Abstract
Recent interest in the non-coding transcriptome has resulted in the identification of large numbers of long non-coding RNAs (lncRNAs) in mammalian genomes, most of which have not been functionally characterized. Computational exploration of the potential functions of these lncRNAs will therefore facilitate further work in this field of research. We have developed a practical and user-friendly web interface called ncFANs (non-coding RNA Function ANnotation server), which is the first web service for functional annotation of human and mouse lncRNAs. On the basis of the re-annotated Affymetrix microarray data, ncFANs provides two alternative strategies for lncRNA functional annotation: one utilizing three aspects of a coding-non-coding gene co-expression (CNC) network, the other identifying condition-related differentially expressed lncRNAs. ncFANs introduces a highly efficient way of re-using the abundant pre-existing microarray data. The present version of ncFANs includes re-annotated CDF files for 10 human and mouse Affymetrix microarrays, and the server will be continuously updated with more re-annotated microarray platforms and lncRNA data. ncFANs is freely accessible at http://www.ebiomed.org/ncFANs/ or http://www.noncode.org/ncFANs/.
INTRODUCTION
Large numbers of long non-coding RNAs (lncRNAs) have been detected in mammalian genome through large-scale analyses of full-length cDNA sequences (1,2). Several lncRNAs such as NRON (3), MEG3 (4), lincRNA-P21 (5) and MALAT-1 (6) have already been well characterized, suggesting that lncRNAs function in a range of biological processes such as imprinting control, cell differentiation, immune response and chromatin modification (7–9). Though lack of conservation does not necessarily imply lack of function (10), the low conservation levels of most lncRNAs is an impediment to functional research. The tens of thousands of mouse lncRNAs were provided by FANTOM3 (11,12), and data on both mouse and human lncRNAs obtained by recent deep-sequencing efforts (13–15) have increased the alertness of the scientific community to the important roles of these transcripts in biological processes (16,17). Guttman et al. (13) identified numerous large intervening non-coding RNAs by chromatin-state maps and assigned functions to these ncRNAs based on the coding-non-coding gene co-expression relationship extracted from custom-designed tiling array data. In spite of such efforts, custom-designed tiling array analysis is expensive and relatively inflexible, and is therefore not a preferred method for lncRNA studies.
Based on high-throughput experiment datasets including microarrays, physical interactions, genetic interactions, and phylogenetic profiles, a number of functional prediction tools have already been designed for protein coding genes, such as N-Browse (18), FunCoup (19) and GeneMANIA (20). However, no such tools have yet been developed for lncRNAs, and it is therefore still a challenging task to mine out the potential functions for this type of molecules. We have recently shown that several thousand probes in the Affymetrix Mouse Genome 430 2.0 array perfectly match sequences of lncRNAs (21). Similarly, Risueno et al. (22) found that 27% of the probes in the HG_U133plus2 array could be remapped to ncRNAs. Furthermore, Michelhaugh et al. (23) used re-annotated Affymetrix U133A and B arrays to demonstrate that five lncRNAs were upregulated in the brains of heroin abusers as compared to matched drug-free control subjects, the results which subsequently could be confirmed by quantitative RT–PCR. We therefore re-annotated the Affymetrix Mouse Genome 430 2.0 Array probes corresponding to both coding and non-coding genes, and constructed a co-expression coding-non-coding (CNC) network based on existing microarray data (21). Applying three widely-used methods of functional prediction, the work showed that lncRNA functions could be reliably predicted by such a co-expression network. Noticing that probes targeting lncRNAs are common in various Affymetrix array platforms, it is of great importance to re-mine the abundance of existing microarray data by similar strategies.
To provide an easy way to re-use the existing microarray data for lncRNA functional annotation, we have developed a practical and user-friendly web interface called ncFANs (non-coding RNA Function ANnotation server), the first web service for annotating lncRNA functions in mouse and human through re-annotation of Affymetrix array data. ncFANs pre-processes the uploaded microarray raw data into expression profiles for both coding and lncRNA genes, and then annotates the functions of lncRNAs, based on the CNC network pipelined according to the aforementioned method (21), or by identification of condition-related differentially expressed lncRNAs in the microarray data.
