Abstract
Bacterial genomes encode a plethora of small RNAs (sRNAs), which are heterogeneous in size, structure and function. Most sRNAs act as post-transcriptional regulators by means of specific base pairing interactions with the 5′-untranslated region of mRNA transcripts, thereby modifying the stability of the target transcript and/or its ability to be translated. Here, we present RNApredator, a web server for the prediction of sRNA targets. The user can choose from a set of over 2155 genomes and plasmids from 1183 bacterial species. RNApredator then uses a dynamic programming approach, RNAplex, to compute putative targets. Compared to web servers with a similar task, RNApredator takes the accessibility of the target during the target search into account, improving the specificity of the predictions. Furthermore, enrichment in Gene Ontology terms, cellular pathways as well as changes in accessibilities along the target sequence can be done in fully automated post-processing steps. The predictive performance of the underlying dynamic programming approach RNAplex is similar to that of more complex methods, but needs at least three orders of magnitude less time to complete. RNApredator is available at http://rna.tbi.univie.ac.at/RNApredator.
INTRODUCTION
Bacterial small RNAs (sRNAs) are very heterogeneous in size, structure and function (1). Despite notable exceptions, most sRNAs act as post-transcriptional regulators by interacting with the 5′-untranslated region of mRNA transcripts (2). Similar to miRNAs in eukaryotes, sRNAs may target more than one mRNA and, conversely, a mRNA may be targeted by more than one sRNA. In contrast to miRNAs, however, sRNAs may cause both down- and upregulation of its target (3–5). This effect depends on the exact location of the interaction region and its effect on the structure of the target mRNA.
Many approaches have been developed to find sRNA targets. BLAST was successfully used to identify targets for micC (6) and istR-1 (7). TargetRNA (8,9) implements a Smith–Waterman (10) recursion scoring the base pairing potential of two RNAs. A slightly more complex model is used by Mandin et al. (11), where base pair stacks are scored according to the standard RNA folding energy model (12,13) and bulge penalties are optimized so that known interactions rank high.
More general approaches to describe RNA–RNA interactions based on the RNA folding energy model and consider the target site accessibility, like IntaRNA (14), RNAup (15,16) or biRNA (17) greatly improved sRNA–target predictions at the cost of an increased computation time.
In this contribution, we present RNApredator, a web server dedicated to the genome-wide prediction of sRNA targets in bacterial genomes. The main machinery used by RNApredator is RNAplex (18,28), a new approach for RNA–RNA interaction search, which has a prediction accuracy similar to that of algorithms that explicitly consider intramolecular structures, but running at least three orders of magnitude faster than RNAup or IntaRNA. In addition to the improved run time, RNApredator offers the user a graphical overview of the accessibility around the target ribosomal binding sites upon sRNA binding, as well as a Gene Ontology enrichment analysis for a set of user selected gene of interest.
DESCRIPTION OF THE WEBSERVER
Functionality of RNApredator
For all annotated mRNAs in the selected target sequences, RNApredator computes several relevant interaction characteristics by launching RNAplex. Thanks to its ability of considering target accessibility, RNAplex reaches
prediction accuracies similar to more complex and computationally much more demanding methods, while being at least three orders of magnitude faster than alternative methods considering target site accessibility (see Supplementary Data for more information). RNApredator is thus applicable to genome-wide sRNA target prediction.
After completing the computation of all candidate sRNA–target interactions, RNApredator returns a list of target sites sorted by the energy of interaction. In addititon, an enrichmenent analysis of GO terms is performed for all or a user-defined subset of the predicted interactions.
Furthermore, the influence of sRNA binding to its target on the accessibility of the ribosomal entry site can be studied with RNAup, predicting whether the sRNA will act as a positive or negative regulator at a particular target site (16).
Other tools
While a large number of tools are available for the prediction of miRNA targets in eukaryotes [for a review see (19)], comparably little effort has been invested to characterize targets of sRNA regulators. At present, the only web server specifically advertized for target prediction in prokaryotes is TargetRNA (8,9), which implements a modification of the Smith–Waterman (10) dynamic programming algorithm that assesses base pairing potential instead of base homology. This is achieved with the help of a custom-tailored scoring system. Alternatively, TargetRNA can also be run with thermodynamic parameters for RNA folding (12,13), at the expense of a run time increased by at least a order of magnitude (8).
