Abstract
Next-generation sequencing (NGS) technologies are making sequence data available on an unprecedented scale. In this context, new catalogs of Single Nucleotide Polymorphism and mutations generated by resequencing studies are usually stored in genome position files (e.g. Variant Call Format, SAMTools pileup, BED, GFF) comprising of large lists of genomic positions, which are difficult to handle by researchers. Here, we present PileLineGUI, a novel desktop application primarily designed for manipulating, browsing and analysing genome position files (GPF), with specific support to somatic mutation finding studies. The developed tool also integrates a new genome browser module specially designed for inspecting GPFs. PileLineGUI is free, multiplatform and designed to be intuitively used by biomedical researchers. PileLineGUI is available at: http://sing.ei.uvigo.es/pileline/pilelinegui.html.
INTRODUCTION
Nowadays, next-generation sequencing (NGS) technologies allow the generation of sequence data on an unprecedented scale. The versatility of NGS-based techniques together with the remarkable reduction in per-base sequencing cost, have encouraged researchers to produce a growing number of DNA resequencing studies. The 1000 Genomes Project leads this trend and represents a notable effort to understand the human variation at a genome-wide scale (1). The goal is to report a comprehensive catalogue of DNA variants corresponding to a number of human populations, expanding the concept of reference human genome from the current haploid and mosaic sequence to a frequency-based model built from population-specific variants. Along the same lines, the detection of novel DNA variants is also an objective for the International Cancer Genome Consortium [ICGC (http://www.icgc.org)]. This project is currently resequencing more than 50 types of cancer providing new collections of tumour-associated genomic mutations with the aim of relating them to the oncogenic processes, tumour evolution and clinical prognosis (2).
The new catalogues of Single Nucleotide Polymorphism (SNPs) and mutations generated by resequencing studies are usually stored in genome position files. Basically, genome position files (GPFs) consist of tab-delimited text files containing information regarding the base reported by the NGS experiment, defined by chromosome name and genomic coordinates. Several standard formats have been recently proposed to organize the information contained in GPFs, such as the Variant Call Format (.vcf) adopted by the 1000 Genomes Project (http://www.1000genomes.org/wiki/Analysis/Variant%20Call%20Format/vcf-variant-call-format-version-40), and the .pileup format (3). More traditional GPFs also include the .bed and .gff formats (http://genome.ucsc.edu/FAQ/FAQformat).
In such a situation, GPFs analysis becomes a critical task for a deeper understanding of the genome and its relation to human diseases. However, given that GPFs usually include large lists of genomic positions, their handling is cumbersome. This situation calls for efficient, practical and intuitive tools to explore, analyse, visualize and annotate DNA variants. To this end, some authors have recently introduced novel, free and useful applications to deal with standard GPFs. For instance, the Annotate Variation (ANNOVAR) toolbox (4) is a collection of Perl scripts to functionally annotate DNA variants detected at gene (or region) level. Among other things, the software can handle several GPFs standard formats and provides tools to estimate the biological impact of DNA variants (i.e. retrieve non-synonymous mutations) from pre-computed Sorting Intolerant From Tolerant (SIFT) scores (5). Nevertheless, ANNOVAR does not implement a genome browser able to explore variants along with the reference genome and its command-line nature could be too difficult for biomedical researchers without computing skills. EagleView (6), Tablet (7), Savant (8), IGV (9), Artemis (10) and MagicViewer (11) offer friendly GUIs together with powerful genomic browsers for visualization of genetic variation and its associated annotation. Nonetheless, common tasks in biomedical studies such as variant pairwise comparisons (i.e. case versus control) or variant comparisons across n GPFs are not supported by these applications. In addition, they do not provide specific support to generate inputs for third-party programs with the goal of predicting the biological impact of DNA variants.
