Abstract
The Fold and Function Assignment System (FFAS) server [Jaroszewski et al. (2005) FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Research, 33, W284–W288] implements the algorithm for protein profile–profile alignment introduced originally in [Rychlewski et al. (2000) Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Science: a Publication of the Protein Society, 9, 232–241]. Here, we present updates, changes and novel functionality added to the server since 2005 and discuss its new applications. The sequence database used to calculate sequence profiles was enriched by adding sets of publicly available metagenomic sequences. The profile of a user’s protein can now be compared with ∼20 additional profile databases, including several complete proteomes, human proteins involved in genetic diseases and a database of microbial virulence factors. A newly developed interface uses a system of tabs, allowing the user to navigate multiple results pages, and also includes novel functionality, such as a dotplot graph viewer, modeling tools, an improved 3D alignment viewer and links to the database of structural similarities. The FFAS server was also optimized for speed: running times were reduced by an order of magnitude. The FFAS server, http://ffas.godziklab.org, has no log-in requirement, albeit there is an option to register and store results in individual, password-protected directories. Source code and Linux executables for the FFAS program are available for download from the FFAS server.
OVERVIEW
The original publication about the Fold and Function Assignment System (FFAS) server (1) introduced the server and suggested optimal strategies for using it for challenging cases of remote homology and protein structure prediction. The FFAS algorithm was described in 2000 (2), and subsequent improvements were described in 2005 (1). Here we review tools and data added to the server and discuss several new applications of FFAS.
Methods for detecting remote homology are most often used to predict protein structures. Three-dimensional (3D) models of protein structures allow identification of functionally relevant residues and, thus, enable applications such as planning of mutagenesis experiments or computational docking of ligand molecules. Alignments between the protein of interest and proteins with known structures make it possible to identify structural domains in multidomain proteins (3), helping design constructs for X-ray crystallography and identify surface residues that may be modified to increase the likelihood of crystallization by the method of surface entropy reduction (SER) (4).
However, detection of remote homology may be a very valuable source of information, even if it does not link the protein of interest to any known structure (5). For instance, the homology between the protein of interest and a functionally annotated protein or protein family often provides a hypothesis about a protein’s function and helps in the planning of experiments. This application of FFAS is becoming more relevant with the rapid growth of protein sequence databases fueled by continued improvements of DNA sequencing techniques, which are increasingly used to probe novel, previously never studied regions of the protein universe (6–8). Recent analyses suggest that despite their novelty, these regions are dominated by very divergent members of known protein families rather than completely new ones (9).
Validation of the method
FFAS is regularly assessed in CASP (10) competitions and continually benchmarked in the LIVEBENCH (11) experiment. In the last available LIVEBENCH evaluation, FFAS is ranked in the top 2–4 of all sequence-based methods (see http://meta.bioinfo.pl/results.pl?comp_name=livebench-2009.2). In addition, FFAS is continuously tested on pairs of proteins of the same fold but from different superfamilies [based on the SCOP (12) database]. The current version of the FFAS algorithm was optimized in 2003 using SCOP v.1.65 and retested in 2009 on representatives of superfamilies that were added to the PDB later and, thus, not used in any training set. The results of this test confirm that FFAS detects more than twice as many cases of the extremely remote homology as PSI-Blast (13) (14 and 5% of pairs, respectively). Detailed results of this benchmark are included in the server’s documentation, available online.
Other profile–profile comparison servers
The sensitivity of profile–profile comparison is now widely recognized, and many Web servers implementing such algorithms are available, including HHPRED (14), COMPASS (15), COMA (16), PHYRE (17), GenThreader (18), FORTE (19) and webPRC (20). A comprehensive review and comparison of these servers and methods is beyond the scope of this publication. Based on our experience, the strengths of FFAS in comparison to other servers include: speed, the large number of profile databases available for searches, password-protected lists of users’ results, the option of processing multiple sequences (from registered accounts), lists of precalculated results, dotplot analysis of local similarities in two profiles, and, last but not least, the longevity and stability of the server, which has been in continuous use for over 10 years now.
