Abstract
Bacterial and fungal secondary metabolism is a rich source of novel bioactive compounds with potential pharmaceutical applications as antibiotics, anti-tumor drugs or cholesterol-lowering drugs. To find new drug candidates, microbiologists are increasingly relying on sequencing genomes of a wide variety of microbes. However, rapidly and reliably pinpointing all the potential gene clusters for secondary metabolites in dozens of newly sequenced genomes has been extremely challenging, due to their biochemical heterogeneity, the presence of unknown enzymes and the dispersed nature of the necessary specialized bioinformatics tools and resources. Here, we present antiSMASH (antibiotics & Secondary Metabolite Analysis Shell), the first comprehensive pipeline capable of identifying biosynthetic loci covering the whole range of known secondary metabolite compound classes (polyketides, non-ribosomal peptides, terpenes, aminoglycosides, aminocoumarins, indolocarbazoles, lantibiotics, bacteriocins, nucleosides, beta-lactams, butyrolactones, siderophores, melanins and others). It aligns the identified regions at the gene cluster level to their nearest relatives from a database containing all other known gene clusters, and integrates or cross-links all previously available secondary-metabolite specific gene analysis methods in one interactive view. antiSMASH is available at http://antismash.secondarymetabolites.org.
INTRODUCTION
Microbial secondary metabolites offer great potential for the development of new medicines. They belong to a wide variety of chemical classes, and many of them have cholesterol-lowering, anti-tumor or antibiotic activities. The rapid decrease in the cost of genome sequencing now allows the discovery of hundreds or even thousands of gene clusters encoding the biosynthetic machinery for these compounds (1). However, laboratory research cannot keep pace with the speed of genomic discovery, as the experimental characterization of each gene cluster is still very laborious. Therefore, effective in silico identification of the most promising targets within genomes is essential for the successful mining of the genomic riches available. Manual annotation is very labor-intensive and time-consuming, leading to incomplete annotations. Automatic annotation of secondary metabolite clusters may enhance accuracy as well as completeness of the annotation. A few in silico methods have been published thus far to automate the analysis of secondary metabolism in bacterial genomes. The first of these was ClustScan (2), which allows the uploading of genomic data to a server for the semi-automatic detection and annotation of polyketide synthase (PKS) and non-ribosomal peptide synthetase (NRPS) gene clusters. Additionally, Anand et al. (3) recently published the SBSPKS toolbox for structure-based PKS analysis. Li et al. (4) constructed the NP.searcher web server, which is specialized in predicting the possible chemical structures resulting from a subset of gene cluster types. Unfortunately, all these tools are largely limited to the analysis of the core genes for type I polyketide (PK) and non-ribosomal peptide (NRP) biosynthesis. Thus far, accessory genes as well as core genes for many other secondary metabolite scaffolds have largely been neglected in computational approaches, even though some very good but also very specific tools are available for bacteriocin (5) and type III PKS (6) detection. For fungal genomes, the SMURF tool (7) has recently become available, which is capable of generating a somewhat more comprehensive list of secondary metabolite biosynthesis gene clusters, but this tool offers little further detailed analysis. CLUSEAN (8) currently offers the most comprehensive analysis by including a full genome annotation, but it is difficult to operate for the non-specialist and requires intensive manual analysis of the output.
Here, we present a software pipeline for secondary metabolite gene cluster identification, annotation and analysis which is comprehensive, rapid and user-friendly (Figure 1). It can be run either from a web server (http://antismash.secondarymetabolites.org/) or as a stand-alone version on a standard desktop computer. It can rapidly detect all known classes of secondary metabolite biosynthesis gene clusters, provide detailed NRPS/PKS functional annotation, and predict the chemical structure of NRPS/PKS products with higher accuracy than existing methods. Additionally, by constructing a database of all currently known secondary metabolite biosynthesis gene clusters throughout the tree of life, we were able to equip the tool with a comparative gene cluster analysis module. In this module, evolutionary similarities between a queried gene cluster and other gene clusters are detected and visualized in order to be able to rapidly infer functions of genes and operons based on homology. Finally, from the genes within this database of gene clusters, we constructed secondary metabolism Clusters of Orthologous Groups (smCOGs). These are used in yet another module to predict and categorize the functions of accessory genes, and to calculate phylogenetic trees for each gene with a seed alignment of its smCOG protein family. Our benchmark results show that our method reliably detects gene clusters of a wide variety of biosynthetic types, and that it is able to significantly enhance manual genome annotations of secondary metabolite biosynthesis.
