Abstract
PILGRM (the platform for interactive learning by genomics results mining) puts advanced supervised analysis techniques applied to enormous gene expression compendia into the hands of bench biologists. This flexible system empowers its users to answer diverse biological questions that are often outside of the scope of common databases in a data-driven manner. This capability allows domain experts to quickly and easily generate hypotheses about biological processes, tissues or diseases of interest. Specifically PILGRM helps biologists generate these hypotheses by analyzing the expression levels of known relevant genes in large compendia of microarray data. Because PILGRM is data-driven, it complements a user’s knowledge and literature analysis with mining of diverse functional genomic data, thereby generating novel predictions that can drive experimental follow-up. This server is free, does not require registration and is available for use at http://pilgrm.princeton.edu.
INTRODUCTION
High-throughput genomic data contain information about diverse processes, tissues and diseases. The application of data-mining algorithms to these large genomic datasets provides great potential for uncovering novel biology, but currently this potential is not often realized because collecting, properly processing and analyzing these data requires substantial computational resources and sophisticated programming knowledge. On the other hand, setting up analyses to address important biological questions and testing novel predictions resulting from such analyses requires detailed experimental knowledge.
Although there are several successful applications of sophisticated computing approaches to diverse functional genomics data collections (1–5), including some that share results through a web site (6–9), currently there is not an easy way for a researcher to set up new analyses and ask specific biological questions by focusing these analyses on a sub-process or tissue of interest. This greatly constrains the utility of the novel predictions, because direct experimental validation for some processes or tissues may be impractical. PILGRM (the platform for interactive learning by genomics results mining) addresses this limitation by allowing its users to generate specific biological hypotheses by directing the supervised analyses of global microarray expression collections simply by defining their own gold standards (lists of genes relevant to a process, disease or tissue). Such an approach puts sophisticated computational tools in the hands of biologists, thereby combining their biological insight with a powerful computational strategy. This flexibility lets users address questions as diverse as their research programs while targeting predictions to experimentally testable pathways, tissues or phenotypes.
Efforts to predict protein function, expression or localization from high-throughput data compendia generally make computational predictions based on annotations from expert-curated literature-derived databases. The limited coverage of these databases constrains bioinformatics strategies that use only database standards. These databases also do not represent unpublished experimental results that may be informative for future experiments. By encouraging and enabling users to define their own standards, PILGRM also alleviates this issue of limited database coverage.
However, PILGRM does not eschew these expert-curated literature-derived databases. Indeed as the successful prior applications of data mining strategies to these compendia have shown, these databases have great value. This is why PILGRM contains extensive collections of data and database-derived gold standards (detailed in Table 1) for Homo sapiens and the model organisms Mus musculus, Rattus norvegicus, Caenorhabditis elegans, Arabidopsis thaliana and Saccharomyces cerevisiae. We automatically process and integrate many sources of gene-annotation in PILGRM. We include the Gene Ontology, which has annotations for a protein’s biological process involvement, localization and biochemical function (10,11), the Plant Ontology, which has annotations for a protein’s role in plant development and anatomy (12), the Saccharomyces Genome Database phenotype annotations, which specify phenotypes observed when genes are knocked out (13) and the Human Protein Reference Database’s Tissue annotations, which provide literature-derived annotations of tissue specific expression, localization and function for human proteins (14). We are adding new databases as they are requested by users. These database annotations provide a convenient starting point for user-defined standards and analyses.
Table 1.
PILGRM contains large data compendia and standards derived from literature-curated databases for each the organisms that it covers
| Experiments | Arrays | Genes | Standards | Unique publications | |
|---|---|---|---|---|---|
| Human | 2392 | 77 473 | 21 702 | 7484 | 32 567 |
| Mouse | 2012 | 31 374 | 24 555 | 6864 | 14 248 |
| Yeast | 117 | 1801 | 6077 | 4231 | 10 134 |
| Arabidopsis | 408 | 5465 | 22 121 | 3929 | 6836 |
| Rat | 440 | 10 376 | 21 416 | 5242 | 14 395 |
| Worm | 53 | 963 | 17 027 | 1782 | 2489 |
The unique publications column shows how many distinct publications are represented in the gold standards pre-loaded in PILGRM for each organism. This table shows the status of these collections as of 31 January 2011.