NOVEL FEATURES
New searchable databases of profiles and precalculated results
The original FFAS server was designed to answer a specific question: ‘Is my protein homologous (and thus structurally similar) to any protein with an already known structure?’ We found out that many users are interested in related, but more general, questions, such as: ‘Does an organism A contain a (putative) member of a protein family B?’ or ‘What percentage of proteins in organism A have detectable homology to known structures or annotated families’. To make answering such questions possible, we added databases of profiles for complete proteomes to the FFAS server (Table 1). In addition to direct searches of profile databases with the FFAS algorithm, a user may search the precalculated FFAS results of comparisons between these proteomes to selected databases of profiles such as PDB (21), SCOP (12), Pfam (22) and COG (23).
Table 1.
Databases used by the FFAS server that were added or significantly modified since 2005 [databases of profiles such as PDB, PfamA, SCOP and COG, added before 2005, are regularly updated; for details, see (1)]
| Database | Sources and preparation of the data |
|---|---|
| Profile preparation database used to calculate sequences profiles | |
| NR85S (sequences) | The NR database from National Center for Biotechnology Information (NCBI) and the following sets of metagenomic sequences: Global Ocean Sampling (GOS) data from the JCVI and CAMERA consortia (6), microbial metagenome samples from the Joint Genome Institute (http://imgweb.jgi-psf.org/cgi-bin/m/main.cgi), human gut metagenome samples from the Hattori Lab (24), the Human Oral Microbiome Database from The Forsyth Institute (http://www.homd.org/index.php), and the human gut dataset from the Meta-HIT consortium (7). All sequences have been clustered at 85% of sequence identity with the CD-HIT program (25). The regions of low complexity have been masked with the SEG program (26). |
| New annotation databases available for profile–profile searches by FFAS | |
| VFDB (profiles) | VFDB: Virulence Factors Database (VFDB) (27) from http://www.mgc.ac.cn/VFs/ |
| HUMSAVAR (profiles) | Human polymorphisms and disease mutations (HUMSAVAR) (28) from (http://www.uniprot.org/docs/humsavar). Proteins containing >1000 residues were split into overlapping fragments of 500 residues. |
| Complete human proteome (profiles) | The set of sequences of canonical isoforms of human proteins have been downloaded from the Uniprot database page of Complete Proteomes (http://www.uniprot.org/taxonomy/complete-proteomes). Proteins containing >600 residues were split into overlapping fragments of 300 residues. Signal peptides predicted with SignalP (29) were removed from all sequences (similarities between signal peptides present in different proteins tend to increase the number of false positives in profile–profile searches). |
| Selected microbial proteomes (pathogens and members of human microbiome) and two eukaryotic proteomes (profiles) | The proteomes of Bacillus anthracis, Borrelia burgdorferi, Bacteroides thetaiotaomicron, Caulobacter crescentus, Chlamydia trachomatis, Escherichia coli, Eubacterium rectale, Helicobacter pylori, Mycoplasma genitalium, Mycoplasma pneumoniae, Mycobacterium tuberculosis, Neisseria meningitidis, Staphylococcus aureus, Saccharomyces cerevisiae, Salmonella typhi, Thermotoga maritima and Yersinia pestis have been downloaded from the NCBI database of complete microbial genomes (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). When multiple strains of the same organism were available, the strain with the most references in the literature was used. Signal peptides predicted with SignalP were removed from all sequences. Proteins containing >1000 residues were split into overlapping fragments of 500 residues. |
Dotplot graphs
The FFAS server returns a single, local–local alignment for each pair of compared sequences, represented by their profiles. Dotplot graphs allow a visual inspection of a the entire landscape of similarity between two proteins being compared, allowing a user to identify regions of similarity not included in the reported alignment, such as repeats, and domains that are present in more than one copy. It also makes it possible to assess the relative reliability (stability) of different sections of the alignment. An element (M, N) of the similarity matrix used in dynamic programming is a profile–profile similarity score of a position M in the first sequence and a position N in the second sequence. Visualization of this matrix as an M by N heat map with a color scale ranging from blue (the highest similarity between N and M) to red (the lowest similarity) is available on the ‘align 2 sequences and dot plot’ tab of the FFAS server.