MATERIALS AND METHODS
Filtering the lncRNA data sets
The mouse lncRNAs based on the mm5 version of mouse genome were downloaded from FANTOM3 database (11), and the human lncRNAs based on the hg19 version of human genome were curated from Vega as given by Ørum et al. (24). We excluded non-coding transcripts with length <200 nt, and converted the FANTOM3 lncRNAs from mm5 to mm9 using the UCSC liftOver tools. To avoid false positives, we assessed the coding potential of the FANTOM3 and Vega lncRNAs by CPC (25), and only retained transcripts classified by CPC as ‘non-coding’ and ‘non-coding (weak)’.
Re-annotation of Affymetrix microarrays
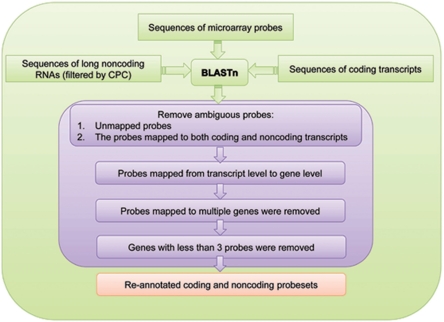
In its current version, ncFANs provides re-annotation of 10 mouse and human Affymetrix arrays, including six 3′-end expression arrays and four whole-genome expression arrays (Table 1). The probes of these arrays were re-annotated to coding and lncRNA genes using a modified pipeline proposed by Liao et al. (21) (Figure 1). The probe sequences were downloaded from the Affymetrix website (http://www.affymetrix.com). The sequences of coding transcripts were obtained from the RefSeq database (26), whereas the mouse and human lncRNA sequences were obtained from the filtered data sets of FANTOM3 and Vega, respectively. In the re-annotated CDF files, coding genes are identified by the corresponding NCBI Entrez Gene ID of RefSeq ID. With respect to Affymetrix arrays with perfect match–mismatch (PM–MM) probe pairs, we only re-annotated the PM probes and retained the corresponding MM probes. The information of gene coverage for each re-annotated CDF file is shown in Table 1.
Table 1.
Gene coverage information for each re-annotated CDF files
| Affymetrix microarray | The coverage of re-annotated CDF files |
||
|---|---|---|---|
| Total number | Coding genes | lncRNA genes | |
| Mouse430_2 | 17 761 | 13 673 | 4088 |
| Mouse430A_2 | 10 424 | 10 132 | 292 |
| MG_U74Av2 | 6649 | 6507 | 142 |
| MG_U74Bv2 | 5359 | 4711 | 648 |
| MG_U74Cv2 | 3132 | 2340 | 792 |
| MoExon-1_0-st-v1 | 39 171 | 19 950 | 19 221 |
| MoGene-1_0-st-v1 | 18 934 | 18 554 | 380 |
| HG_U133plus2 | 13 273 | 13 028 | 245 |
| HuExon-1_0-st-v2 | 20 544 | 18 943 | 1601 |
| HuGene-1_0-st | 15 800 | 15 785 | 15 |
Figure 1.
The detailed pipeline for re-annotation of Affymetrix array probes.
Construction of the coding-non-coding (CNC) co-expression network
Based on the re-annotated expression profiles containing both coding and lncRNA genes, the co-expression relationship of each gene pair was estimated by the Pearson Correlation Coefficient or the Spearman Rank Correlation Coefficient. The P-values of the correlation coefficients for all-against-all gene pairs were adjusted using the Bonferroni multiple testing correction. Co-expression relationships with significant P-values within a given upper percentile were retained as edges and coding and lncRNA genes were entered as nodes in the CNC network.
Functional annotation of lncRNAs based on CNC network characteristics
Based on the CNC network, ncFANs predicts functions for the lncRNAs using three methods (i.e. module-based, hub-based and co-location-based) as described by Liao et al. (21). In the module-based method, ncFANs employs the Markov cluster algorithm (MCL) with default parameters to identify modules of co-expressed genes in the CNC network. Based on the hypothesis that co-expressed modules often represent functional units (e.g. molecular complexes or pathways) whose genes have similar functions, the lncRNAs within a co-expressed module are then assigned functions that enriched among the coding genes in the same module. In the hub-based method, the hub lncRNAs are selected by a user-defined cut-off for the node degree, and the functions that are enriched among its immediate coding gene neighbours will be assigned to the lncRNA. In the co-location method, the functions of an lncRNA are annotated with its co-expressed, co-located coding gene partners defined by a given threshold for the distance between the loci on the chromosome (e.g. 10 kb). The lncRNAs were assigned with the functions of the coding genes within the same chromosome region, under the hypothesis that lncRNAs are commonly involved in the same biological processes as its adjacent upstream and downstream co-expressed coding genes (21,27). In the current version of ncFANs, the functional types include Gene Ontology (28) and KEGG pathways (29) terms, and the statistical significance of the functional enrichment is evaluated by the hypergeometric test.