IntaRNA (14,20) also allows to search sRNA-mRNAs duplexes with a more realistic energy model (12,13) and an increased specificity owing to the inclusion of target and query secondary structures information. It can be used to be employed for target search in bacterial genomes. There is also a web server based on RNAup (21) available. Thanks to its unapproximated energy model, RNAup allows to more precisely describe the thermodynamics of mRNA–sRNA interactions than with RNAplex. Still the high run time of RNAup as well as the inability of the RNAup webserver to handle more than a pair of sequences at a time, makes it unpractical for genome-wide target search.
Input
RNApredator takes as input a single sRNA sequence consisting of lower or uppercase [A,T,C,G,U] letters, where T is automatically converted into U. The targets of this sequence can be searched against the ensemble of plasmids/chromosomes referred by a NCBI taxonomy ID or a specific plasmid/chromosome referred by a NCBI accession number. Currently, 1183 bacterial species are available, encompassing a total of 2155 chromosomes and plasmids. Alternatively, the species of interest can be chosen from a taxonomic tree.
Once the desired genome has been selected, a sRNA sequence should be entered in the sRNA sequence field. The target search is launched after the predict button has been pressed. Targets are searched for each annotated gene, including 5′- and 3′-UTR. The 5′-UTR and 3′-UTR regions are defined as the 200 nt regions directly up and downstream of the coding sequence. subsectionOutput after submission of the sRNA RNApredator returns the target predictions. The results should be similar to the accessibility-based RNAplex and better than RNAplex without accessibility information. Still different parameters used to compute accessibility profiles from RNAplfold leads to different accessibilities and consequently to different RNAplex results. In case of the sRNA micA in Escherichia coli (NC_000913), RNApredator needs ∼5 min to finish the computation, scanning the full coding sequence and 200 nt upstream of the start codon. TargetRNA needed 40 s for the whole genome, processing each coding sequence from 20 nt upstream of the translation start and 30 nt downstream with a seed length set to 1 and G:U pairs allowed.
The IntaRNA web server is much slower, as it takes 3 h to finish the computation, under the supplementary constraint that for each gene only subsequences of up to 500 nt can be searched.
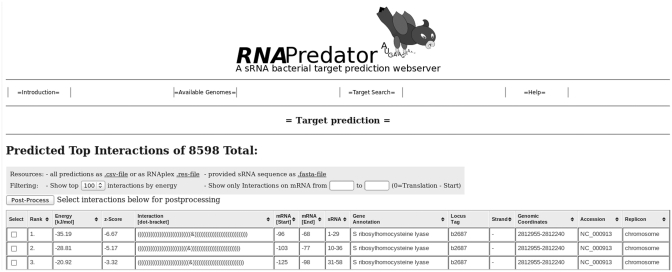
The web server outputs a table of the 100 most stable duplexes found by RNAplex (see Figure 1). Each line of the table contains the energy of interaction, i.e. the raw hybridization energy corrected for the opening energies on both the target and the sRNA sequences, the corresponding Z-score, which is useful for comparing interactions involving different sRNAs, the duplex structure in dot-bracket format, the start and end of the duplex on the target and query sequences, gene annotation, the NCBI accession number, genomic coordinates, as well as the type of replicon where the gene is found (chromosome/plasmid). Results can be sorted by all duplex characteristics, on the exception of the hybrid structure.
Figure 1.
Results page. After the target search is completed, RNApredator presents the list of the 100 best interactions. Each line contains the rank, total energy of interaction, corresponding Z-score, duplex structure in dot-bracket format, interaction coordinates on the sRNA and mRNA, gene annotation, locus tag, strand, genomic coordinates of the target, NCBI accession number as well as the type of replicon the target is located on. Results can be filtered based on the coordinates of the target locations (for up to 500 interactions). Moreover, it is possible to limit the displayed interactions to the 25, 50, 75, 100, 500 best interactions. Finally, the complete results table can be downloaded in .csv or raw RNAplex format.