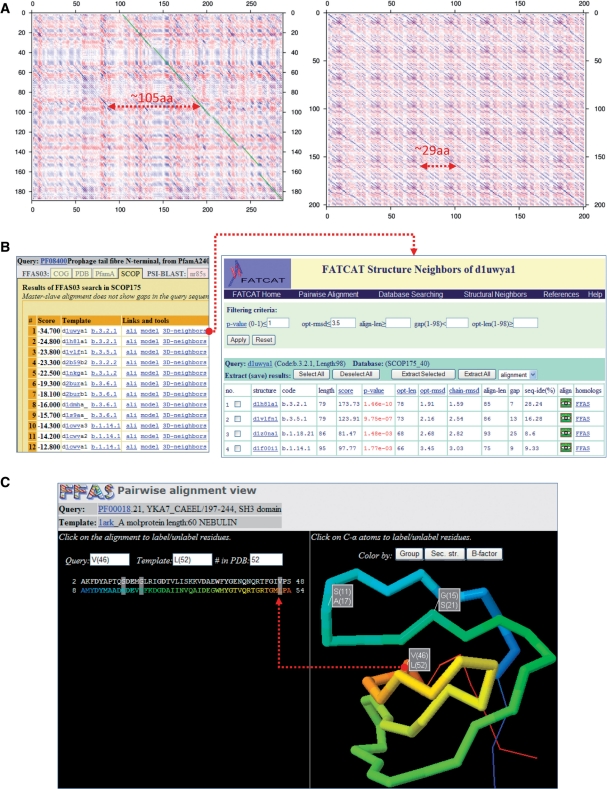
The interface allows modification of the averaging window used in preparation of dotplot graphs. The averaging radius of 0 corresponds to the visualization of the original profile–profile similarity matrix used to calculate the FFAS alignment; using non-zero values often enhances regions of local similarity. An optimal alignment returned by FFAS can also be displayed on the graph as a series of diagonal lines. This feature can be used to determine whether there are any regions of similarity between two proteins that are not included in the standard alignment [See example in Figure 1A. The presence of regions of high similarity (diagonal blue lines) not overlapping with actual alignments (series of green lines) often indicates the presence of a sequence repeat or duplicated domain].
Figure 1.
Examples of novel features of the FFAS server. (A) Dotplot graphs generated with the new FFAS tool. Left panel: the dotplot graph of a leucine-rich repeat region of the human NACHT protein compared to itself. Right panel: the dotplot graph visualizing similarity between C-terminal parts of SusE and SusF proteins from Bacteroides thethaiotaomicron. Arrows indicate the estimated lengths of repeats in NACHT LRRs and the lengths of repeated (homologous) domains in the alignment of SusE with SusF. (B) FFAS results are now linked to a database of structural similarities calculated with FATCAT. These links can be used to evaluate structural consistency of FFAS results. In this example, the fact that two different folds are aligned with the same query (Prophage tail fibre N-terminal domain) is explained by a list of structural neighbors that shows that a Prealbumin-like fold (b.3 code in SCOP) and an Immunoglobulin-like beta-sandwich (b.1 code in SCOP) are structurally similar despite being classified as separate folds. (C) 3D alignment viewer allows quick inspection of the alignment as ‘projected’ on a template structure (labeling of residues in a Jmol viewer is synchronized with alignment labeling).
ProtMod modeling tools
The FFAS server provides links to the ProtMod modeling server, which allows building 3D protein models with the SCWRL (30) algorithm. The modeling job on the ProtMod server can be launched via model links, displayed next to the alignments with templates from the PDB and the SCOP databases. Clicking on such a link sends the alignment between the query and the modeling template to the ProtMod server. On the ProtMod input page, a user can select the model type and the modeling program that will be used. Two model types are available: all-atom models, in which all sidechains of a modeling template are replaced according to the FFAS alignment, and ‘mixed models’ with truncated residue sidechains. ‘Mixed’ models are intended to be used in phasing of X-ray crystallography data by molecular replacement (MR), especially in cases in which a modeling template is only remotely homologous to the protein of interest (query) (31).