Figure 1.
Outline of the pipeline for genomic analysis of secondary metabolites. Genes are extracted or predicted from the input nucleotide sequence, and gene clusters are identified with signature gene pHMMs. Subsequently, several downstream analyses can be performed: NRPS/PKS domain analysis and annotation, prediction of the core chemical structure of PKSs and NRPSs, ClusterBlast gene cluster comparative analysis, and smCOG secondary metabolism protein family analysis. The output is visualized in an interactive XHTML web page, and all details are stored in an EMBL file for additional analysis and editing in a genome browser. A Microsoft Excel file with an overview of all detected gene clusters and their details is also generated.
METHODS AND IMPLEMENTATION
File and options input
The input front end of the antiSMASH web server allows uploading of sequence files of a variety of types (FASTA, GBK, or EMBL files). Alternatively, a GenBank/RefSeq accession number can be provided, which is used by the web server to automatically obtain the associated file from GenBank. If the user chooses to use a FASTA input file, gene prediction is performed by Glimmer3 (9)—using its long-orfs tool to construct a gene model based on the input sequence itself—or by GlimmerHMM (10) when eukaryotic input data is submitted. Before starting the antiSMASH analysis run, the user can select the gene cluster types he or she wants to search for. Additionally, he can select which of the downstream analysis modules to include. For those users who, e.g. work with proprietary data, a stand-alone version with a Java graphical user interface is available with the same input options as the web version. Finally, expert users may choose to directly run the Python-based pipeline program from the command line in order to batch analyze a larger number of inputs.
Detection of secondary metabolite biosynthesis gene clusters
Using the HMMer3 tool (http://hmmer.janelia.org/), the amino acid sequence translations of all protein-encoding genes are searched with profile Hidden Markov Models (pHMMs) based on multiple sequence alignments of experimentally characterized signature proteins or protein domains (proteins, protein subtypes or protein domains which are each exclusively present in a certain type of biosynthetic gene clusters). Using both existing pHMMs (5,11–13) and new pHMMs from seed alignments, we constructed a library of models specific for type I, II and III PK, NRP, terpene, lantibiotic, bacteriocin, aminoglycoside/aminocyclitol, beta-lactam, aminocoumarin, indole, butyrolactone, ectoine, siderophore, phosphoglycolipid, melanin and aminoglycoside biosynthesis signature genes. Additionally, we constructed a number of pHMMs specific for false positives, such as the different types of fatty acid synthases which show homology to PKSs. The final detection stage operates a filtering logic of negative and positive pHMMs and their cut-offs. The logic is based on knowledge of the minimal core components of each gene cluster type taken from the scientific literature. The cut-offs were determined by manual studies of the pHMM results when run against the NCBI non-redundant (nr) protein sequence database (ftp://ftp.ncbi.nlm.nih.gov/blast/db). All technical details on the pHMM library and the detection rules are available in Supplementary Tables S1 and S2, respectively.
Gene clusters are defined by locating clusters of signature gene pHMM hits spaced within <10 kb mutual distance. To include flanking accessory genes, gene clusters are extended by 5, 10 or 20 kb on each side of the last signature gene pHMM hit, depending on the gene cluster type detected. As a consequence of this greedy methodology, gene clusters that are spaced very closely together may be merged into ‘superclusters’. These gene clusters are indicated in the output as ‘hybrid clusters’; they may either represent a single gene cluster which produces a hybrid compound that combines two or more chemical scaffold types, or they may represent two separate gene clusters which just happen to be spaced very closely together.