In this context, here we introduce PileLineGUI, a novel desktop application specifically intended for effectively handling and browsing GPFs generated in DNA-seq experiments. PileLineGUI is open software, multi-platform and designed to be intuitively used by biomedical researchers. The application includes all the core functionalities implemented in the PileLine command-line toolbox (12) and a new interactive genome browser to explore DNA variants. PileLineGUI implements a number of operations for filtering, searching, annotating and analysing DNA variants, with specific support for generating out-files that can be subsequently used by third-party software such as SIFT (5), Polyphen2 (13) and FireStar (14) specialized in prediction of biological impact of DNA variants.
PileLineGUI supports pair-wise comparisons versus the reference and is able to generate four different types of specific variants lists: (i) variants in case, but not in control, (ii) variants in control, but not in the case, (iii) common variants in both case and control and (iv) discrepant variants in both case and control. It also allows comparisons across n GPFs at genomic coordinate or whole-gene level. This functionality may be of particular interest to detect a common mutation (or SNP) across several individuals sharing a particular phenotype. PileLineGUI supports Variant Cell Format (VCF), SAMTools pileup, GFF and BED standard files, together with the FASTA format used by the genome browser for reference genome visualization.
DATA INPUT
All the input files required by the PileLineGUI tool are standard formats. The software reads and visualizes inputs in .pileup and .vcf for single-nucleotide files (each line contains information for a single genome position), as well as .bed and .gff for intervals files (each line defines a continuous interval in the genome, i.e. a gene). Moreover, input GP files can be also BGZF-compressed in order to alleviate the disk requirements while keeping the possibility to manage them efficiently.
The built-in genome browser works with the standard SAMTools indexed FASTA files (.fai) and currently supports all previous GP files for tracks, including their compressed form.
GP FILES MANAGEMENT AND ANNOTATION
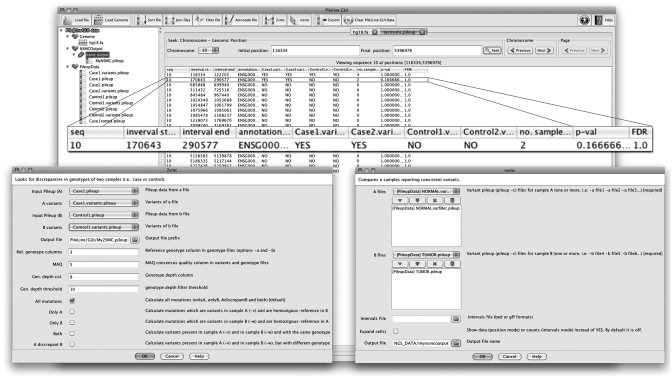
PileLineGUI allows the user to load and browse GP files via a user-friendly interface that can be used to instantly search for a given range within a sequence. Figure 1 shows a screenshot of the GP file browser.
Figure 1.
PileLineGUI GP file browser interface. Standard GP files may be easily explored and analysed using this tool. In this example, the output of the nsmc functionality is shown. Note that the second row corresponds to a variant consistently detected in cases, but not in controls.
In addition, PileLineGUI provides a set of basic processing functions for general purposes. Two single-nucleotide GP files (A and B) can be joined together in order to match the information associated to the same genomic position in both A and B. Moreover, the join function allows the user to include unmatched lines of A (left outer join) or B (right outer join). If the user is only interested in specific intervals of a given GP file (e.g. genes, dbSNP, custom targets, etc.), a single-nucleotide GP file can be filtered using the intervals specified in an intervals GP file. The filter function also allows the user to perform ‘inverse’ filters, for example, to filter-out all entries, which are known SNPs in dbSNP. Similarly to the filter function, the user can annotate each position in any GP file using an intervals file.