Links to the database of structural similarities
In FFAS searches against the SCOP database, a user can easily check the consistency of structural predictions by comparing SCOP classification codes of predicted homologs. Usually, all SCOP domains aligned with a specific region of a query protein belong to the same fold. If this is not the case (SCOP domains aligned with a specific query region belong to two or more different folds), it often indicates possible problems with the prediction. However, some SCOP folds share partial structural similarity and, thus, the fact that they both appear on the list of FFAS hits for the same protein does not have to indicate inconsistencies in the prediction. We addressed this issue by providing the results of the FATCAT structural alignment program (32), which are displayed next to the alignments with template SCOP databases (see example in Figure 1B).
3D alignment viewer
The alignment viewer available via ali links displayed by individual hits on the FFAS results page (Figure 1C) allows quick visualization of a query–template alignment and ‘projects’ the alignment onto the template structure if the structure is available (for comparisons to the PDB and SCOP database) using a Jmol (33) viewer plug-in. The pairwise alignment viewer was expanded to allow quick identification of pairs of aligned residues in the alignment and in the 3D structure. By clicking on any of the residues in the 3D structural view or on the alignment, a user can highlight residues in the alignment and, at the same time, label these residues in the 3D view (Figure 1C).
Technical improvements, parallelization and availability of the program
The increase in the number and size of databases of profiles used by the FFAS server made it necessary to increase the program’s execution speed. This was achieved by several technical improvements: introduction of a binary format of profile databases (speeding up loading of the databases), parallelization and optimization of the FFAS program using options provided by the Intel(R) Fortran Compiler, and installation of the FFAS server on a dedicated 12-node Linux cluster using dual quad-core CPUs per node. The combined effect of these updates (with the largest impact from parallelization enabled by a new generation of multi-core CPUs) was a reduction of execution times by an order of magnitude, despite significant increase of both the size and the number of the annotation databases. The source code of all programs included in the FFAS suite and accompanying Perl scripts and Linux executables are now available for download from the FFAS server (‘Download’ tab).
Server output
Adding more searchable databases and tools to the server required a significant reorganization of the FFAS server’s interface, which is now displayed in a ‘tab’ view. Server output shows a ‘master–slave’ alignment of sequences represented in a database of profiles with the query sequence. (In a master–slave format, gaps in the query sequence are omitted.) Individual query–template alignments can be displayed by clicking ali links on the results page. The ProtMod modeling tool is available via model links. A user can also display FFAS results for each template profile by clicking follow links. The follow feature often allows detection of very remote similarities by finding a protein or protein domain that is similar to both the query and the template. However, one has to make sure that the same region of an ‘intermediate’ protein domain is aligned to both proteins.
NOVEL APPLICATIONS
Novel modeling and alignment analysis tools are intended to help in protein structure prediction, which remains the most popular application of the FFAS server. It is noteworthy that structural predictions are increasingly used to aid experimental structure determination. At the same time, adding full proteomes of several organisms as searchable profile databases should help in another, increasingly frequent application of FFAS, i.e. using remote homology to link newly sequenced proteins to better annotated proteins or protein families.
Discovery of new domains in eukaryotic proteins
Dividing proteins into structural domains is a relatively straightforward task if it is possible to align them with homologous proteins of known structures (which are often already parsed into domains in resources such as SCOP). However, this task becomes increasingly difficult when homology is very weak. In such cases, remote homology prediction tools such as FFAS are in many cases the only source of complete alignment with known structures that allow determination of domain boundaries.