Selection of differentially expressed lncRNAs
ncFANs provides two validated, widely-used methods, Fold-Change and Student’s t-test, for selecting differentially expressed lncRNAs. In the Fold-Change method, the expression values for each lncRNA are averaged across the samples in each experimental condition and their ratios are used to rank the lncRNAs. The Fold-Change is used in combination with the t-test. The lncRNA, within a user-defined higher fold change with a t-test P-value <0.05, is considered as differentially expressed. In the latter method, the statistical significance of the expression values for each lncRNA is estimated by the t-test after adjustment with the Benjamini and Hochberg correction for multiple comparisons. The cut-off for the t-test P-values is set by the user.
WEB SERVER DESCRIPTION
The system overview of ncFANs
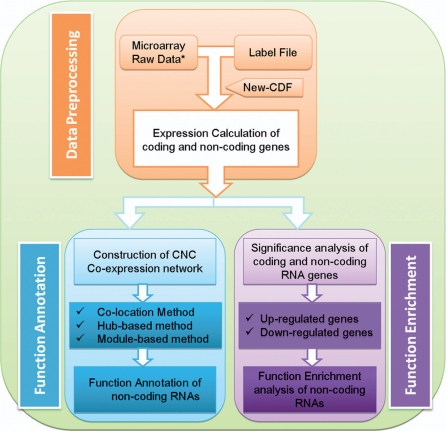
ncFANs is a freely available web server for functional annotation of lncRNAs, and is available at http://www.ebiomed.org/ncFANs/ or http://www.noncode.org/ncFANs/. So far, ncFANs includes 10 Affymetrix arrays for two organisms (mouse and human). ncFANs is comprised of two major parts. Part I deals with microarray data pre-processing, and Part II concerns lncRNA functional annotation. The workflow of ncFANs is shown in Figure 2.
Figure 2.
The workflow of ncFANs. Asterisk for the microarray raw data larger than 100 M, Part I of ncFANs, ‘Microarray Data Pre-processing’, should be implemented on user’s side by using the local pre-processing programs provided by ncFANs.
In the ‘Microarray Data Pre-processing’ step (Part I), the Affymetrix array raw data uploaded by the user are processed into gene expression data (for both coding and lncRNA genes) using the corresponding re-annotated Affymetrix CDF files. In its current version, ncFANs provides re-annotated CDFs for seven mouse arrays and three human arrays. In future ncFANs versions, re-annotated CDF files for more chip types will be included, and the coding and lncRNA genes data will be updated with the development of genome information. ncFANs provides two widely-used methods, RMA and MAS5.0, for processing the raw data of Affymetrix ‘PM-MM’ probe-designed microarrays. As an optional step, the expression of coding and lncRNA genes under an experimental condition could also be evaluated by MAS5CALLS. For the PM-only probe-designed microarrays (e.g. the exon array) ncFANs only provides RMA since MAS5.0 depends on MM probes which are absent in these arrays.
ncFANs applies two strategies to annotate the lncRNAs with potential functions (Part II): ‘Functional Annotation’ based on co-expression network characteristics, and ‘Functional Enrichment’ based on differential lncRNA expression in the microarrays. In the former strategy, ncFANs constructs a CNC network based on the expression profiles derived from Part I. The co-expression relationships are evaluated by statistical correlation coefficients. The parameters for constructing the CNC networks will be decided by the user. As outlined above, potential GO term or KEGG pathway functions are assigned to the lncRNAs based on the CNC network characteristics (using three different methods, including module-based, hub-based and co-location-based) and user-defined cut-offs. The ‘Functional Enrichment’ strategy is suitable for case-control experiment data, and applies common statistical methods such as Fold-Change and t-test to identify lncRNAs with significantly differential expression under the given experimental conditions.