SOMATIC MUTATIONS FINDING
PileLineGUI includes two additional modules to find somatic mutations. Working with SAMTools pileup files, the two samples somatic mutations calling (2smc) function compares two samples (A and B) of an individual (e.g. healthy against tumour sample). Each sample is defined by (i) a variants-only file and (ii) a complete sequencing file (including non-variant positions). The 2smc operation generates 4 GP files as output, including (i) variants found in A, which were sequenced as non-variants in B, (ii) variants found in B, which were sequenced as non-variants in A, (iii) variants found in A and B and (iv) variants found in A and B, but with a discrepant genotype. For example, if A represents a tumour sample and B a healthy one from the same individual, variants found only in A are candidates for being considered as somatic mutations (not present in the germ line).
In addition, the n samples somatic mutation calling (nsmc) function allows the user to assess the reproducibility of several 2smc experiments. For example, if we have several samples, we could first extract the non germ-line mutations with 2smc and then use nsmc to check, in which samples each mutation is found. Further, if we have two groups of samples (e.g. the tumour samples plus a control group), nsmc provides a Fisher’s exact test over each mutation in order to arrive at the significance of the reproducibility. Furthermore, nsmc can also compare samples at the single-nucleotide level or using genomic intervals (e.g. gene or exon level). In this sense, it is sometimes more interesting to see whether tumour samples contain mutations in several genes of interest, regardless of their exact positions.
Finally, a pileup file containing a list of candidate somatic mutations can be exported to several mutation consequence assessment packages like SIFT, Polyphen and Firestar.
EXPLORING GENOME POSITION FILES USING THE GENOME BROWSER
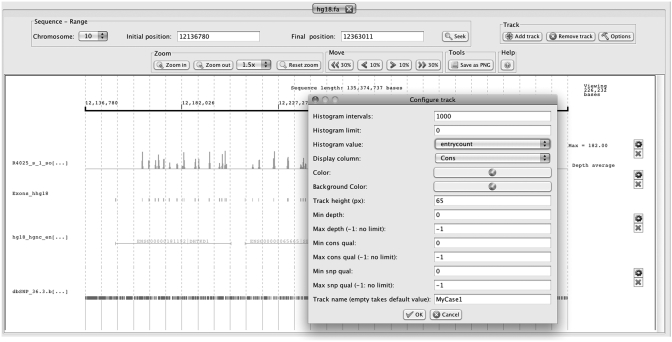
PileLineGUI incorporates a powerful genome browser for GP files including multiple track support, fully interactive zoom and image export facilities. The browser loads a genome via a standard SAMTools FASTA index (.fai) and allows the user to add several GP files as individual tracks. Single-nucleotide GP files (.pileup and .vcf) are rendered in the genome by displaying the value of a custom column at the corresponding genome position. Moreover, pileup files can be dynamically filtered by depth, SNP quality and consensus quality. If the zoom level is low (showing a very large range of bases), the browser plots a configurable histogram to see the distribution of occurrences in the GP file. Intervals GP files (.bed and .gff) are rendered as segments. If there are overlapping segments, they are plotted at different heights in the track. An example of the genome browser can be seen in Figure 2.
Figure 2.
PileLineGUI browser interface has been designed to be customizable and includes multiple track support. In this example, the first track (in blue) represents the sequencing depth along a genomic region in chromosome 10. Since data correspond to a full-exome experiment, depth peaks match to exon positions depicted in the second track (green). The third track (black) shows four Ensembl genes located in this particular genomic region. dbSNP annotations are available in the last track (red).
IMPLEMENTATION
PileLineGUI is implemented using the Java programming language on top of the PileLine tools (14), which can be seen as the command-line version of this software. The graphical user interface of PileLineGUI was developed using the AIBench application framework (15). The compatibility with SAMTools BGZF compressed files was included in PileLine by using the Java ports provided by the SAMTools project (http://picard.sourceforge.net/, http://samtools.sourceforge.net/tabix.shtml).
In addition, PileLineGUI implements a new genome browser as a separate plugin to facilitate its later inclusion and reuse in other projects. Moreover, this component was carefully designed in order to add full support to other types of tracks (e.g. a BAM track including aligned reads is also planned).
