Abstract
The Genome-Linked Application for Metabolic Maps (GLAMM) is a unified web interface for visualizing metabolic networks, reconstructing metabolic networks from annotated genome data, visualizing experimental data in the context of metabolic networks and investigating the construction of novel, transgenic pathways. This simple, user-friendly interface is tightly integrated with the comparative genomics tools of MicrobesOnline [Dehal et al. (2010) Nucleic Acids Research, 38, D396–D400]. GLAMM is available for free to the scientific community at glamm.lbl.gov.
INTRODUCTION
As the volume of genomic, experimental and metabolic network data increases, so has the need for clean, unobtrusive methods for visualizing and contextualizing these data. With this in mind, we have developed the Genome-Linked Application for Metabolic Maps (GLAMM). GLAMM provides a unified web interface for visualizing metabolic networks, reconstructing metabolic networks from annotated genome data or custom user-defined networks, visualizing experimental data in the context of metabolic networks and investigating the construction of novel, transgenic pathways.
Other web resources (1–6) such as the KEGG Atlas, iPath, Pathway Projector, MetaCyc and Reactome offer similar, web-based mapping-style interfaces, but GLAMM also incorporates an interface for biological retrosynthesis (7–9), visualization of thousands of publicly accessible experimental or other user-defined data in the context of metabolic pathways, and integration with MicrobesOnline (10). This integration provides GLAMM users access to MicrobesOnline’s powerful comparative phylo-genomic and functional genomic tools and a database of nearly 2000 prokaryotic and fungal genomes, allowing rapid analysis of genome context, regulon discovery and so on.
GLAMM was developed using the GoogleTM Web Toolkit (GWT, http://code.google.com/webtoolkit/) for the client UI and server implementation. The underlying maps are Scalable Vector Graphics (SVG) documents rendered in real time on the client side in a GWT widget, with UI components and event handling provided by the GWT. Both of these technologies have the advantage of consistent cross-browser support, as well as a highly optimized execution path, with JavaScript and SVG rendered by the browser’s own internal implementations. As such, GLAMM will only work with browsers that support both JavaScript and SVG (e.g. Firefox, Chrome and Safari). This implementation performs well for thousands of on-screen elements on a typical personal computer.
In addition to a client-side interface, we have implemented a server that is integrated with MicrobesOnline. The GLAMM server communicates with the client via highly modularized and separable XML. The client can request any combination of pathways, reactions, genes and compounds. It also can request functional data, currently gene expression data, but data associated with reactions (e.g. flux) and metabolites (e.g. concentrations) will be supported in the near future. We chose to create a new, lightweight XML format that only included the features needed by the interface rather than employ an existing format such as SBML (11) or BioPAX (12) in order to minimize the data necessary to transfer and because we needed to add support for features not already captured by SBML or BioPAX. We expect to support export to BioPAX and perhaps SBML in the future.
UNDERLYING METABOLIC NETWORK
We have developed a method for aggregating and normalizing compound, reaction and pathway data from several different metabolic databases. We chose to first focus our attention on combining KEGG (13), MetaCyc (4) and the compound and reaction databases provided for the Escherichia coli iJR904 model (6). We also included reconciliation of metabolites with PubChem (14) and ChEBI (15). The database aggregation and normalization code is general enough to accommodate information from any similar database with the addition of a compatible parser with an eye toward inclusion of custom pathways, such as those found in organisms of interest to bioremediation and bio-fuel production.
Compounds and reactions were extracted from flat-file representations of the databases and converted to a normal form. For compounds, this normal form includes information such as common name, mass, formula, SMILES representation (16), InChI representation (17), compound name synonyms and external references to the compound in other databases. Similarly, for reactions, the normal form includes a normalized form of the balanced reaction equation, a human-readable reaction definition, external references to this reaction in other databases, E.C. numbers (18) (if applicable), KEGG RPAIR role information for the reactants and products and the KEGG pathway to which this reaction belongs. The normalized format is flexible enough to be expanded as custom reactions are introduced.