Input
ncFANs requires two inputs: Affymetrix array raw data (CEL files) and the label file. The CEL files should be compressed into one zip file for uploading. The label file should consist of two columns which are tab-delimited, the first column containing the CEL file names and the second column the corresponding label for each CEL file in numeric format. The label file must be uploaded in zip compressed format. In addition, if the size of the uploaded microarray raw data is >100 M, the raw data should be processed on the user’s side using local pre-processing programs which can be downloaded from the Download page. In this situation, the user should upload the gene expression profile and its corresponding label file directly to ncFANs for the calculations in Part II. In order to gain a stable statistical significance, it is suggested that for the ‘Functional Annotation’ strategy in Part II, the total sample size of the input data should be five or more, and for the ‘Functional Enrichment’ strategy in Part II, each class of input data should include at least five samples.
Output
The output of ncFANs will be shown in the ‘Results’ web page, including the pre-processed gene expression profile and the functional annotation results. Employing the relevant re-annotated CDF file(s), the uploaded microarray raw data will be pre-processed into gene expression profile by the methods selected by the user, and the pre-processed gene expression profiles (containing expression values for both coding and lncRNA genes) can be downloaded from the results page.
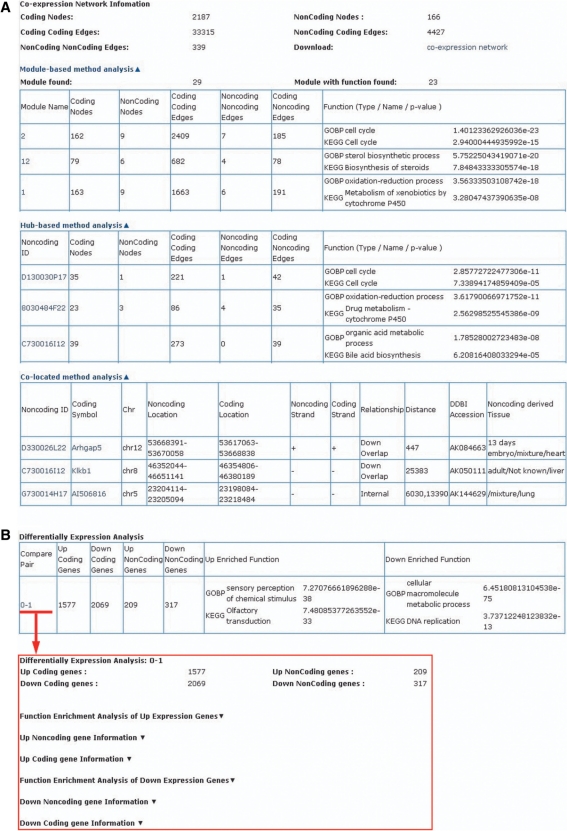
The output mode of the lncRNA functional annotations will depend on which of the two alternative functional annotation strategies (Part II) that has been employed. For the ‘Functional Annotation’ strategy, the potential functions of lncRNAs annotated by the three methods (module-based, hub-based and co-location based) will be shown with P-values calculated by the hypergeometric test. Moreover, several details, such as the size and members of each co-expression module, will also be shown. For each lncRNA, information concerning DDBJ Accession number, genomic location, derived tissues and neighbouring coding genes will be included, while for each coding gene, its gene symbol and description are shown. In addition, the whole CNC network will be exported to the user in the Cytoscape file format for graphical representation. For the ‘Functional Enrichment’ strategy, the output will show lncRNAs with significantly differential expression under the given experimental conditions. Furthermore, the coding genes will be grouped according to their expressional changes under the same experimental conditions. Significantly enriched functions in each group will be shown, thus enabling comparison to the relevant differentially expressed lncRNAs. An example of the data output is shown in Figure 3. The re-annotated CDF files and local pre-processing programs are available to users from the ncFANs download page.
Figure 3.
The output of ncFANs for Affymetrix Mouse Genome 430 2.0 Array using the example data sets. (A) The output from the ‘Functional Annotation’ strategy for a subset of GSE6998. (B) The output from the ‘Functional Enrichment’ strategy for a subset of GSE11990.