For prokaryotic proteins without detectable similarity to any known structures or annotated domains, it is oftentimes possible to propose putative domain boundaries based on conserved blocks in multiple sequence alignment of homologous sequences. For eukaryotic proteins, it is usually much more challenging because of the presence of multiple domains and long regions of structural disorder and low complexity that regularly surround structural domains. These factors frequently cause ‘profile contamination’ (34,35) that can diminish or bias a sequence conservation ‘signal’ from a structural domain. Besides remote homology detection algorithms, sequence profiles are used in local structure prediction methods such as programs for predicting secondary structure and structural disorder. As a result, ‘profile contamination’ not only interferes with remote homology detection and makes it impossible to notice conserved blocks corresponding to structural domains, but also introduces noise into secondary structure and disorder predictions. This problem can be alleviated by dividing the sequence of a protein of interest into overlapping fragments and submitting them separately to profile-based prediction servers, such as FFAS, or secondary structure services. In our experience, it is useful to try at least two different sets of such fragments of different lengths (for instance, 500 and 300 amino acid). If any such fragment corresponds to a structural domain, it should be possible to predict its secondary structure and sometimes even detect homology to known protein structures or annotated protein families, which is oftentimes impossible when a full protein sequence is used. In the current implementation, we applied this procedure to proteomes stored on the FFAS server, where all proteins longer than a specific threshold are divided into shorter overlapping fragments (Table 1).
Detection of internal repeats and alternative alignment variants
Dotplot graphs described in the previous section allow detection of internal repeats in protein sequences and alternative variants of alignments between two proteins. Profile–profile dotplot graphs are expected to be more sensitive than traditional sequence–sequence graphs. However, as is the case with all profile-based methods, they may be prone to profile contamination. Because of this, dotplot analysis of repeats should be done in parallel with a full analysis of a protein and splitting a protein sequence into (predicted) structural domains. Then, detection of internal repeats should be performed again for individual domains to see whether results remain consistent.
Aiding protein crystallography
Protein crystallization remains the main bottleneck in structure determination by X-ray crystallography, and remote homology detection by servers such as FFAS can address at least two aspects of this problem. Our participation in a structural genomics center gives us a unique opportunity to test these applications of FFAS on real-life examples, but we would like to note that other accurate alignment methods can also be used for these purposes.
Construct design
Protein crystallization often depends on the design of a proper crystallization construct (36)—a fragment of a protein sequence that corresponds to one or more structural domains. While prokaryotic proteins can routinely be crystallized in full length, eukaryotic proteins usually require nontrivial construct design. The problem of construct design is directly related to the problem of detecting structural domains described in the previous paragraph. Alignment with a known structure is a potential source of information about optimal construct boundaries, especially if a protein region is aligned with a complete protein structure or a complete domain. It is important to note that protein sequences longer than 500 amino acid should be split into putative domains before submitting them to FFAS. Thus, construct design with FFAS is often an iterative process in which approximate domain boundaries are improved in subsequent searches. FFAS predictions are extensively used to design protein constructs at the Joint Center for Structural Genomics and first structures based on these constructs have already been solved.
Prediction of exposed residues for surface engineering
It is known that sidechains involved in contacts between different protein molecules in the crystal have a significant impact on the proteins’ ability to crystallize, and by performing site-directed mutagenesis of these residues, one can significantly improve their likelihood of crystallization (37). The candidate residues for such mutations can be proposed by a method of SER (4). The application of SER is greatly facilitated if it is known which high-entropy sidechains are exposed to the solvent. Information about solvent exposure can be derived from 3D models of proteins, and by detecting remote homology to known structures, FFAS may reduce the number of mutations that need to be tested.
Modeling for MR
Solving the phase problem remains a bottleneck in X-ray crystallography of proteins. The MR method addresses this problem by calculating phase information from a predicted 3D model. The success of MR strongly depends on the accuracy of this model. By finding modeling templates for proteins without close similarity to known structures, FFAS extends the applicability of MR. For instance, over 70 protein structures have been solved at the Joint Center of Structural Genomics using models based on FFAS alignments, including 17 with <30% sequence identity to their modeling templates (31). A detailed description of strategies of MR phasing with FFAS models has been described by our group previously (31,38).
