background distribution of occurrences of the novel motif, utilizing contextual information to help screen out the inevitable spurious chance matches.
We recently made our powerful de novo SLiM discovery tool, SLiMFinder (16), available as a web server (19). To aid interpretation of SLiMFinder results, we made a new tool available, SLiMSearch, which allows users to search protein data sets with user-defined motifs, including motif prediction output from SLiMFinder (20). SLiMSearch utilized the same sequence context assessment as SLiMFinder, enabling results to be masked or ranked based on the important biological indicators of sequence conservation and structural disorder (10,21) and features the same SLiMChance algorithm for assessing statistical overrepresentation of SLiM occurrences, correcting for biases introduced by evolutionary relationships within the data (16). Like SLiMFinder, SLiMSearch was optimized for small protein data sets. In this article, we describe a complementary server, SLiMSearch 2.0, which is optimized for searches of a whole proteome.
SLiMSearch 2.0 replaces SLiMChance data set probabilities with individual likelihoods for each motif instance that permit the ranking of many motif occurrences and helps separate putative functional instances from the background of stochastic occurrences. A comprehensive study of the Eukaryotic Linear Motif (ELM) database by Fuxreiter et al. (22) found that SLiMs are more likely to be found in disordered regions, while Chica et al. (9) found that conserved motifs are more likely to be true positives. Our previous work with both discovery of new instances of known motifs and of novel motifs shows that, motifs in disordered regions and conserved motifs are typically (but not always) more likely to be true positives (10). Therefore, we encourage the use of an optional disorder filter and we present the results ranked according to conservation. Enrichment scores for motif counts are calculated (i) versus reversed/shuffled variants of the motif, (ii) for Gene Ontology (GO) terms (23) and (iii) for known BioGRID interactors of individual hub proteins (24). In addition to identifying individual occurrences of known motifs, therefore, SLiMSearch 2.0 can indicate possible functional significance for entirely novel motifs. Input, output and results visualizations are fully compatible with our existing SLiM analysis web servers, SLiMDisc (25), CompariMotif (18), SLiMFinder (19) and SLiMSearch 1.0 (20), providing a suite of integrated tools for analysing these biologically important sequence features.
THE SLIMSEARCH 2.0 ALGORITHM
SLiMSearch 2.0 performs a motif regular expression search against a proteome allowing restriction of considered sequences to set of proteins or a given GO term. Features include annotation of overlapping sequence annotation and calculation of global and local motif statistics and attributes.
Pre-formatted database
To speed up motif attribute calculations, pre-computed databases for each proteome are used. The current release has only Human UniProt release v1.37 (Aug 2010) (26); however, more model proteomes will be added as data is computed. Two pre-computed conservation scores are calculated for each protein in the proteome, a column-based tree-weighted conservation score (WCS) (9) and a relative local conservation (RLC) metric (10). Homologues for each sequence are identified using a BLAST search against a database of 70 complete EnsEMBL proteomes (Ensembl 59, October 2010, 69 Metazoan proteomes and Saccharomyces cerevisiae) (27) and orthologues are predicted using GOPHER (default options) (25). Predicted orthologues are aligned by MAFFT (28) and used to calculate conservation scoring metrics on a residue-by-residue basis. Disorder scores for each residue are calculated using IUPred (default options) (21).
Several features of interest are also preformatted for rapid querying: (i) Domain data from Pfam (29); (ii) structure data from PDB (30); (iii) experimentally validated motifs from the ELM database (2); and (iv) SNP and modification data from UniProt annotation (26).
Scoring
The IUPred disorder score, IUP, of the motif is calculated as the mean disorder score across the defined (non-wildcard) residues. The WCS of a motif is calculated similarly. SLiMSearch 2.0 extends the RLC score to return a probability. Based on the assumption, consistent with empirical observation, that the RLC scores for a residue are normally distributed (10), the RLC of a residue is converted into a probability, P(RLC), using the Gaussian Cumulative Distribution Function (CDF). The relative conservation probability of a motif, P, the probability of each residue of a motif having its given RLC or higher can be calculate as the product of the P(RLC) for each residue within the motif. A significance value, P(cons), representing the probability of a given motif having that P-value or lower by chance, can then be calculated for the motifs P-value using the CDF of the uniform product distribution [Equation (1)]. Thus, the P(cons) statistic provides a useful measure of how likely it is that this motif will have the observed degree of local conservation (or higher) by chance. Note that it does not provide any indication of the probability of the motif itself, which is best inferred from the enrichment values.
| (1) |
P(cons) is the probability of a given motif having that P-value or higher by chance, calculated as the CDF of the uniform product distribution, i.e. the distribution of the product of n uniform distributions, where n is the number of non-wildcard positions in the motif, P is the relative conservation probability of a motif and Γ is the incomplete gamma function.