Computational implementation
After a job is launched, the user is redirected to a job status page that automatically reloads to report the progress status until the job is finished. Alternatively, users are asked to provide an email address to which their results will be forwarded when the job is finished. The time cost of Part I depends on the number of chips. The processing of a raw data set of 17 chips with default parameters requires 198.65 s on our Linux-cluster. With default parameters for Part II, ncFANs needs 78.06 s to construct a co-expression network from an expression profile with 15 290 genes and 17 experimental points. The time cost of functional annotation with a 68 044-edge network and default parameters is 42.24 s.
CONCLUSION
ncFANs is the first web tool that performs functional annotation of lncRNAs based on a coding-non-coding gene co-expression network constructed from the re-annotated microarray data. In addition, ncFANs also provides access to pre-processed expression profiles and re-annotated CDF files, which enables users to integrate systematic analysis with various types of other high-throughput biological data sets. In the future, ncFANs will include more array platforms of Affymetrix and other corporations such as Agilent, Illumina and Invitrogen. With the development of genome information, the re-annotated CDF files will be updated by integrating the lncRNA information from FANTOM3 and Vega with several other available databases such as NONCODE (30), RNAdb (31) and lncRNAdb (32). Besides, ncFANs will improve the methods of functional annotation by integrating various upcoming high-throughput lncRNA data sets, such as physical interaction data, genetic interaction data and RNA structure data. In summary, ncFANs introduces a highly useful tool for re-use of the abundant existing microarray data. We strongly believe that ncFANs will be an informative data analysis tool for the scientific community, and will provide valuable information regarding the role of lncRNAs in biological processes.
FUNDING
Knowledge Innovation Program of the Chinese Academy of Sciences (KSCX2-EW-R-01); 2010 Innovation Program of Beijing Institutes of Life Science, the Chinese Academy of Sciences; National Natural Science Foundation of China (No. 31071137, 31000586, 30970623); International Science and Technology Cooperation Projects (2010DFA31840, 2010DFB33720). Funding for open access charge: National Natural Science Foundation of China (No. 31071137).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Jinwu Wang for helping us to design the style of the website. The authors also thank Dr Lei Kong for collecting the microarray data sets from GEO for us. The authors wish to thank the executive editor and two anonymous referees for their constructive advice and comments to improve this web server.
REFERENCES
- 1.Okazaki Y, Furuno M, Kasukawa T, Adachi J, Bono H, Kondo S, Nikaido I, Osato N, Saito R, Suzuki H, et al. Analysis of the mouse transcriptome based on functional annotation of 60,770 full-length cDNAs. Nature. 2002;420:563–573. doi: 10.1038/nature01266. [DOI] [PubMed] [Google Scholar]
- 2.Ota T, Suzuki Y, Nishikawa T, Otsuki T, Sugiyama T, Irie R, Wakamatsu A, Hayashi K, Sato H, Nagai K, et al. Complete sequencing and characterization of 21,243 full-length human cDNAs. Nat. Genet. 2004;36:40–45. doi: 10.1038/ng1285. [DOI] [PubMed] [Google Scholar]
- 3.Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza-Blanc P, Hogenesch JB, Schultz PG. A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science. 2005;309:1570–1573. doi: 10.1126/science.1115901. [DOI] [PubMed] [Google Scholar]
- 4.Zhou Y, Zhong Y, Wang Y, Zhang X, Batista DL, Gejman R, Ansell PJ, Zhao J, Weng C, Klibanski A. Activation of p53 by MEG3 non-coding RNA. J. Biol. Chem. 2007;282:24731–24742. doi: 10.1074/jbc.M702029200. [DOI] [PubMed] [Google Scholar]
- 5.Huarte M, Guttman M, Feldser D, Garber M, Koziol MJ, Kenzelmann-Broz D, Khalil AM, Zuk O, Amit I, Rabani M, et al. A large intergenic noncoding RNA induced by p53 mediates global gene repression in the p53 response. Cell. 2010;142:409–419. doi: 10.1016/j.cell.2010.06.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene. 2003;22:8031–8041. doi: 10.1038/sj.onc.1206928. [DOI] [PubMed] [Google Scholar]
- 7.Taft RJ, Pang KC, Mercer TR, Dinger M, Mattick JS. Non-coding RNAs: regulators of disease. J. Pathol. 2010;220:126–139. doi: 10.1002/path.2638. [DOI] [PubMed] [Google Scholar]