Compounds present in multiple databases are resolved into single entries by comparing the external reference IDs (e.g. PubChem) and merging normalized entries if a match is found. For consistency, KEGG common names, masses and formulae take precedence over those from other databases. We are continuing to investigate schemes for normalizing reactions, a more complicated endeavor as a consequence of the numerous similar but non-identical names given to reactants, products and secondary metabolites and which are included in the definition of each reaction (e.g. the inclusion of chirality information, different protonation states, polymers, etc.).
The data aggregation and normalization code is written entirely in object-oriented Perl and therefore can be run on almost any platform. This will no doubt change, as we intend to develop a fully automatic update and reconciliation mechanism. While the individual databases we have incorporated are curated, there remain some reactions that do not always account for mass balance or possess other eccentricities. Regrettably, it is beyond the scope of this project to rectify those issues, but we will update our imported network as improved data becomes available.
AUTOMATED METABOLIC RECONSTRUCTION
GLAMM uses the gene annotations in MicrobesOnline to automatically reconstruct the metabolic networks of almost 2000 organisms. It combines MicrobesOnline’s E.C. assignments [derived from hits to TIGRFAMs (19), KEGG annotations and orthologs from reference genomes] with the E.C. number to reaction ID mappings from the public databases aggregated in GLAMM. Taken together, these mappings loosely determine the set of reactions available for a given organism. We recognize that automated E.C. assignments based solely on homology to a gene family are limited and by no means comparable to that of dedicated reconstruction pipelines such as ModelSEED (20) or manually curated reconstructions (6). GLAMM therefore supports custom, user-uploaded reconstructions (see below) and will support reconstructions from other databases in the future.
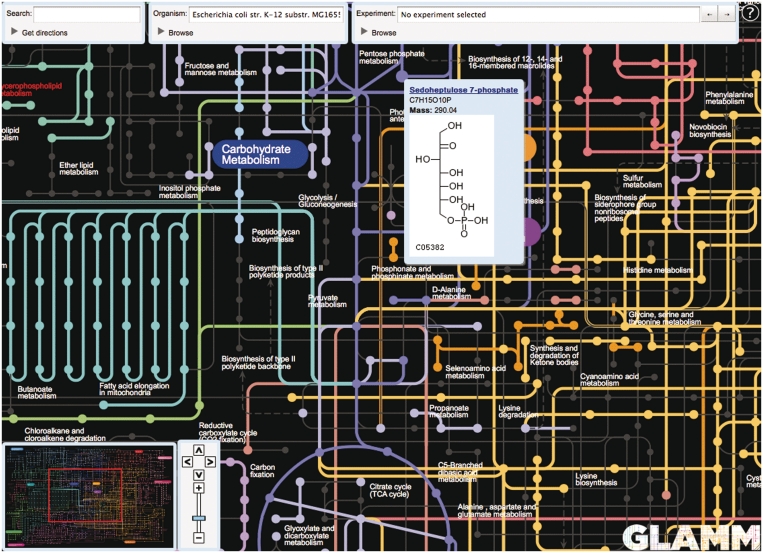
When the user selects a host organism, GLAMM prunes the set of reaction edges in the global map to only include those reactions available to that organism (Figure 1). The remaining reaction edges on the displayed map are grayed out. Based on the connectivity information supplied with the map, GLAMM also prunes compound nodes that have no reactions associated with them. This not only yields the metabolic reaction network, but also the set of all compounds endogenous to the host, within the constraints of the displayed map which is, by necessity, a subset of the actual metabolic network of known chemical transformations.
Figure 1.
Metabolic reconstruction of E. coli K12 substr. MG1655 with metabolite information for Sedoheptulose 7-phosphate in a pop-up window. Reactions with genes identified in the reconstruction are shown in color, missing reactions in gray.
There are obvious limitations to this technique, including the incompleteness of E.C. assignments for genes and that E.C. numbers often specify a broad class of reactions and therefore may not be substrate-specific. We aim to overcome these limitations in the future by augmenting the MicrobesOnline database with direct gene to reaction mappings (e.g. using KEGG orthologs.)