Even though most of the sRNAs act in vicinity of the 5′-end of the target RNA, there are growing evidences that sRNA may exert their effects by binding also in the coding sequence region (22,23). In order to concentrate on the region of interest, the user can filter the duplexes by setting a position filtering (for up to 500 interactions) on the target sites coordinates. Further filtering is achieved by limiting the number of returned duplex to 25, 50 and 75. If desired, the user can increase the number of displayed interactions to 500 or to the complete results returned by RNAplex.
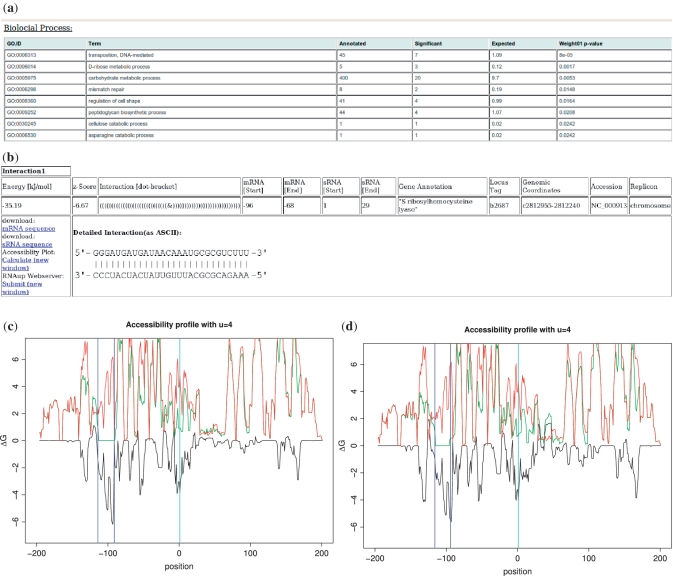
The left-most column allows the user to select genes of interest for further post-processing (Figure 2b), in particular the analysis of the accessibility around the target site for the bound (green line in Figure 2c and d) and unbound target (red line). These accessibility profiles are computed with RNAup. This adds important information since many sRNAs regulate their targets by changing the accessibility of the ribosomal binding site (5,24). Therefore, the difference in the accessibility before and after binding (black line), the position of the start codon (cyan vertical line) as well as the boundaries of the target site (blue vertical line) are displayed, see Figure 2c and d. In the case of the RprA-rpoS and DsrA-rpoS duplexes (bottom left and right of Figure 2), for instance, the interactions take place 100 nt upstream of the start codon, but increase the accessibility of the region around the start codon (Figure 2c and d). Both interactions lead to a reduction of up to 4 kcal/mol of the opening energy around the start codon, leading to a strong upregulation of rpoS (5,24).
Figure 2.
Post-processing page: detailed information about interactions selected on the results page. (a) The upper part of the three GO term class tables, i.e. biological process, molecular function or cellular component. The 20 most significant GO terms are shown in a separate table. Each line of the table contains the GO term ID, human readable term, total number of annotated genes linked to the GO term, number of targets selected linked to the GO term, expected number of targets linked to the GO term and P-value of the GO term enrichment. (b)The selected interactions are shown in detail; Relevant duplex characteristics are recapitulated and a graphical representation of the duplex structure is shown. mRNA and sRNA sequences can be downloaded in .fasta format. The Calculate-link enables the user to get a plot of the opening energy for all stretches of 4 nt for the region around the start codon before (red line) and after (green line) the sRNA binding. The accessibility difference is shown in (black line). [(c) rpoS-RprA, (d) rpoS-DsrA]. The 5′-end of the start codon is represented with a cyan line and the interaction site with two blue lines. Recalculation of the duplex is possible by using the Calculate-link to RNAup web server.