For example, a researcher studying breast cancer progression may be interested in identifying novel candidate genes involved in breast cancer progression while avoiding genes that appear relevant simply because they are expressed in mammary epithelium (i.e. genes discoverable by a simple correlation analysis). This researcher can take advantage of both custom standards and the included database annotations in PILGRM. Setting up such an analysis without PILGRM would require that he download the full collection of over 70 000 gene expression experiments for human, develop appropriate data processing, normalization and integration methods, and set up a machine-learning framework for the analysis. He would then have to download the HPRD database to identify genes known to be expressed in the mammary epithelium, in addition to creating his custom standard of genes involved in breast cancer progression.
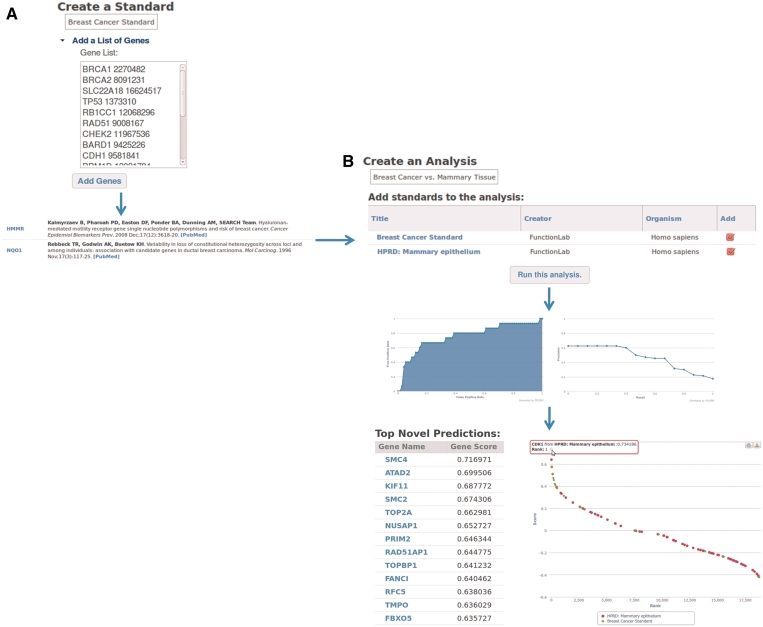
In contrast, this analysis takes minutes in PILGRM: Figure 1 shows the steps that this user performs during the preparation and interpretation of this analysis. First, the researcher develops a gold standard for genes involved in breast cancer through his own expertise and a literature search (Figure 1A). The PILGRM server allows each link between a gene and a gold standard to be associated with PubMed identifiers and these publication-annotated links are included in a downloadable document (PDF format) describing each analysis that is made available to the user (such a document can be used for additional record keeping by the users, to inform a Materials and methods section, or directly as Supplementary Data in publications resulting from this analysis). Second, he creates an analysis and pairs this breast cancer standard with the HPRD mammary epithelium standard included in PILGRM (Figure 1B). As a final step, the researcher runs this analysis and both metrics for the machine-learning results and novel predictions are returned by PILGRM.
Figure 1.
The flow of a PILGRM analysis that uses one custom standard and a pre-loaded standard to discover genes related to breast cancer progression while excluding general mammary epithelium genes. (A) The process of creating a standard and adding genes (here shown with optional PubMed IDs) to it. (B) The process of setting up and running an analysis. The breast cancer standard from (A) is combined with the HPRD mammary epithelium standard that is pre-loaded into PILGRM. The breast cancer standard is a positive and the mammary epithelium is a negative (here both are shown together for clarity). The analysis is run and standards quantitative performance metrics and novel predictions are provided to the user.