CUSTOM METABOLIC RECONSTRUCTION
GLAMM also provides a mechanism for uploading custom metabolic networks. Initially, this is in the form of tab-delimited files containing gene ID to E.C. number or gene ID to reaction ID mappings. Eventually we aim to support SBML and BioPAX specified pathways directly. The default metabolic reconstruction for any organism in MicrobesOnline may be downloaded, modified and re-uploaded.
GLAMM FEATURE HIGHLIGHTS
Metabolites and metabolic pathways
The current GLAMM global view presents the KEGG Atlas map, but can be updated with any metabolic map using a standard format that we have designed. The resultant visualized map is pannable and zoomable as typical of web-mapping applications. Compounds are represented as nodes on the map. Reactions, along with their corresponding genes in the organism-specific metabolic network reconstructions, are represented as edges. Clicking on the nodes presents a popup window (Figure 1) containing the compound’s name, its formula, its mass and a structural diagram, if available. Similarly, clicking on the edges of the map presents a pop-up window containing the reaction’s human-readable definitions, its E.C. numbers and the number of genes corresponding to those E.C. numbers in the target organism. The global map also contains textual labels for the various sub-pathways, and clicking on those labels presents pop-up windows containing schematic representations of the more detailed KEGG pathway maps. All pop-ups include links back to the corresponding pathways, genes or metabolites in MicrobesOnline.
Route finding and retrosynthesis
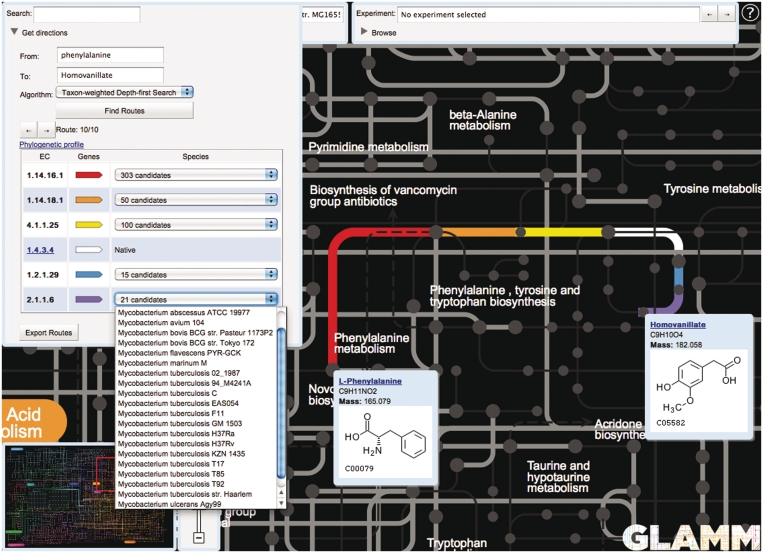
For convenience, we have included a search dialog box that re-centers the map around any compound, reaction or gene name specified by the user. Additionally, the global view will allow the user to ‘get directions’ in finding optimal pathways between a starting metabolite and a desired target metabolite (Figure 2). In the event of ambiguous compound search results, often due to the presence of multiple isomers on the map (e.g. glucose may specify α-d-glucose or β-d-glucose,) a disambiguation popup will appear, allowing the user to specify the desired compound. Suggested pathways may offer routes for retrosynthesis and traverse all annotated organisms or otherwise conceivable reaction steps using a variety of appropriate pathway/gene set cost functions, returning the necessary genes to add to the host in order to complete the pathway from the chassis network to the target molecule. The routes are overlaid on top of the main map view, and all non-participating reactions are grayed out. If a host organism is selected, the E.C. number links to MicrobesOnline for candidate genes and retrosynthetic pathways are enabled in order to facilitate further examination with its powerful comparative systems biology tools, including gene trees, genome context and operon predictions, functional residue alignments, basic structural models and functional expression data. These tools are provided with the intent of developing a mutually consistent set of genes for introducing the pathway into the host organism.