NRPS/PKS domain architecture analysis
NRPS/PKS domain architectures are analyzed (Figure 2) using another pHMM library comprising existing models (8,11–15) as well as newly constructed models specific for NRPS/PKS protein domains and functional/phylogenetic subgroups of these domains (Supplementary Table S3).
Figure 2.
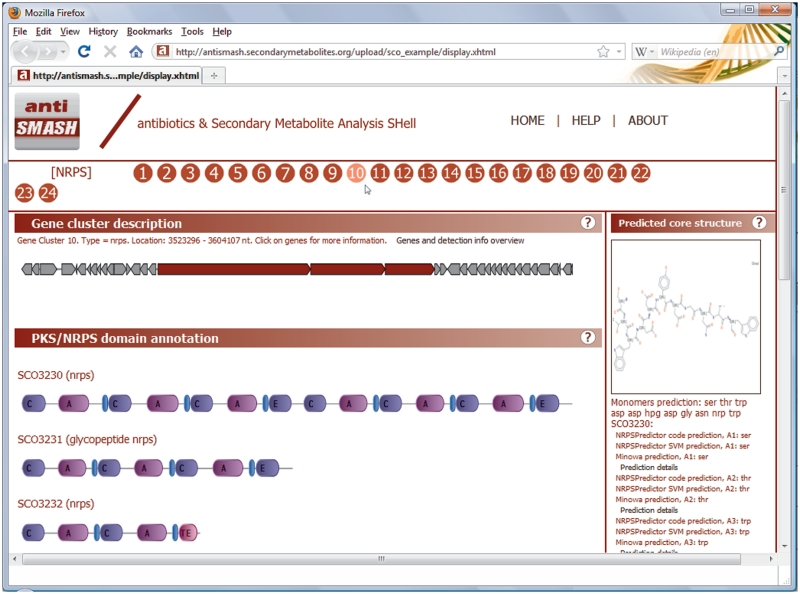
Interactive XHTML visualization of results. The numbers below the banner represent the gene clusters that were detected, the type of which is shown to the left of them at mouse-over. Once a gene cluster has been selected, the ‘Gene cluster description’ tab will display an SVG image with all genes within the approximate gene cluster, with the detected signature genes displayed in red. Locus tags appear on mouse-over, and on clicking a gene a small panel pops up with annotation information and cross-links to other web services. If PKS/NRPS proteins are encoded in the gene cluster, their domain annotations are given in the ‘PKS/NRPS domain annotation’ tab. More detailed domain annotation information and cross-links are provided on mouse-over. In the ‘Predicted core structure’ tab, a prediction of the core chemical structure is given for PKS or NRPS gene clusters based on the predictions displayed below it. All tabs contain a wide range of links to pop-ups which further detail the prediction information.
Conserved motifs within key PKS and NRPS domains are also detected using the pHMMs described earlier in the CLUSEAN package (8), and are written to the detailed downloadable EMBL output. PKS/NRPS gene names are annotated according to the domains and domain subtypes that the genes contain (e.g. ‘hybrid NRPS-PKS’, ‘enediyne PKS’, ‘glycopeptide NRPS’, ‘trans-AT PKS’, etc.).
Substrate specificity, stereochemistry and final structure predictions
Substrate specificity prediction of PKS and NRPS modules, based on the active sites of their respective acyltransferase (AT) and adenylation (A) domains, is performed by various available methods. PKS AT domain specificities are predicted using a 24 amino acid signature sequence of the active site (16), as well as with pHMMs based on the method of Minowa et al. (17), which is also used to predict co-enzyme A ligase domain specificities. NRPS A domain specificities are predicted using both the signature sequence method and the support-vector machines-based method of NRPSPredictor2 (18,19), and using the method of Minowa et al. (17). Finally, all predictions are integrated into a consensus prediction by a majority vote. Ketoreductase domain-based stereochemistry predictions for PKSs (2) are performed as well. An estimate of the biosynthetic order of PKS/NRPS modules is predicted based on PKS docking domain sequence residue matching [for type I modular PKSs, (3)] or assumed colinearity, and a final predicted core chemical structure is generated as a SMILES string (20), i.e. a unique text description of the chemical structure, and visualized in a picture file (Figure 2). To increase the reliability of the core structure prediction, monomers for which there was no consensus in the predictions are represented as generic amino acids or ketides with unspecified R-groups.