Enrichment scores
Enrichment scores for motif counts are calculated for the input motif against the reverse of the motif and a randomly shuffled variant of the motif. The score is a simple quotient, where the input motif count is the divisor and the shuffled or reverse motif count is the dividend. Enrichment scores for each GO term and BioGRID interaction hub protein are calculated versus the expectation provided by the whole proteome, i.e. the number of motif occurrences in proteins with that GO term/interaction partner divided by the expected number of proteins, which is the total number of proteins with the motif multiplied by the proportion of the proteome with that GO term/interaction partner. Enrichment significance is calculated using the Fisher’s exact test. Counts are normalized for independence by clustering highly similar proteins based on UniRef50 groups.
THE SLIMSEARCH 2.0 WEBSERVER
The SLiMSearch 2.0 server is available at http://bioware.ucd.ie/slimsearch2.html. The website is free and open to all and there is no login requirement. The purpose of the web server is to allow researchers to identify novel occurrences of user-defined SLiMs in a set of sequences. A rapid pattern matching search is first performed to identify all occurrences of the motif in the proteome (or a defined subset). Pre-formatted databases are then used to rapidly extract scores and sequence features for each occurrence before enrichment scores are calculated. Interactive output and visualizations permit easy exploration of returned occurrences of the motif and their sequence context. These features of the web server are described in more detail in the following sections.
Input
A motif to be compared against the search is the sole compulsory input. The motif should be expressed as a regular expression using single letter amino acid codes (e.g. R.LF or RxLF but not Arg–x–Leu–Phe). The format allows for ambiguity (i.e. positions that can be any residue from a set of residues, e.g. [ILV] meaning any aliphatic residue), flexibility (e.g. ‘.{1,3}’, meaning a wildcard position between 1 and 3 residues in length), termini definition (where ^ is the N-terminus and $ is the C-terminus) and conditional motifs (e.g. (motif1)|(motif2) meaning motif1 or motif2). Two optional filtering options are also available, restricting the protein search space to a subset of a proteome: by GO term (in the format GO:0005868) to restrict the search to a particular ontology and similarly, to a set of proteins by UniProt accessions. For clarity, example inputs are available above each entry box on the input page of the web server.
Submitting jobs
Once input has been determined, clicking ‘Submit job’ will enter the run queue. Run times will vary according to input data size, motif complexity and the current load of the server but are generally in the order of a few seconds. Users can either wait for their jobs to run or bookmark the page and return to it later, although jobs are deleted after 21 days. The web server can also be run directly using a URL containing the motif to be searched and (optionally) a list of UniProt IDs.
Output
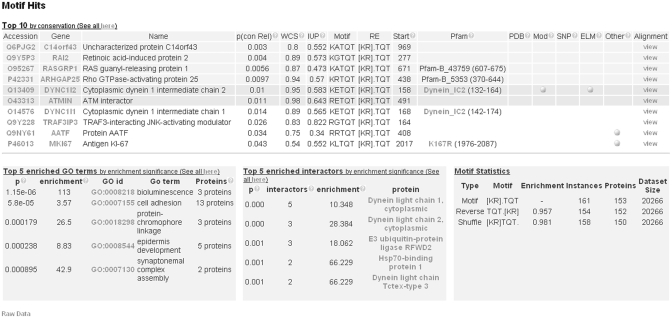
The main output is a table of motif instances annotated with attributes including: (i) conservation and disorder statistics; (ii) overlapping feature, such as Pfam domains, PDB structures, SNPs and modifications; and (iii) overlapping experimentally validated motifs (Figure 1). In addition, alignments of 100 amino acid regions overlapping each motif occurrence can be visualized. Discovered motifs are not filtered, therefore all instances are returned. By default, motifs are ranked based on P(cons). Several additional tables are also returned: GO terms which are enriched for the motif; hub proteins where the interactors are enriched for the motif and motif count statistics. All results are returned as tab-delimited files and in a more visually appealing html format. Initially, an overview of the most interesting instances and enrichments are returned. More detailed data are available and can be sorted by each attribute. Instance data can be also filtered based on IUPred mean disorder score, IUP.
Figure 1.
Main results page. In addition to individual statistics for the top ten motif occurrences, the default results page displays motif counts, GO enrichment and protein interactor enrichment for all occurrences. If no enriched GO terms and/or interactors have been found, these sections will be blank.