By combining custom standards with appropriate literature-curated databases and sophisticated machine learning of the Support Vector Machine (SVM) classifier implemented in PILGRM, this researcher discovers genes relevant to his research and saves time without compromising the flexibility or quality of his data-driven predictions. These relevant genes behave similarly (e.g. through co-expression) to the genes defined as interesting by the user (the positive standard) in informative experimental conditions. The machine-learning approach automatically identifies the conditions that best differentiate positive standard genes from those in the negative standard (genes with properties that the user wishes to avoid in new predictions). PILGRM provides both novel predictions and high-quality interactive visualizations of analysis results for the researcher to explore.
PILGRM’s main features are as follows.
A flexible interface that encourages user-defined data-driven analyses that answer diverse questions of biological interest including those outside the scope of common databases.
Regularly updated compendia of uniformly processed genomic data for human and common model organisms.
Regularly updated gold standards for tissue, function and development from common sources (GO, PO, HPRD, etc.) that make setting up analyses quick and easy.
User-set levels of access control (public, hidden, private) for standards and analyses, allowing users to include unpublished results in PILGRM.
SYSTEM DESCRIPTION
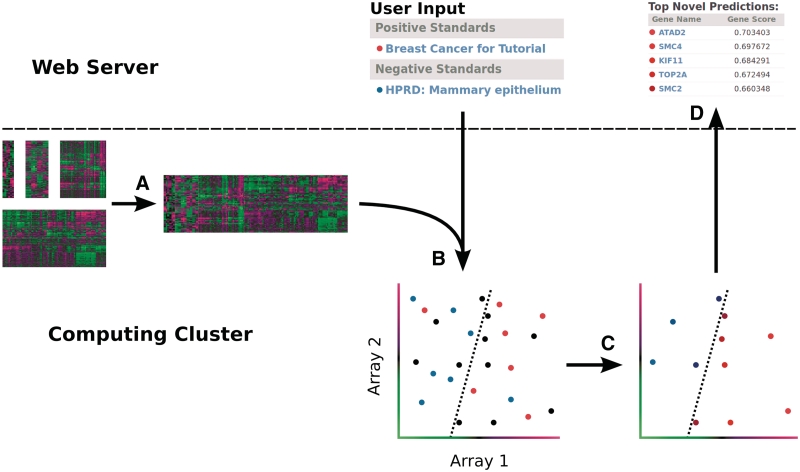
Each PILGRM analysis begins with an important biological question defined by the user. The user translates this question into appropriate gold standards, thereby defining the corresponding machine-learning problem. Gold standards are structured as positives (which represent genes like those that the user is seeking) and negatives (which represent genes with properties the user wants to exclude) and can be drawn from databases or developed by the user. These standards are added to an analysis that is run by the user. PILGRM then classifies all other genes in the organism of interest with a machine-learning algorithm that employs the user-provided positive and negative standards, thereby generating novel predictions. This process is summarized in Figure 2 and discussed in detail in Supplementary Data S1.
Figure 2.
(A) This diagram shows the flow of each PILGRM analysis. We pre-process separate datasets into a gene-expression compendium for each organism. (B) The user provides positive and negative standards (either input by the user or from common databases) and the data are labeled with these standards. The SVM algorithm identifies the maximum-margin hyperplane (here a dotted line for the two-arrays in this example, but in practice this is a plane in very high-dimensional space) that best separates the positive (red) and negative (blue) standards by gene expression. Unlabeled genes (black) are then ranked by their distance to this plane (C), and the ranked list is returned to the user as predictions (D). The user is also provided detailed evaluation plots based on cross-validation (Figure 3).