CONCLUSION
GPFs analyses are critical for a deeper understanding of the genome and its relation to human diseases. However, given that GPFs usually include large lists of genomic positions, they are often difficult to handle systematically by researchers. This situation calls for efficient, practical and intuitive tools to explore, analyse, visualize and annotate DNA variants.
Here, we present PileLineGUI, a desktop application to efficiently handle, browse and analyse genome position files focusing on somatic mutation finding. By also including a new interactive genome browser, this tool is useful for wet-lab users demanding fast and easy-to-use local applications able to analyze huge amounts of data coming from next-generation sequencing studies.
Further work aims at (i) extending the somatic mutation calling functions (2smc and nsmc) to handle VCF files, since the SAMTools pileup format is now deprecated and (ii) include more standard file formats to the genome browser, such as BAM.
PileLineGUI is licensed under the terms of LGPLv3 (GNU Lesser General public License, version 3) and it is available at: http://sing.ei.uvigo.es/pileline/pilelinegui.html. Full documentation can be found at: http://sing.ei.uvigo.es/pileline/index.php/GUI_Documentation.
FUNDING
Spanish Ministry of Science and Innovation, the Plan E from the Spanish Government and the European Union from the ERDF (TIN2009-14057-C03-02); University of Vigo (to M.R.-J., pre-doctoral fellowship). Funding for open access charge: Spanish Ministry of Science and Innovation, the Plan E from the Spanish Government and the European Union from the ERDF (TIN2009-14057-C03-02).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We want to thank all the beta testers, especially those from the Lymphoma group at the Spanish National Cancer Research Centre (CNIO) in Madrid.
REFERENCES
- 1.1000 Genomes Project Consortium. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.International Cancer Genome Consortium. International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar P, Henikoff S, Ng PC. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protocols. 2009;4:1073–1081. doi: 10.1038/nprot.2009.86. [DOI] [PubMed] [Google Scholar]
- 6.Huang W, Marth G. EagleView: a genome assembly viewer for next-generation sequencing technologies. Genome Res. 2008;18:1538–1543. doi: 10.1101/gr.076067.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Milne I, Bayer M, Cardle L, Shaw P, Stephen G, Wright F, Marshall D. Tablet—next generation sequence assembly visualization. Bioinformatics. 2009;26:401–402. doi: 10.1093/bioinformatics/btp666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fiume M, Williams V, Brook A, Brudno M. Savant: genome browser for high-throughput sequencing data. Bioinformatics. 2010;26:1938–1944. doi: 10.1093/bioinformatics/btq332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Robinson JT, Thorvaldsdóttir H, Winckler W, Guttman M, Lander ES, Getz G, Mesirov JP. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 11.Hou H, Zhao F, Zhou L, Zhu E, Teng H, Li X, Bao Q, Wu J, Sun Z. MagicViewer: integrated solution for next-generation sequencing data visualization and genetic variation detection and annotation. Nucleic Acids Res. 2010;38:W732–W736. doi: 10.1093/nar/gkq302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Glez-Peña D, Gómez-López G, Reboiro-Jato M, Fdez-Riverola F, Pisano DG. PileLine: a toolbox to handle genome position information in next-generation sequencing studies. BMC Bioinformatics. 2011;12:31. doi: 10.1186/1471-2105-12-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nat. Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.López G, Valencia A, Tress ML. Firestar: prediction of functionally important residues using structural templates and alignment reliability. Nucleic Acids Res. 2007;35:W573–W577. doi: 10.1093/nar/gkm297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glez-Peña D, Reboiro-Jato M, Maia P, Díaz F, Fdez-Riverola F. AIBench: a rapid application development framework for translational research in biomedicine. Comput. Methods Programs Biomed. 2010;98:191–203. doi: 10.1016/j.cmpb.2009.12.003. [DOI] [PubMed] [Google Scholar]