FUNDING
The maintenance and development of FFAS server is funded by National Institute of Health (grant GM087218). Funding for open access charge: National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank all members of Godzik’s Lab and the JCSG team for useful comments and extensive testing of the server.
REFERENCES
- 1.Jaroszewski L, Rychlewski L, Li Z, Li W, Godzik A. FFAS03: a server for profile–profile sequence alignments. Nucleic Acids Res. 2005;33:W284–W288. doi: 10.1093/nar/gki418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rychlewski L, Jaroszewski L, Li W, Godzik A. Comparison of sequence profiles. Strategies for structural predictions using sequence information. Protein Sci. 2000;9:232–241. doi: 10.1110/ps.9.2.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peti W, Page R. Strategies to maximize heterologous protein expression in Escherichia coli with minimal cost. Protein Expr. Purif. 2007;51:1–10. doi: 10.1016/j.pep.2006.06.024. [DOI] [PubMed] [Google Scholar]
- 4.Goldschmidt L, Cooper DR, Derewenda ZS, Eisenberg D. Toward rational protein crystallization: a Web server for the design of crystallizable protein variants. Protein Sci. 2007;16:1569–1576. doi: 10.1110/ps.072914007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.George RA, Spriggs RV, Bartlett GJ, Gutteridge A, MacArthur MW, Porter CT, Al-Lazikani B, Thornton JM, Swindells MB. Effective function annotation through catalytic residue conservation. Proc. Natl Acad. Sci. USA. 2005;102:12299–12304. doi: 10.1073/pnas.0504833102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yooseph S, Sutton G, Rusch DB, Halpern AL, Williamson SJ, Remington K, Eisen JA, Heidelberg KB, Manning G, Li W, et al. The Sorcerer II Global Ocean Sampling expedition: expanding the universe of protein families. PLoS Biol. 2007;5:e16. doi: 10.1371/journal.pbio.0050016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wu D, Hugenholtz P, Mavromatis K, Pukall R, Dalin E, Ivanova NN, Kunin V, Goodwin L, Wu M, Tindall BJ, et al. A phylogeny-driven genomic encyclopaedia of Bacteria and Archaea. Nature. 2009;462:1056–1060. doi: 10.1038/nature08656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jaroszewski L, Li Z, Krishna SS, Bakolitsa C, Wooley J, Deacon AM, Wilson IA, Godzik A. Exploration of uncharted regions of the protein universe. PLoS Biol. 2009;7:e1000205. doi: 10.1371/journal.pbio.1000205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kryshtafovych A, Venclovas C, Fidelis K, Moult J. Progress over the first decade of CASP experiments. Proteins. 2005;61(Suppl. 7):225–236. doi: 10.1002/prot.20740. [DOI] [PubMed] [Google Scholar]
- 11.Rychlewski L, Fischer D. LiveBench-8: the large-scale, continuous assessment of automated protein structure prediction. Protein Sci. 2005;14:240–245. doi: 10.1110/ps.04888805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soding J, Biegert A, Lupas AN. The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 2005;33:W244–W248. doi: 10.1093/nar/gki408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sadreyev RI, Tang M, Kim BH, Grishin NV. COMPASS server for homology detection: improved statistical accuracy, speed and functionality. Nucleic Acids Res. 2009;37:W90–W94. doi: 10.1093/nar/gkp360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Margelevicius M, Laganeckas M, Venclovas C. COMA server for protein distant homology search. Bioinformatics. 2010;26:1905–1906. doi: 10.1093/bioinformatics/btq306. [DOI] [PubMed] [Google Scholar]
- 17.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat. Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 18.Lobley A, Sadowski MI, Jones DT. pGenTHREADER and pDomTHREADER: new methods for improved protein fold recognition and superfamily discrimination. Bioinformatics. 2009;25:1761–1767. doi: 10.1093/bioinformatics/btp302. [DOI] [PubMed] [Google Scholar]