To better apprehend the function of the sRNA of interest, RNApredator provides an enrichment analysis of GO terms in the set of selected targets. For each GO categories (Biological Process, Molecular Function, Cellular Component), the 20 highest enriched terms are returned in tabular format. Besides the GO-ID, annotated term, total number of genes linked to this GO-ID, total number of predicted targets linked to this GO-ID, number of expected linked targets as well as the P-value are returned. The results can be classified by any of the above characteristics.
Finally, the post-processing page shows in greater details the relevant characteristics of the duplex (ascii string) and allows to download the sequences of the target and sRNA by following the mRNA and sRNA sequence link, respectively.
Implementation details
RNApredator was implemented in Perl 5. It uses the javascript library jQuery jquery.com to allow sorting of the results table. Computation of the accessibility profiles in the post-processing steps is performed with the help of the RNAup program. RNApredator relies on different databases. The bacterial genomes were downloaded from NCBI ftp://ftp.ncbi.nlm.nih.gov/genomes/Bacteria, while taxonomy data were retrieved from NCBI ftp://ftp.ncbi.nih.gov/pub/taxonomy. All available bacterial GO term flatfiles, which are necessary for the GO term enrichment analysis were downloaded from ftp://ftp.ebi.ac.uk/pub/databases/GO/goa/proteomes. The computation of the GO term enrichment is based upon these files and an R-script based on the TopGO (25) library.
The most time consuming step in the interaction prediction is the computation of accessibilities along the bacterial genome. In order to speed up the calculation, we have precomputed the accessibility profiles for all genomes using RNAplfold (26,27).
BENCHMARK
RNApredator was benchmarked against TargetRNA for a set of 30 interactions retrieved from the literature. For each experimentally confirmed interaction, the number of better scoring interactions was computed for both prediction tools. The ranking procedure only considered interactions predicted to be located between position −150 and 100 and −30 and 20 relative to the start codon, respectively [see Table 1. 73% of the interactions (22) ranked higher in RNApredator than in TargetRNA]. TargetRNA was used with an hybridization length of 1, with allowed G:U pairs and with a P-value threshold set to 100. The sRNA was always characterized as a new sequence. It should be noted that TargetRNA thermodynamic energy scoring was not able to return any result. For this reason, the benchmark/hlreports only the results for the sequence-based energy scoring.
Table 1.
Summary of TargetRNA and RNApredator ranking of 30 experimentally confirmed interactions
| Genome | Species | sRNA | mRNA | Gene | TargetRNA | RNApredator |
|---|---|---|---|---|---|---|
| NC_000964 | B.s. | FsrA | sdhC | BSU28450 | NF(NF) | 153(83) |
| NC_011601 | E.c.O | OmrA | ompR | b3405 | NF(NF) | 436(49) |
| NC_011601 | E.c.O | OmrA | ompT | b0565 | NF(NF) | 712(93) |
| NC_011601 | E.c.O | OmrB | ompR | b3405 | NF(31) | 312(39) |
| NC_011601 | E.c.O | OmrB | ompT | b0565 | NF(NF) | 210(13) |
| NC_000913 | E.c.K. | CyaR | ompX | b0814 | NF(NF) | 495(86) |
| NC_000913 | E.c.K. | CyaR | yqaE | b2666 | NF(NF) | 541(97) |
| NC_000913 | E.c.K. | DsrA | hns | b1237 | 52(6) | 8(4) |
| NC_000913 | E.c.K. | FnrS | metE | b3829 | 5(8) | 120(37) |
| NC_000913 | E.c.K. | FnrS | sodB | b1656 | 24(21) | 615(192) |
| NC_000913 | E.c.K. | GcvB | cycA | b4208 | 37(5) | 41(10) |
| NC_000913 | E.c.K. | IstR | tisB | b4405 | 2(NF) | NF(NF) |