Secondary metabolite clusters of orthologous groups
In order to rapidly annotate the accessory genes surrounding the detected core signature genes in the various types of secondary metabolite biosynthesis gene clusters, we constructed a database of all gene clusters contained in the latest NCBI nt database (15 February 2011). To do so, pHMMs described above were used to detect all secondary metabolite biosynthesis gene cluster signature genes in the nr database. The accession numbers of all hits meeting the described cut-offs were extracted and used to download the corresponding GenPept files. If the taxonomy identifier included ‘bacteria’ or ‘fungi’, the nucleotide source accession number was extracted. The corresponding nucleotide GenBank files were then downloaded as well, and cross-checked for presence of the queried protein accession number. For each nucleotide GenBank file, gene clusters were detected as described above. Amino acid sequences of all genes contained within the gene clusters were written to a FASTA file with headers containing key information, and a summary of all detected gene clusters (nucleotide accession, nucleotide description, cluster number, cluster type, protein accession numbers) was written to a text file. To construct the smCOGs, clustering of all gene cluster proteins was performed using OrthoMCL (21), and consensus annotations were manually assigned based on the frequencies of the five most prevalent annotations of each smCOG in GenBank. For each smCOG, a seed alignment was created from 100 randomly picked sequences using MUSCLE 3.5 (22), and a pHMM of each smCOG was generated based on the conserved core of each alignment (Supplementary Figure S1). Within the antiSMASH software pipeline, the smCOG pHMMs are used for functional annotation of all accessory genes within the gene clusters. After assignment of an smCOG to a gene—based on the highest-scoring pHMM on its sequence above a certain e-value threshold—the predicted protein sequence is aligned to the smCOG seed alignment, and a rough neighbor-joining phylogenetic tree is calculated using FastTree 2 (23) and visualized with TreeGraph 2 (24) (Supplementary Figure S1).
ClusterBlast comparative gene cluster analysis
Secondary metabolite biosynthesis gene clusters are highly modular, and their genes are transferred frequently from one gene cluster to another during evolution (25,26). Therefore, when trying to obtain a functional understanding of a gene cluster, it is highly beneficial to be able to compare it with (parts of) other gene clusters which show similarity to it and which may have been characterized experimentally. In order to facilitate this, we applied our annotated database of gene clusters to link up protein sequences with their parent gene clusters and create a comparison tool—based on the most recent BLAST+ implementation (27)—which ranks gene clusters by similarity to a queried gene cluster. Clusters are sorted first based on an empirical similarity score S = h + H + s + S + B, in which h is the number of query genes with a significant hit, H is the number of core query genes with a significant hit, s is the number of gene pairs with conserved synteny, S is the number of gene pairs with conserved synteny involving a core gene, and B is a core gene bonus (three points given when at least one core gene has a hit in the subject cluster). If the similarity scores are equal, the hits are subsequently ranked based on the cumulative BlastP bit scores between the gene clusters. This feature enables a rapid assessment of the comparative genomics for each annotated cluster (Figure 3).
Figure 3.
Example of ClusterBlast alignment of gene clusters homologous to the query gene cluster. In this case, the ten best hits to the calcium-dependent antibiotic NRPS gene cluster from Streptomyces coelicolor A3(2) are displayed. Homologous genes (BLAST e-value < 1E-05; 30% minimal sequence identity; shortest BLAST alignment covers over >25% of the sequence) are given the same colors. The ‘select gene cluster alignment’ drop-down menu provides links to one-by-one gene cluster alignments to each gene cluster hit. In the one-by-one gene cluster alignments, PubMed and/or PubChem links are provided for gene clusters associated with a known compound.