Users need to consider two separate lines of evidence when assessing the significance or otherwise of the findings presented. First, the motif enrichment over the reversed and shuffled sequences gives an indication to what extent the motifs that are provided occur by chance. If a motif occurs 40 times and the reverse occurs 20 times, this means that we expect that about half of the observed instances are false positives (assuming no negative selection on randomly occurring motifs). The user can then scroll down the list of occurrences, and investigate the conservation values, to form a judgement regarding which motifs are most likely to be true positives. Assuming a typical mammalian motif, it would be expected in this case that the 20 least conserved motifs are most likely to be false positives and the 20 most conserved are most likely to be true positives. In many cases, the enrichment may be relatively modest; P(cons) only provides guidance, rather than proof, regarding the likelihood that a given motif occurrence is a true positive.
Example analysis
The web server incorporates a full example for searching the human proteome with the manually curated, experimentally validated, Dynein Light Chain binding motif ([KR].TQT; ELM entry LIG_Dynein_DLC8_1 (2)). A full walkthrough for this data set is provided in the help pages and fully interactive example output is also provided. Example proteome restrictions by sequence (the three curated human occurrences of LIG_Dynein_DLC8_1) and GO term (cytoplasmic dynein complex) can also be loaded at the front page.
Getting help
SLiMSearch 2.0 is supported by an extensive help section, including a quickstart guide and walkthrough with screenshots. Example input files are provided and example input data can be loaded into the input forms. Fully interactive example output (corresponding to running the example input with default parameters) is clearly linked from the help pages (See ‘Example analysis’ section).
FUTURE WORK
Currently, only human proteome searches are available but other proteomes will be added with time. A selection of model organisms will be added in the near future.
CONCLUSION
There are many sources of de novo motifs, including experimental approaches such as mutagenesis and peptide arrays or phage display. With recent developments in experimental technologies for determining protein–protein interaction networks and computational techniques for predicting interaction motifs from them, the number of putative SLiMs is likely to increase dramatically in the next few years. SLiMSearch 2.0 represents a valuable tool for the annotation of such motifs. In addition to de novo motifs, the server is useful for finding candidate occurrences of established SLiMs, including those found in motif databases such as ELM (2) and MiniMotif Miner (3). Often, the definition of these motifs is not conclusive and so there are also times when it is useful to search using a specific variant or a relaxed motif definition. For many known SLiMs, we currently only have annotated occurrences for a restricted set of taxonomic groups (2) but, due to their short and degenerate nature, they often evolve convergently (31). As the number of full proteomes continues to increase, the SLiMSearch 2.0 server will enable the identification of SLiMs in new taxa, helping to shed light on the breadth and depth of functional SLiMs. The SLiMSearch 2.0 server is available at: http://bioware.ucd.ie/slimsearch2.html.
FUNDING
Science Foundation Ireland (08/IN.1/B1864) and the University of Southampton; European Molecular Biology Laboratory [EMBL Interdisciplinary Postdoc (EIPOD) fellowship to N.E.D.] Funding for open access charge: The University of Southampton.
Conflict of interest statement. None declared.
REFERENCES
- 1.Diella F, Haslam N, Chica C, Budd A, Michael S, Brown NP, Trave G, Gibson TJ. Understanding eukaryotic linear motifs and their role in cell signaling and regulation. Front. Biosci. 2008;13:6580–6603. doi: 10.2741/3175. [DOI] [PubMed] [Google Scholar]
- 2.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rajasekaran S, Balla S, Gradie P, Gryk MR, Kadaveru K, Kundeti V, Maciejewski MW, Mi T, Rubino N, Vyas J, et al. Minimotif miner 2nd release: a database and web system for motif search. Nucleic Acids Res. 2009;37:D185–D190. doi: 10.1093/nar/gkn865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ramu C. SIRW: A web server for the Simple Indexing and Retrieval System that combines sequence motif searches with keyword searches. Nucleic Acids Res. 2003;31:3771–3774. doi: 10.1093/nar/gkg546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Castro E, Sigrist CJ, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res. 2006;34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gutman R, Berezin C, Wollman R, Rosenberg Y, Ben-Tal N. QuasiMotiFinder: protein annotation by searching for evolutionarily conserved motif-like patterns. Nucleic Acids Res. 2005;33:W255–W261. doi: 10.1093/nar/gki496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32:D138–D141. doi: 10.1093/nar/gkh121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sigrist CJ, Cerutti L, de Castro E, Langendijk-Genevaux PS, Bulliard V, Bairoch A, Hulo N. PROSITE, a protein domain database for functional characterization and annotation. Nucleic Acids Res. 2010;38:D161–D166. doi: 10.1093/nar/gkp885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chica C, Labarga A, Gould CM, Lopez R, Gibson TJ. A tree-based conservation scoring method for short linear motifs in multiple alignments of protein sequences. BMC Bioinformatics. 