| NC_000913 | E.c.K. | MicA | phoP | b1130 | 80(23) | 57(10) |
| NC_000913 | E.c.K. | MicC | ompC | b2215 | 2(5) | 2(2) |
| NC_000913 | E.c.K. | MicF | ompF | b0929 | 43(5) | 2(2) |
| NC_000913 | E.c.K. | OmrA | gntP | b4321 | NF(NF) | 79(17) |
| NC_000913 | E.c.K. | OmrB | csgD | b1040 | 50(NF) | 2(NF) |
| NC_000913 | E.c.K. | RseX | ompC | b2215 | 98(NF) | 504(238) |
| NC_000913 | E.c.K. | RyhB | iscS | b2530 | NF(NF) | 123(30) |
| NC_000913 | E.c.K. | RyhB | sodB | b1656 | 24(21) | 184(52) |
| NC_000913 | E.c.K. | SgrS | ptsG | b1101 | NF(NF) | 5(1) |
| NC_003210 | L.m. | LhrA | lmo085 | lmo0850 | NF(NF) | 31(NF) |
| NC_002505 | V.c. | MicX | vca0620 | vca0620 | NF(34) | 48(7) |
| NC_002505 | V.c. | Qrr1 | luxO | vca1021 | NF(NF) | 196(44) |
| NC_002505 | V.c. | Qrr1 | vca0939 | vca0939 | NF(NF) | 5(NF) |
| NC_002505 | V.c. | Qrr2 | luxO | vca0620 | NF(NF) | 12(NF) |
| NC_002505 | V.c. | Qrr2 | vca0939 | vca0939 | NF(NF) | 3(NF) |
| NC_002505 | V.c. | Qrr3 | vca0939 | vca0939 | NF(NF) | 4(NF) |
| NC_002505 | V.c. | Qrr4 | vca0939 | vca0939 | NF(NF) | 4(NF) |
| NC_002505 | V.c. | VrrA | tcpA | vca0838 | 35(NF) | 246(71) |
The first column contains the NCBI accession ID of the species, the species name is indicated in the second column. The third and fourth columns contain the sRNA and mRNA gene tag, the fifth column shows the locus tag and the sixth and seventh columns contain the rank of the interaction for TargetRNA and RNApredator. In the last two columns, the number in parenthesis corresponds to the rank when the target search is constrained to a region located 30 nt upstream and 20 nt downstream of the start codon, while the other numbers correspond to the rank for the region spanning 150 nt upstream and 100 nt downstream of the start codon. NF stands for not found (TargetRNA does not return targets with a rank >100, and RNApredator hits also contain suboptimal interactions) B.s. is Bacillus subtilis subsp. subtilis str. 168, E.c.O is Escherichia coli O127:H6 str. E2348/69, E.c.K. is Escherichia coli str. K-12 substr. MG1655, L.m. is Listeria monocytogenes EGD-e and V.c. is Vibrio cholerae O1 biovar El Tor str. N16961.
The RNAup web server was not used in the benchmark as it is designed to give an in-depth understanding of the thermodynamics of a sRNA–mRNA interaction, rather than searching genome wide for putative targets. Furthermore, the important time complexity of RNAup algorithm impede it to return putative targets in a reasonable amount of time (see Supplementary Data). Still the users of RNApredator can use RNAup to study interactions of interest during the post-processing step of RNApredator.
DISCUSSION
RNApredator is a freely available web server that facilitates the search for putative sRNA targets in bacterial genomes. Predictions from RNApredator reach the accuracy of more complex methods like RNAup, IntaRNA or biRNA, while saving at least three orders of magnitude of CPU time. This allows to search for sRNA targets in bacterial species in a few minutes, compared to hours or days for IntaRNA or RNAup, respectively.
Unique features of the RNApredator web server are the post-processing steps. The computation of accessibility changes of the target upon sRNA binding may help in deciding whether the target will be up- or downregulated. The GO term enrichment allows to further filter the targets in order to select genes that belong to the group of highly enriched terms.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Austrian GEN-AU projects ‘bioinformatics integration network III ’ and ‘ regulatory ncRNAs ’ (in part).
Conflict of interest statement. None declared.