Genome-wide BLAST and Pfam analysis and prediction of potential unknown secondary metabolite biosynthesis gene cluster types
To facilitate further thorough manual genome analysis, antiSMASH has also been linked up to the whole-genome BLAST and Pfam analysis modules from the previously published CLUSEAN framework (8). The CLUSEAN results are integrated into an EMBL output file. Furthermore, as unknown biosynthetic gene cluster types are likely to exist which may be missed by the antiSMASH gene cluster detection module, the Pfam results are also used to predict genomic regions with a high probability of constituting secondary metabolite biosynthesis gene clusters in a more generalized fashion than the signature genes pHMMs method. For this, the genome sequence is converted to a string of predicted Pfam domains which is fed to a hidden Markov model (P. Cimermancic et al., manuscript in preparation) with transitions between a gene cluster state and a rest-of-the-genome state. This model was trained on Pfam domain frequencies from a set of 473 cloned gene clusters (gene cluster state) and from the set of ∼1100 genomes currently in the JGI IMG database (rest-of-the-genome state). The result of this analysis is visualized in a PNG graph.
Output and visualization
All pipeline analysis results are visualized in a user-friendly interactive XHTML page (Figure 2), which can be used to browse through the different gene clusters. For PKS and NRPS gene clusters, the predicted core chemical structures are shown as images. Gene cluster maps are drawn with scalable vector graphics (SVGs), to which interactive on-click and mouse-over functions are added through JavaScript to provide annotation information, pipeline result scores, and BLAST hyperlinks. Detected signature genes on which the gene cluster identification is based are shown in a distinct color. ClusterBlast results are displayed in a similar way, as aligned gene cluster maps in which genes with mutual BLAST hits are given identical colors. Additionally, available at the bottom right of the page, fully annotated EMBL output files provide the user with the additional possibility to browse their genome in a genome browser such as Artemis (28).
RESULTS
Compared to previous software, the pipeline described here is uniquely comprehensive: it integrates all previously published analysis types into one tool and adds valuable novel functionalities (Table 1).
Table 1.
Comparison of different software tools for secondary metabolite biosynthesis analysis
| Software | Open-source & stand-alone available | Covers full tree of life | NRPS/PKS detection | NRPS/PKS detailed functional domain annotation | NRP/PK core structure prediction | Detection of other biosynthetic classes | Gene cluster border prediction | Comparative gene cluster analysis | Prediction of all secondary metabolite-like genomic regions |
|---|---|---|---|---|---|---|---|---|---|
| ClustScan | + | + | + | + | ± | ||||
| CLUSEAN | + | + | + | ||||||
| NP.searcher | + | + | + | + | |||||
| SBSPKS | + | + | + | ||||||
| SMURF | + | ± | + | ||||||
| antiSMASH | + | + | + | + | + | + | + | + | + |
Comparison of functionalities of currently existing programs or software packages for secondary metabolite biosynthesis analysis.