- 19.Tomii K, Akiyama Y. FORTE: a profile-profile comparison tool for protein fold recognition. Bioinformatics. 2004;20:594–595. doi: 10.1093/bioinformatics/btg474. [DOI] [PubMed] [Google Scholar]
- 20.Brandt BW, Heringa J. webPRC: the Profile Comparer for alignment-based searching of public domain databases. Nucleic Acids Res. 2009;37:W48–W52. doi: 10.1093/nar/gkp279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berman HM, Bhat TN, Bourne PE, Feng Z, Gilliland G, Weissig H, Westbrook J. The Protein Data Bank and the challenge of structural genomics. Nat. Struct. Biol. 2000;7(Suppl.):957–959. doi: 10.1038/80734. [DOI] [PubMed] [Google Scholar]
- 22.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kurokawa K, Itoh T, Kuwahara T, Oshima K, Toh H, Toyoda A, Takami H, Morita H, Sharma VK, Srivastava TP, et al. Comparative metagenomics revealed commonly enriched gene sets in human gut microbiomes. DNA Res. 2007;14:169–181. doi: 10.1093/dnares/dsm018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]
- 26.Wootton JC, Federhen S. Analysis of compositionally biased regions in sequence databases. Methods Enzymol. 1996;266:554–571. doi: 10.1016/s0076-6879(96)66035-2. [DOI] [PubMed] [Google Scholar]
- 27.Yang J, Chen L, Sun L, Yu J, Jin Q. VFDB 2008 release: an enhanced web-based resource for comparative pathogenomics. Nucleic Acids Res. 2008;36:D539–D542. doi: 10.1093/nar/gkm951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yip YL, Famiglietti M, Gos A, Duek PD, David FP, Gateau A, Bairoch A. Annotating single amino acid polymorphisms in the UniProt/Swiss-Prot knowledgebase. Hum. Mutat. 2008;29:361–366. doi: 10.1002/humu.20671. [DOI] [PubMed] [Google Scholar]
- 29.Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- 30.Krivov GG, Shapovalov MV, Dunbrack RL., Jr Improved prediction of protein side-chain conformations with SCWRL4. Proteins. 2009;77:778–795. doi: 10.1002/prot.22488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Schwarzenbacher R, Godzik A, Grzechnik SK, Jaroszewski L. The importance of alignment accuracy for molecular replacement. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1229–1236. doi: 10.1107/S0907444904010145. [DOI] [PubMed] [Google Scholar]
- 32.Ye Y, Godzik A. FATCAT: a web server for flexible structure comparison and structure similarity searching. Nucleic Acids Res. 2004;32:W582–W585. doi: 10.1093/nar/gkh430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McMahon B, Hanson RM. A toolkit for publishing enhanced figures. J. Appl. Crystallogr. 2008;41:811–814. doi: 10.1107/S0021889808015616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bork P, Koonin EV. Predicting functions from protein sequences–where are the bottlenecks? Nat. Genet. 1998;18:313–318. doi: 10.1038/ng0498-313. [DOI] [PubMed] [Google Scholar]
- 35.Gonzalez MW, Pearson WR. Homologous over-extension: a challenge for iterative similarity searches. Nucleic Acids Res. 2010;38:2177–2189. doi: 10.1093/nar/gkp1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graslund S, Nordlund P, Weigelt J, Hallberg BM, Bray J, Gileadi O, Knapp S, Oppermann U, Arrowsmith C, Hui R, et al. Protein production and purification. Nat. Methods. 2008;5:135–146. doi: 10.1038/nmeth.f.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Derewenda ZS. Rational protein crystallization by mutational surface engineering. Structure. 2004;12:529–535. doi: 10.1016/j.str.2004.03.008. [DOI] [PubMed] [Google Scholar]
- 38.Schwarzenbacher R, Godzik A, Jaroszewski L. The JCSG MR pipeline: optimized alignments, multiple models and parallel searches. Acta Crystallogr. D Biol. Crystallogr. 2008;64:133–140. doi: 10.1107/S0907444907050111. [DOI] [PMC free article] [PubMed] [Google Scholar]