Figure 2.
Route finding and retrosynthesis ‘Getting directions’ between the metabolites l-Phenylalanine and Homovanillate using E. coli K12 substr. MG1655 as the host organism. Both endogenous (white) and exogenous reactions (colored) are shown, including the species names for the source of candidate genes for the transgenic steps in the pathway.
Experimental data visualization
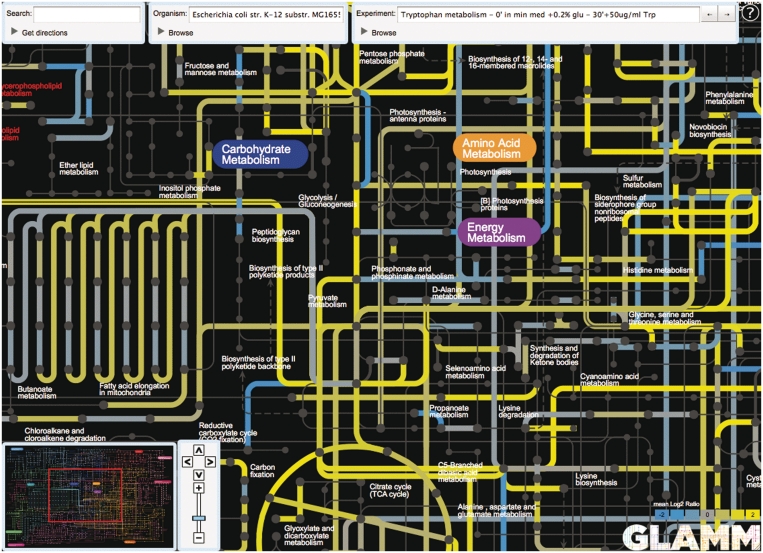
Additionally, the global view can be used to visualize any data as an ‘overlay’, including *omics data such gene expression, protein levels, flux, source organism for a given reaction in a synthetic network, kinetic and thermodynamic parameters, optimal paths between metabolites and so on (Figure 3). For example, *omics data will permit the user to analyze the global behavior of the network when challenged by stressful conditions or particular nutrient levels and to identify key pathways that are either directly involved in target molecule synthesis or may otherwise impact metabolic engineering.
Figure 3.
Experimental data visualization. Overlay of expression data collected during a metabolism experiment on E. coli K12 substr. MG1655. The reactions corresponding to upregulated genes are shown in yellow, reactions corresponding to downregulated genes are shown in blue.
Custom data overlay
In addition to public experimental data available on MicrobesOnline, the user may upload tab-delimited files with a list of genes and numerical data values for those genes. Similar to the downloadable metabolic reconstructions, one may also download experimental data sets that contain gene names consistent with metabolic reconstructions, to which new data values may be applied.
Future directions
GLAMM will continue to be developed to support additional data types and custom display of data associated with reactions and metabolites. Additional bounds on retrosynthesis pathways, as well as longer pathways will be implemented to permit the user to require routes that pass through or avoid user-defined intermediates, that maximize or minimize use of particular cofactors, that maximize predicted flux and so on. Source code will be made available freely for academic research.
FUNDING
Office of Biological and Environmental Research (BER) of the US Department of Energy (DOE) Office of Science under Contract No. DE-AC02-05CH11231 with the E.O. Lawrence Berkeley National Laboratory (LBNL) (to Joint BioEnergy Institute, JBEI); Office of Biological and Environmental Research in the US DOE Office of Science with American Recovery and Reinvestment Act (ARRA) funding to Oak Ridge National Laboratory (ORNL) (to ‘Knowledgebase R&D’ project performed at LBNL) administered by UT-Battelle, LLC, for the US Department of Energy under contract DE-AC05-00OR22725 (to ORNL). Funding for open access charge: Office of Biological and Environmental Research (to JBEI), of the US DOE Office of Science under Contract No. DE-AC02-05CH11231 with the E.O. Lawrence Berkeley National Laboratory (LBNL).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would also like to thank Thanya Suwansawad for the design of the GLAMM logo.