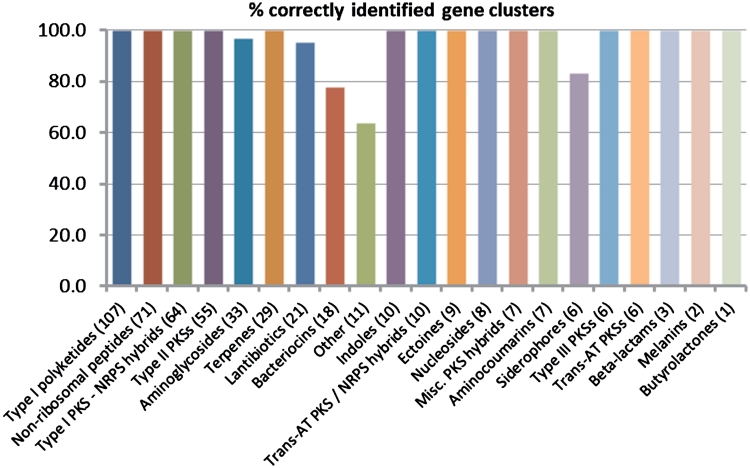
In order to measure the accuracy of the gene cluster predictions, we performed two independent benchmark evaluations of the method. First, we collected the sequences of cloned gene clusters of known compounds of biosynthetic types by searching both the GenBank/RefSeq databases and the scientific literature with a range of different keywords. From the resulting set of 484 cloned gene cluster GenBank files, 473 (97.7%) were correctly identified by antiSMASH, and 468 (96.7%) were given exactly the same annotation by antiSMASH as by the articles describing their experimental characterization (Figure 4 and Supplementary Table S4). In order to test for false positives as well, we also benchmarked the method on five well-annotated genomes from different taxonomic groups. Besides genomes of three different actinomycetes (the organisms on which the tool is likely to be used most often) these included a Proteobacterium (Pseudomonas fluorescens Pf-5) and a fungus (Aspergillus fumigatus Af293). In the five genomes, 97.3% of all 111 annotated gene clusters were detected by antiSMASH (Figure 5 and Supplementary Table S5). Under closer scrutiny, two of the three gene clusters that were missed by antiSMASH appeared to lack a complete set of genes associated with biosynthesis of a known chemical scaffold. More interestingly, 35 additional gene clusters were detected (31.5%) which had been missed during initial genome annotation and which after close inspection all appeared to have a high probability of being actual biosynthetic gene clusters. The cluster types that appeared to be frequently missed during the annotation of these genomes appeared to be butyrolactones (eight gene clusters missed), terpenes (seven gene clusters missed), NRPSs/PKSs (six gene clusters missed) and lantibiotics (five gene clusters missed), which suggests that the computational approach used can yield improvements even in finding gene clusters of common biosynthetic types.
Figure 4.
Benchmark results on a set of 473 cloned secondary metabolite biosynthesis gene clusters found in the GenBank nucleotide database. The numbers behind the names of the biosynthetic types indicate how many gene clusters of that type were in the benchmark set.
Figure 5.
Benchmark results on five genome sequences. All except three annotated gene clusters from the five genome publications were detected; two of these annotated gene clusters (SGR5285-SGR5295 in Streptomyces griseus and Strop_3244-Strop_3253 in Salinispora tropica) appeared to lack core genes for biosynthesis of a known secondary metabolite chemical scaffold. The one certain gene cluster which was not detected was a small gene cluster for the biosynthesis of hydrogen cyanide from Pseudomonas fluorescens Pf-5.
We also compared the performance of antiSMASH with other existing tools. No similarly comprehensive tools are available, but NP.searcher and SMURF each offer automated gene cluster detection for a small subset of the cluster types detected by antiSMASH (NP.searcher detects bacterial NRPS/PKS gene clusters, and SMURF detects fungal NRPS, PKS, and dimethylallyl tryptophan synthase gene clusters). Our analysis of the results of these tools on four bacterial and two fungal genomes (Supplementary Table S6), respectively, showed that antiSMASH and SMURF performed equally well (both detect 74 gene clusters, with 93.4% overlap). Compared to NP.searcher, antiSMASH detected significantly more (47 versus 31, i.e. 51.6% more) NRPS/PKS gene clusters, while all NP.searcher-detected gene clusters were also picked up by antiSMASH. The gene clusters that were detected by antiSMASH but not by NP.searcher were all small NRPS-like or PKS-like gene clusters. None of the three tools gave predictions that were clear false positives, except one SMURF detection of a probable fatty acid synthase (GenBank ID CAP98191.1) labeled as PKS.