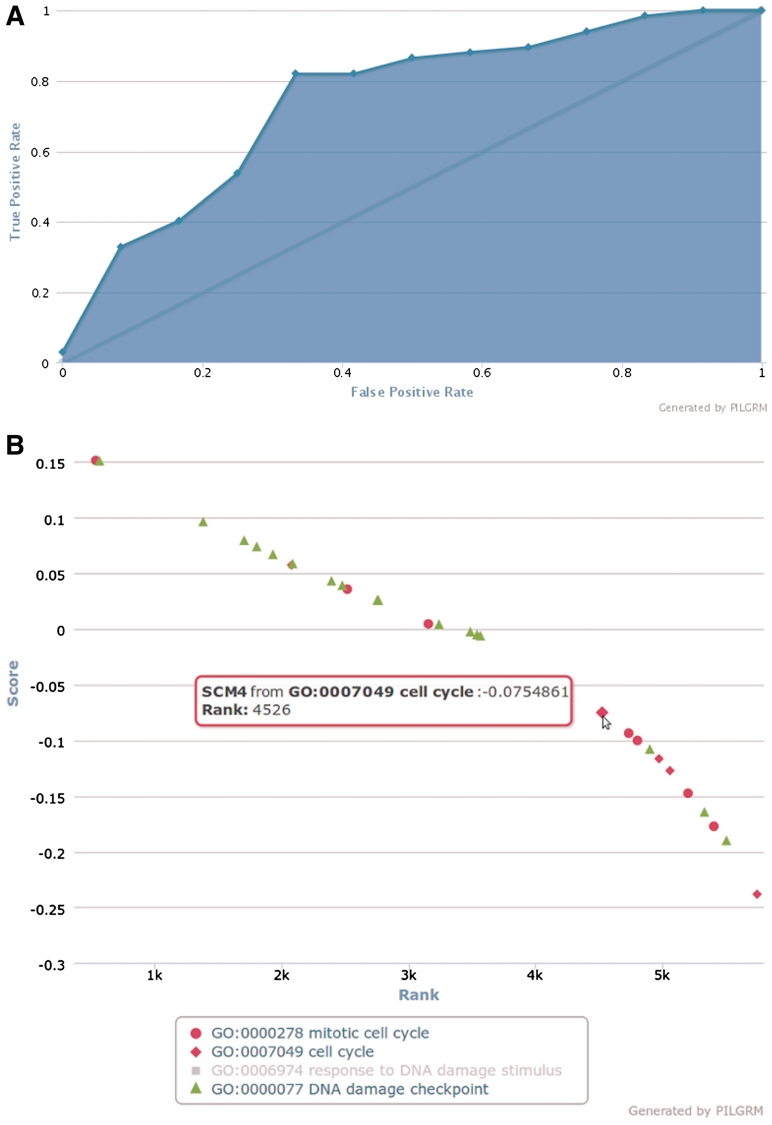
In addition to novel predictions, the user is provided with interactive visualizations of standard quantitative metrics for evaluating results of classification algorithms including the area under the curve (AUC), a figure showing the precision-recall trade-off, and a figure comparing the true positive rate and false positive rate (shown in Figure 3A). PILGRM provides this high-quality results visualization with cross-platform JavaScript that is accessible without proprietary plugins in all modern web browsers. JavaScript also allows for interactive figures that provide additional information on mouseover (as with the mouseover display of genes from each standard shown in Figure 3B). This interactivity allows researchers to more fully understand how each gene in a standard is classified. Users also have the option of including validation standards that are also shown on this figure. Validation standards are not used for classification and can be used to highlight genes of interest or to further assess prediction quality. All these results figures can be exported to JPG, PNG, SVG or PDF for easy inclusion in reports and publications. Additionally, the web server is capable of producing a document for each analysis that provides a detailed explanation of the methods, data and results specific to a user’s analysis. This document is formatted as a PDF and is intended as Supplementary Data for molecular biology manuscripts informed by a PILGRM analysis.
Figure 3.
An example of figures produced by PILGRM. (A) The true positive rate at various false positive rates for the case study of yeast DNA-damage repair. The area under the curve, shown in blue, is 0.7189 for this analysis and the performance of a random classifier is shown by the grey line. (B) Illustrations how PILGRM figures are highly interactive. In this visualization, the rank and score from PILGRM are plotted for each gene in the positive (red) and negative (green) standards. Moving the mouse over a point shows which gene it represents. Clicking on a standard toggles it between shown and hidden (here GO:0006974 has been hidden).
Our server employs SVMs for classification. Specifically we use the linear SVM implementation from SVMperf (15). We have evaluated other implementations (including polynomial and RBF kernels) and linear SVM offers classification performance that is better or comparable to more complex forms often at substantially faster speed (16). Our server handles running the analysis, parameter selection and cross validation. The analyses are run on a high-performance computing cluster in the Lewis–Sigler Institute for Bioinformatics at Princeton University.