2008;9:229. doi: 10.1186/1471-2105-9-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Davey NE, Shields DC, Edwards RJ. Masking residues using context-specific evolutionary conservation significantly improves short linear motif discovery. Bioinformatics. 2009;25:443–450. doi: 10.1093/bioinformatics/btn664. [DOI] [PubMed] [Google Scholar]
- 11.Via A, Gould CM, Gemund C, Gibson TJ, Helmer-Citterich M. A structure filter for the Eukaryotic Linear Motif Resource. BMC Bioinformatics. 2009;10:351. doi: 10.1186/1471-2105-10-351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jonassen I, Collins JF, Higgins DG. Finding flexible patterns in unaligned protein sequences. Protein Sci. 1995;4:1587–1595. doi: 10.1002/pro.5560040817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Neduva V, Russell RB. DILIMOT: discovery of linear motifs in proteins. Nucleic Acids Res. 2006;34:W350–W355. doi: 10.1093/nar/gkl159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Davey NE, Shields DC, Edwards RJ. SLiMDisc: short, linear motif discovery, correcting for common evolutionary descent. Nucleic Acids Res. 2006;34:3546–3554. doi: 10.1093/nar/gkl486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Edwards RJ, Davey NE, Shields DC. SLiMFinder: A Probabilistic Method for Identifying Over-Represented, Convergently Evolved, Short Linear Motifs in Proteins. PLoS ONE. 2007;2:e967. doi: 10.1371/journal.pone.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lieber DS, Elemento O, Tavazoie S. Large-scale discovery and characterization of protein regulatory motifs in eukaryotes. PLoS ONE. 2010;5:e14444. doi: 10.1371/journal.pone.0014444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Edwards RJ, Davey NE, Shields DC. CompariMotif: quick and easy comparisons of sequence motifs. Bioinformatics. 2008;24:1307–1309. doi: 10.1093/bioinformatics/btn105. [DOI] [PubMed] [Google Scholar]
- 19.Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMFinder: a web server to find novel, significantly over-represented, short protein motifs. Nucleic Acids Res. 2010;38:W534–W539. doi: 10.1093/nar/gkq440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davey NE, Haslam NJ, Shields DC, Edwards RJ. SLiMSearch: a webserver for finding novel occurrences of short linear motifs in proteins, incorporating sequence context. In: Dijkstra TMH, Tsivtsivadze E, Marchiori E, Heskes T, editors. Pattern Recognition in Bioinformatics, Lecture Notes in Bioinformatics. Vol. 6282. Berlin: Springer; 2010. pp. 50–61. [Google Scholar]
- 21.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 22.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stark C, Breitkreutz BJ, Chatr-Aryamontri A, Boucher L, Oughtred R, Livstone MS, Nixon J, Van Auken K, Wang X, Shi X, et al. The BioGRID Interaction Database: 2011 update. Nucleic Acids Res. 2011;39:D698–D704. doi: 10.1093/nar/gkq1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Davey NE, Edwards RJ, Shields DC. The SLiMDisc server: short, linear motif discovery in proteins. Nucleic Acids Res. 2007;35:W455–W459. doi: 10.1093/nar/gkm400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.The UniProt Consortium. Ongoing and future developments at the Universal Protein Resource. Nucleic Acids Res. 2011;39:D214–D219. doi: 10.1093/nar/gkq1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flicek P, Amode MR, Barrell D, Beal K, Brent S, Chen Y, Clapham P, Coates G, Fairley S, Fitzgerald S, et al. Ensembl 2011. Nucleic Acids Res. 2011;39:D800–D806. doi: 10.1093/nar/gkq1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K, Toh H. Recent developments in the MAFFT multiple sequence alignment program. Brief. Bioinform. 2008;9:286–298. doi: 10.1093/bib/bbn013. [DOI] [PubMed] [Google Scholar]
- 29.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rose PW, Beran B, Bi C, Bluhm WF, Dimitropoulos D, Goodsell DS, Prlic A, Quesada M, Quinn GB, Westbrook JD, et al. The RCSB Protein Data Bank: redesigned web site and web services. Nucleic Acids Res. 2011;39:D392–D401. doi: 10.1093/nar/gkq1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Davey NE, Trave G, Gibson TJ. How viruses hijack cell regulation. Trends Biochem. Sci. 2010;36:159–169. doi: 10.1016/j.tibs.2010.10.002. [DOI] [PubMed] [Google Scholar]