DISCUSSION AND CONCLUSIONS
antiSMASH not only provides a unique integration of previously widely dispersed tools, but it also achieves very high accuracy in its individual cluster annotations, which are enhanced by unique novel analyses such as BLAST-based gene cluster alignments and secondary metabolite COG phylogenetic trees for accessory genes. As the field of synthetic biology is opening up new ways to study these gene clusters in a high-throughput fashion (29), antiSMASH will enable experimental researchers to quickly pinpoint those gene clusters most interesting for further study, and swiftly collect secondary metabolite BioBricks for the (re-)design of gene clusters. Moreover, the new comparative analyses that antiSMASH offers provide unprecedented possibilities to interpret the functions of both complete gene clusters and their particular genes in their evolutionary context. The approaches developed are likely to soon allow global analysis of all small molecule biosynthesis gene clusters throughout the tree of life, so that we can acquire a more and more comprehensive understanding of how nature itself designs novel bioactive compounds.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Dutch Technology Foundation STW, which is the applied science division of NWO and the Technology Programme of the Ministry of Economic Affairs (STW 10463); GenBioCom program of the German Ministry of Education and Research (BMBF) (grant 0315585A); Rosalind Franklin Fellowship, University of Groningen (to E.T.); NWO-Vidi Fellowship (to R.B.); NIH DP2 Award (OD007290) (to M.A.F.); Travel grant from the Boehringer Ingelheim Fonds (to M.H.M.). Funding for open access charge: STW (STW 10463).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Mike Li for kindly providing a script for the conversion of strings of amino acid and polyketide residues into SMILES strings. The authors thank Marc Röttig and Oliver Kohlbacher for providing NRPSpredictor2.
REFERENCES
- 1.Walsh CT, Fischbach MA. Natural products version 2.0: connecting genes to molecules. J. Am. Chem. Soc. 2010;132:2469–2493. doi: 10.1021/ja909118a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Starcevic A, Zucko J, Simunkovic J, Long PF, Cullum J, Hranueli D. ClustScan: an integrated program package for the semi-automatic annotation of modular biosynthetic gene clusters and in silico prediction of novel chemical structures. Nucleic Acids Res. 2008;36:6882–6892. doi: 10.1093/nar/gkn685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Anand S, Prasad MV, Yadav G, Kumar N, Shehara J, Ansari MZ, Mohanty D. SBSPKS: Structure based sequence analysis of polyketide synthases. Nucleic Acids Res. 2010;38:W487–W496. doi: 10.1093/nar/gkq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li MH, Ung PM, Zajkowski J, Garneau-Tsodikova S, Sherman DH. Automated genome mining for natural products. BMC Bioinformatics. 2009;10:185. doi: 10.1186/1471-2105-10-185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.de Jong A, van Heel AJ, Kok J, Kuipers OP. BAGEL2: Mining for bacteriocins in genomic data. Nucleic Acids Res. 38:W647–W651. doi: 10.1093/nar/gkq365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mallika V, Sivakumar KC, Jaichand S, Soniya EV. Kernel based machine learning algorithm for the efficient prediction of type III polyketide synthase family of proteins. J. Integr. Bioinform. 2010;7:143. doi: 10.2390/biecoll-jib-2010-143. [DOI] [PubMed] [Google Scholar]
- 7.Khaldi N, Seifuddin FT, Turner G, Haft D, Nierman WC, Wolfe KH, Fedorova ND. SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 2010;47:736–741. doi: 10.1016/j.fgb.2010.06.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Weber T, Rausch C, Lopez P, Hoof I, Gaykova V, Huson DH, Wohlleben W. CLUSEAN: a computer-based framework for the automated analysis of bacterial secondary metabolite biosynthetic gene clusters. J. Biotechnol. 2009;140:13–17. doi: 10.1016/j.jbiotec.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 9.Delcher AL, Bratke KA, Powers EC, Salzberg SL. Identifying bacterial genes and endosymbiont DNA with glimmer. Bioinformatics. 2007;23:673–679. doi: 10.1093/bioinformatics/btm009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20:2878–2879. doi: 10.1093/bioinformatics/bth315. [DOI] [PubMed] [Google Scholar]