PILGRM currently contains data and standards for six organisms (human and the model organisms yeast, worm, mouse, rat and arabidopsis as detailed in Table 1) and additional organisms are added upon request. The data are processed uniformly and in a manner robust to diverse platforms and many experimental biases. As an example, for Affymetrix data compendia all supplementary CEL files available in the Gene Expression Omnibus (17) are downloaded, and their probes mapped to Entrez GeneIDs using the Entrez BrainArray CustomCDF (18). All arrays are processed within their experiment (GEO series) using the affy (19) R package from Bioconductor (20). Expression values are summarized with the medianpolish (21) method after RMA background correction (21) and quantile normalization (22). At this point, experiment sets with five or fewer arrays are combined into a single set of arrays. Genes are then normalized within experiments and combined for learning using our open-source C++ Sleipnir library for computational functional genomics (23).
Data compendia are updated monthly through an automated but supervised pipeline. Each new analysis is assigned to the current organism-specific data compendium when it is created. When data are updated, existing analyses are not affected. Users can, at the granularity of individual analyses, elect to have PILGRM re-perform their exact analysis using the most current data compendium.
Because experimenters can include their own unpublished experimental results as part of their custom gold standards and because PILGRM predictions are used to direct follow-up bench experiments, PILGRM offers multiple levels of access control. Analyses may be completely public, which allows anyone to view the analysis. These are shown in lists of analyses on the site. Analyses may also be hidden. Hidden analyses and standards are not shown in lists on the site and are accessible only through a user-defined web address. With registration, analyses may be made private. This is the highest level of protection and prohibits access by anyone other than the analysis owner. Registration is simple, completely optional (the only PILGRM capability that needs registration is making analyses completely private) and requires only a username, working email address, and password.
PILGRM provides step-by-step tutorials for creating standards and running analyses. Optional example input, which builds a hypothetical analysis of genes relevant to breast cancer but not mammary tissue in general (one step of this analysis is shown in Figure 4), is provided for these tutorials. Standards and analyses created during the tutorials can then be used outside of the tutorial framework.
Figure 4.
PILGRM contains step-by-step tutorials that familiarize users with the system. Optional example input is provided for each tutorial. The optional example represents an analysis of breast cancer progression that avoids genes that appear relevant simply because they are expressed in mammary epithelium.
The PILGRM server is a flexible tool that biologists can use to develop data-driven predictions of gene properties directly relevant to their experimental questions in under an hour. Regularly updated data compendia and database-derived gold standards insure that PILGRM remains current. Its user-defined access control lets researchers include unpublished findings to iteratively improve prediction quality without compromising novel findings. PILGRM gives expert biologists a chance to use their expertise to mine large scale genomic compendia quickly and easily.
CASE STUDY: YEAST DNA-DAMAGE REPAIR
PILGRM’s capabilities are perhaps best illustrated in a case study. This case study represents a researcher interested in identifying novel candidate genes that are involved in DNA-damage repair while excluding genes only generally related to cell cycle control. The first step of a PILGRM analysis is to determine what the positive and negative standards should be. The positive standard should represent DNA-damage repair genes. In this case, the researcher uses a PILGRM-provided positive standard of yeast genes with direct experimental annotations to GO:0006794 (response to DNA-damage stimulus) and GO:0000077 (DNA-damage checkpoint). The negative standard should represent cell-cycle-related genes. She elects to use a negative standard containing yeast genes with direct experimental annotations to GO:0000278 (mitotic cell cycle) and GO:0007049 (cell cycle); this standard is also included in PILGRM (as are all GO-based standards). Although in this case study the analysis uses only standards from the Gene Ontology’s biological process ontology, researchers are free to customize these standards or add additional ones for their own analyses.