- 8.Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev. 2009;23:1494–1504. doi: 10.1101/gad.1800909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat. Rev. Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- 10.Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends Genet. 2006;22:1–5. doi: 10.1016/j.tig.2005.10.003. [DOI] [PubMed] [Google Scholar]
- 11.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 12.RIKEN Genome Exploration Research Group and Genome Science Group (Genome Network Project Core Group) and the FANTOM Consortium. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, Nishida H, Yap CC, Suzuki M, et al. Antisense transcription in the mammalian transcriptome. Science. 2005;309:1564–1566. doi: 10.1126/science.1112009. [DOI] [PubMed] [Google Scholar]
- 13.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, Thomas K, Presser A, Bernstein BE, van Oudenaarden A, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc. Natl Acad. Sci. USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, He S, Liu C, Ru S, Zhao H, Yang Z, Yang P, Yuan X, Sun S, Bu D, et al. MicroRNA regulation of messenger-like noncoding RNAs: a network of mutual microRNA control. Trends Genet. 2008;24:323–327. doi: 10.1016/j.tig.2008.04.004. [DOI] [PubMed] [Google Scholar]
- 17.Babbitt CC, Fedrigo O, Pfefferle AD, Boyle AP, Horvath JE, Furey TS, Wray GA. Both noncoding and protein-coding RNAs contribute to gene expression evolution in the primate brain. Genome Biol. Evol. 2010;2:67–79. doi: 10.1093/gbe/evq002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kao HL, Gunsalus KC. Browsing multidimensional molecular networks with the generic network browser (N-Browse) Current Proc Bioinformatics. 2008 doi: 10.1002/0471250953.bi0911s23. Chapter 9, Unit 9 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Alexeyenko A, Sonnhammer EL. Global networks of functional coupling in eukaryotes from comprehensive data integration. Genome Res. 2009;19:1107–1116. doi: 10.1101/gr.087528.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Warde-Farley D, Donaldson SL, Comes O, Zuberi K, Badrawi R, Chao P, Franz M, Grouios C, Kazi F, Lopes CT, et al. The GeneMANIA prediction server: biological network integration for gene prioritization and predicting gene function. Nucleic Acids Res. 2010;38:W214–W220. doi: 10.1093/nar/gkq537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liao Q, Liu C, Yuan X, Kang S, Miao R, Xiao H, Zhao G, Luo H, Bu D, Zhao H, et al. Large-scale prediction of long non-coding RNA functions in a coding-non-coding gene co-expression network. Nucleic Acids Res. 2011;39:3864–3878. doi: 10.1093/nar/gkq1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Risueno A, Fontanillo C, Dinger ME, De Las Rivas J. GATExplorer: genomic and transcriptomic explorer; mapping expression probes to gene loci, transcripts, exons and ncRNAs. BMC Bioinformatics. 2010;11:221. doi: 10.1186/1471-2105-11-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Michelhaugh SK, Lipovich L, Blythe J, Jia H, Kapatos G, Bannon MJ. Mining Affymetrix microarray data for long non-coding RNAs: altered expression in the nucleus accumbens of heroin abusers. J. Neurochem. 2011;116:459–466. doi: 10.1111/j.1471-4159.2010.07126.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ørom UA, Derrien T, Beringer M, Gumireddy K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q, et al. Long noncoding RNAs with enhancer-like function in human cells. Cell. 2010;143:46–58. doi: 10.1016/j.cell.2010.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kong L, Zhang Y, Ye ZQ, Liu XQ, Zhao SQ, Wei L, Gao G. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007;35:W345–W349. doi: 10.1093/nar/gkm391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pruitt KD, Tatusova T, Maglott DR. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D61–D65. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mondal T, Rasmussen M, Pandey GK, Isaksson A, Kanduri C. Characterization of the RNA content of chromatin. Genome Res. 2010;20:899–907. doi: 10.1101/gr.103473.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He S, Liu C, Skogerbo G, Zhao H, Wang J, Liu T, Bai B, Zhao Y, Chen R. NONCODE v2.0: decoding the non-coding. Nucleic Acids Res. 2008;36:D170–D172. doi: 10.1093/nar/gkm1011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang KC, Stephen S, Dinger ME, Engstrom PG, Lenhard B, Mattick JS. RNAdb 2.0–an expanded database of mammalian non-coding RNAs. Nucleic Acids Res. 2007;35:D178–D182. doi: 10.1093/nar/gkl926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]