- 11.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Letunic I, Doerks T, Bork P. SMART 6: Recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yadav G, Gokhale RS, Mohanty D. Towards prediction of metabolic products of polyketide synthases: An in silico analysis. PLoS Comput. Biol. 2009;5:e1000351. doi: 10.1371/journal.pcbi.1000351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ansari MZ, Sharma J, Gokhale RS, Mohanty D. In silico analysis of methyltransferase domains involved in biosynthesis of secondary metabolites. BMC Bioinformatics. 2008;9:454. doi: 10.1186/1471-2105-9-454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rausch C, Hoof I, Weber T, Wohlleben W, Huson DH. Phylogenetic analysis of condensation domains in NRPS sheds light on their functional evolution. BMC Evol. Biol. 2007;7:78. doi: 10.1186/1471-2148-7-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yadav G, Gokhale RS, Mohanty D. Computational approach for prediction of domain organization and substrate specificity of modular polyketide synthases. J. Mol. Biol. 2003;328:335–363. doi: 10.1016/s0022-2836(03)00232-8. [DOI] [PubMed] [Google Scholar]
- 17.Minowa Y, Araki M, Kanehisa M. Comprehensive analysis of distinctive polyketide and nonribosomal peptide structural motifs encoded in microbial genomes. J. Mol. Biol. 2007;368:1500–1517. doi: 10.1016/j.jmb.2007.02.099. [DOI] [PubMed] [Google Scholar]
- 18.Röttig M, Medema MH, Blin K, Weber T, Rausch C, Kohlbacher O. NRPSpredictor2: A web server for predicting NRPS adenylation domain specificity. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr323. doi: 10.1093/nar/gkr323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rausch C, Weber T, Kohlbacher O, Wohlleben W, Huson DH. Specificity prediction of adenylation domains in nonribosomal peptide synthetases (NRPS) using transductive support vector machines (TSVMs) Nucleic Acids Res. 2005;33:5799–5808. doi: 10.1093/nar/gki885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Weininger D. SMILES, a chemical language and information system. 1. introduction to methodology and encoding rules. J. Chem. Inf. Comput. Sci. 1988;28:31–36. [Google Scholar]
- 21.Li L, Stoeckert CJ, Jr, Roos DS. OrthoMCL: Identification of ortholog groups for eukaryotic genomes. Genome Res. 2003;13:2178–2189. doi: 10.1101/gr.1224503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Price MN, Dehal PS, Arkin AP. FastTree 2–approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5:e9490. doi: 10.1371/journal.pone.0009490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stover BC, Muller KF. TreeGraph 2: combining and visualizing evidence from different phylogenetic analyses. BMC Bioinformatics. 2010;11:7. doi: 10.1186/1471-2105-11-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fischbach MA, Walsh CT, Clardy J. The evolution of gene collectives: how natural selection drives chemical innovation. Proc. Natl Acad. Sci. USA. 2008;105:4601–4608. doi: 10.1073/pnas.0709132105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Donadio S, Sosio M, Stegmann E, Weber T, Wohlleben W. Comparative analysis and insights into the evolution of gene clusters for glycopeptide antibiotic biosynthesis. Mol. Genet. Genomics. 2005;274:40–50. doi: 10.1007/s00438-005-1156-3. [DOI] [PubMed] [Google Scholar]
- 27.Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rutherford K, Parkhill J, Crook J, Horsnell T, Rice P, Rajandream MA, Barrell B. Artemis: Sequence visualization and annotation. Bioinformatics. 2000;16:944–945. doi: 10.1093/bioinformatics/16.10.944. [DOI] [PubMed] [Google Scholar]
- 29.Medema MH, Breitling R, Bovenberg R, Takano E. Exploiting plug-and-play synthetic biology for drug discovery and production in microorganisms. Nat. Rev. Microbiol. 2011;9:131–137. doi: 10.1038/nrmicro2478. [DOI] [PubMed] [Google Scholar]