The researcher runs the analysis using PILGRM’s yeast gene expression compendium, which consists of all S. cerevisiae expression (GDS) datasets from GEO. The PILGRM data processing pipeline (invisible to the user), has already done all the pre-processing for this analysis: the supplied probe identifiers were mapped to Entrez identifiers; each array was normalized with a Fisher Z-transform; genes were normalized with experiments and combined for learning using our Sleipnir library for computational functional genomics (23). In total this compendium of S. cerevisiae GDS datasets from GEO contains 1801 arrays from 117 different experiments covering 6077 Entrez gene identifiers as of 31 January 2011.
She then can interactively interpret the results of her analysis. She sees an AUC visualization and is informed that the area under the curve for this analysis is 0.7189 (Figure 3A). She also can examine the list of novel predictions, with link-outs to appropriate model organism databases to provide gene-specific information for each prediction. In this case, the top novel prediction is the gene YMR090W, which SGD (24) lists as a putative protein with unknown function. This gene is not essential (25) and is up-regulated in response to the fungicide mancozeb in a proteome-wide screen (26). Mancozeb has been shown, in rats, to induce single strand breaks in a dose-dependent manner (27). Thus, in this case study PILGRM discovers a potentially relevant gene not previously associated with DNA-damage repair that has promising experimental support. Such analysis would take a researcher a total of 15 min to perform using PILGRM, including all analysis setup and definition of gold standards. This complete analysis is available at http://pilgrm.princeton.edu/analysis/view/case-study-yeast-dna-damage-response/.
DISCUSSION
PILGRM is a user-friendly exploratory tool for expert biologists who wish to use current knowledge and genome-wide experimental data to guide the design of future experiments. The extensive pre-loaded data collections and literature-based standards from common databases make it easy for researchers to start using the system. PILGRM is being actively developed, and we will continue adding capabilities based upon user requests. Currently we are working to include RNA-Seq data and developing an interface to allow users to perform an analysis on a user-defined subset of the data compendium. This web server’s flexibility allows biologists to customize analyses that address-specific questions of interest within diverse topics such as protein function, tissue-specific gene expression and cellular localization by employing computing approaches for data-driven generation of accurate hypotheses. PILGRM thus brings sophisticated machine-learning methods applied to enormous gene expression compendia into the lab of any researcher, enabling data-driven experiment direction complementary to traditional knowledge-based discovery provided by existing databases.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Science Foundation (NSF) CAREER (award DBI-0546275); National Institutes of Health (grant R01 GM071966); National Institute of General Medical Sciences (NIGMS) Center of Excellence (grant P50 GM071508); National Cancer Institute (NCI) iNRSA T32 CA005928. Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to acknowledge Barbara Mirel for usability testing, Maria Chikina for helpful discussions and programming assistance and Lars Ailo Bongo for automating the process of updating functional genomics data. Our anonymous reviewers provided many helpful subscriptions for both the article and the PILGRM interface.
REFERENCES
- 1.Hess DC, Myers CL, Huttenhower C, Hibbs MA, Hayes AP, Paw J, Clore JJ, Mendoza RM, Luis BS, Nislow C, et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009;5:e1000407. doi: 10.1371/journal.pgen.1000407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hibbs MA, Myers CL, Huttenhower C, Hess DC, Li K, Caudy AA, Troyanskaya OG. Directing experimental biology: a case study in mitochondrial biogenesis. PLoS Comput. Biol. 2009;5:e1000322. doi: 10.1371/journal.pcbi.1000322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Faith JJ, Hayete B, Thaden JT, Mogno I, Wierzbowski J, Cottarel G, Kasif S, Collins JJ, Gardner TS. Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles. PLoS Biol. 2007;5:e8. doi: 10.1371/journal.pbio.0050008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Harsha HC, Kandasamy K, Ranganathan P, Rani S, Ramabadran S, Gollapudi S, Balakrishnan L, Dwivedi SB, Telikicherla D, Selvan LD, et al. A compendium of potential biomarkers of pancreatic cancer. PLoS Med. 2009;6:e1000046. doi: 10.1371/journal.pmed.1000046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nielsen HB, Mundy J, Willenbrock H. Functional Associations by Response Overlap (FARO), a functional genomics approach matching gene expression phenotypes. PLoS ONE. 2007;2:e676. doi: 10.1371/journal.pone.0000676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chikina MD, Huttenhower C, Murphy CT, Troyanskaya OG. Global prediction of tissue-specific gene expression and context-dependent gene networks in Caenorhabditis elegans. PLoS Comput. Biol. 2009;5:e1000417. doi: 10.1371/journal.pcbi.1000417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tedder PM, Bradford JR, McConkey GA, Bulpitt AJ, Westhead DR. PlasmoPredict: a gene function prediction website for Plasmodium falciparum. Trends Parasitol. 2010;26:107–110. doi: 10.1016/j.pt.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 8.Yan H, Venkatesan K, Beaver JE, Klitgord N, Yildirim MA, Hao T, Hill DE, Cusick ME, Perrimon N, Roth FP, et al. A genome-wide gene function prediction resource for Drosophila melanogaster. PLoS One. 2010;5:e12139. doi: 10.1371/journal.pone.0012139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beaver JE, Tasan M, Gibbons FD, Tian W, Hughes TR, Roth FP. FuncBase: a resource for quantitative gene function annotation. Bioinformatics. 2010;26:1806–1807. doi: 10.1093/bioinformatics/btq265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Gene Ontology’s Reference Genome Project. A unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avraham S, Tung CW, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, et al. The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res. 2008;36:D449–D454. doi: 10.1093/nar/gkm908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human Protein Reference Database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Joachims T. Proceedings of the 12th ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, 2006. New York, NY, USA: 2006. Training linear SVMs in linear time; pp. 217–226. [Google Scholar]
- 16.Barutcuoglu Z, Schapire RE, Troyanskaya OG. Hierarchical multi-label prediction of gene function. Bioinformatics. 2006;22:830–836. doi: 10.1093/bioinformatics/btk048. [DOI] [PubMed] [Google Scholar]
- 17.Barrett T, Troup DB, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, et al. NCBI GEO: archive for functional genomics data sets–10 years on. Nucleic Acids Res. 2011;39:D1005–D1010. doi: 10.1093/nar/gkq1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dai M, Wang P, Boyd AD, Kostov G, Athey B, Jones EG, Bunney WE, Myers RM, Speed TP, Akil H, et al. Evolving gene/transcript definitions significantly alter the interpretation of GeneChip data. Nucleic Acids Res. 2005;33:e175. doi: 10.1093/nar/gni179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 20.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, Speed TP. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 22.Bolstad BM, Irizarry RA, Astrand M, Speed TP. A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics. 2003;19:185–193. doi: 10.1093/bioinformatics/19.2.185. [DOI] [PubMed] [Google Scholar]
- 23.Huttenhower C, Schroeder M, Chikina MD, Troyanskaya OG. The Sleipnir library for computational functional genomics. Bioinformatics. 2008;24:1559–1561. doi: 10.1093/bioinformatics/btn237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cherry JM, Adler C, Ball C, Chervitz SA, Dwight SS, Hester ET, Jia Y, Juvik G, Roe T, Schroeder M, et al. SGD: Saccharomyces Genome Database. Nucleic Acids Res. 1998;26:73–79. doi: 10.1093/nar/26.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Giaever G, Chu AM, Ni L, Connelly C, Riles L, Veronneau S, Dow S, Lucau-Danila A, Anderson K, Andre B, et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- 26.Santos PM, Simoes T, Sa-Correia I. Insights into yeast adaptive response to the agricultural fungicide mancozeb: a toxicoproteomics approach. Proteomics. 2009;9:657–670. doi: 10.1002/pmic.200800452. [DOI] [PubMed] [Google Scholar]
- 27.Calviello G, Piccioni E, Boninsegna A, Tedesco B, Maggiano N, Serini S, Wolf FI, Palozza P. DNA damage and apoptosis induction by the pesticide Mancozeb in rat cells: involvement of the oxidative mechanism. Toxicol. Appl. Pharmacol. 2006;211:87–96. doi: 10.1016/j.taap.2005.06.001. [DOI] [PubMed] [Google Scholar]