REFERENCES
- 1.Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36:W423–W426. doi: 10.1093/nar/gkn282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I, Yamada T, Kanehisa M, Bork P. iPath: interactive exploration of biochemical pathways and networks. Trends Biochem. Sci. 2008;33:101–103. doi: 10.1016/j.tibs.2008.01.001. [DOI] [PubMed] [Google Scholar]
- 3.Kono N, Arakawa K, Ogawa R, Kido N, Oshita K, Ikegami K, Tamaki S, Tomita M. Pathway Projector: Web-Based Zoomable Pathway Browser Using KEGG Atlas and Google Maps API. PLoS ONE. 2009;4:e7710. doi: 10.1371/journal.pone.0007710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Croft D, O’Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schellenberger J, Park JO, Conrad TC, Palsson BØ. BiGG: a Biochemical Genetic and Genomic knowledgebase of large scale metabolic reconstructions. BMC Bioinformatics. 2010;11:213. doi: 10.1186/1471-2105-11-213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Prather KLJ, Martin CH. De novo biosynthetic pathways: rational design of microbial chemical factories. Curr. Opin. Biotechnol. 2008;19:468–474. doi: 10.1016/j.copbio.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 8.Henry CS, Broadbelt LJ, Hatzimanikatis V. Discovery and analysis of novel metabolic pathways for the biosynthesis of industrial chemicals: 3-hydroxypropanoate. Biotechnol. Bioeng. 2010;106:462–473. doi: 10.1002/bit.22673. [DOI] [PubMed] [Google Scholar]
- 9.Faulon JL, Carbonell P. Reaction network generation. In: Faulon JL, Bender A, editors. Handbook of Chemoinformatics Algorithms. Chapman & Hall/CRC Series in Mathematical & Computational Biology; 2010. [Google Scholar]
- 10.Dehal PS, Joachimiak MP, Price MN, Bates JT, Baumohl JK, Chivian D, Friedland GD, Huang KH, Keller K, Novichkov PS, et al. MicrobesOnline: an integrated portal for comparative and functional genomics. Nucleic Acids Res. 2010;38:D396–D400. doi: 10.1093/nar/gkp919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hucka M, Finney A, Sauro HM, Bolouri H, Doyle JC, Kitano H, Arkin AP, Bornstein BJ, Bray D, Cornish-Bowden A, et al. The systems biology markup language (SBML): a medium for representation and exchange of biochemical network models. Bioinformatics. 2003;19:524–531. doi: 10.1093/bioinformatics/btg015. [DOI] [PubMed] [Google Scholar]
- 12.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D’Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2011;39:D38–D51. doi: 10.1093/nar/gkq1172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, Mcnaught A, Alcántara P, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weininger D. SMILES, a chemical language and information system. 1. Introduction to methodology and encoding rules. J. Chem. Inform. Model. 1988;28:31. [Google Scholar]
- 17.Stein SE, Heller SR, Tchekhovskoi D. Proceedings of the 2003 International Chemical Information Conference (Nimes) Malmesbury, UK: Infornortics; 2003. An open standard for chemical structure representation: the IUPAC chemical identifier; pp. 131–143. [Google Scholar]
- 18.Webb EC. Enzyme nomenclature 1992: recommendations of the Nomenclature Committee of the International Union of Biochemistry and Molecular Biology on the nomenclature and classification of enzymes. San Diego: International Union of Biochemistry and Molecular Biology by Academic Press; 1992. [Google Scholar]
- 19.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Henry CS, DeJongh M, Best AA, Frybarger PM, Linsay B, Stevens RL. High-throughput generation, optimization, and analysis of genome-scale metabolic models. Nat. Biotechnol. 2010;28:977–982. doi: 10.1038/nbt.1672. [DOI] [PubMed] [Google Scholar]