REFERENCES
- 1.Vogel J, Sharma CM. How to find small non-coding RNAs in bacteria. Biol. Chem. 2005;386:1219–1238. doi: 10.1515/BC.2005.140. [DOI] [PubMed] [Google Scholar]
- 2.Livny J, Brencic A, Lory S, Waldor MK. Identification of 17 pseudomonas aeruginosa sRNAs and prediction of sRNA-encoding genes in 10 diverse pathogens using the bioinformatic tool sRNApredict2. Nucleic Acids Res. 2006;34:3484–3493. doi: 10.1093/nar/gkl453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lease RA, Belfort M. A trans-acting RNA as a control switch in escherichia coli: Dsra modulates function by forming alternative structures. Proc. Natl Acad. Sci. USA. 2000;97:9919–9924. doi: 10.1073/pnas.170281497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lease RA, Cusick ME, Belfort M. Riboregulation in escherichia coli: Dsra RNA acts by RNA:RNA interactions at multiple loci. Proc. Natl Acad. Sci. USA. 1998;95:12456–12461. doi: 10.1073/pnas.95.21.12456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Majdalani N, Chen S, Murrow J, St. John K, Gottesman S. Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol. Microbiol. 2001;39:1382–1394. doi: 10.1111/j.1365-2958.2001.02329.x. [DOI] [PubMed] [Google Scholar]
- 6.Chen S, Zhang A, Blyn LB, Storz G. MicC, a second small RNA regulator of Omp protein expression in escherichia coli. J. Bacteriol. 2004;186:6689–6697. doi: 10.1128/JB.186.20.6689-6697.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vogel J, Argaman L, Wagner EG, Altuvia S. The small RNA istr inhibits synthesis of an sos-induced toxic peptide. Curr. Biol. 2004;14:2271–2276. doi: 10.1016/j.cub.2004.12.003. [DOI] [PubMed] [Google Scholar]
- 8.Tjaden B, Goodwin SS, Opdyke JA, Guillier M, Fu DX, Gottesman S, Storz G. Target prediction for small, noncoding RNAs in bacteria. Nucleic Acids Res. 2006;34:2791–2802. doi: 10.1093/nar/gkl356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tjaden B. TargetRNA: a tool for predicting targets of small RNA action in bacteria. Nucleic Acids Res. 2008;36:109–113. doi: 10.1093/nar/gkn264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 11.Mandin P, Repoila F, Vergassola M, Geissmann T, Cossart P. Identification of new noncoding RNAs in listeria monocytogenes and prediction of mRNA targets. Nucleic Acids Res. 2007;35:962–974. doi: 10.1093/nar/gkl1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuker M, Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981;9:133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathews DH, Burkard ME, Freier SM, Wyatt JR, Turner DH. Predicting oligonucleotide affinity to nucleic acid targets. RNA. 1999;5:1458–1469. doi: 10.1017/s1355838299991148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Busch A, Richter AS, Backofen R. IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics. 2008;24:2849–2856. doi: 10.1093/bioinformatics/btn544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mückstein U, Tafer H, Hackermüller J, Bernhart SH, Stadler PF, Hofacker IL. Thermodynamics of RNA-RNA binding. Bioinformatics. 2006;22:1177–1182. doi: 10.1093/bioinformatics/btl024. [DOI] [PubMed] [Google Scholar]
- 16.Mückstein U, Tafer H, Bernhart SH, Hernandez-Rosales M, Vogel J, Stadler PF, Hofacker IL. Translational control by RNA-RNA interaction: Improved computation of RNA-RNA binding thermodynamics. In: Elloumi M, Köng J, Linial M, Murphy RF, Schneider K, Toma C, editors. Bioinformatics Research and Development. Vol. 13 of Communications in Computer and Information Science, pp. 114–127. Springer, Berlin Heidelberg, 2008. [Google Scholar]
- 17.Chitsaz H, Salari R, Sahinalp SC, Backofen R. A partition function algorithm for interacting nucleic acid strands. Bioinformatics. 2009;25:365–373. doi: 10.1093/bioinformatics/btp212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tafer H, Hofacker IL. RNAplex: a fast tool for RNA-RNA interaction search. Bioinformatics. 2008;24:2657–2663. doi: 10.1093/bioinformatics/btn193. [DOI] [PubMed] [Google Scholar]
- 19.Thomas M, Lieberman J, Lal A. Desperately seeking microRNA targets. Nat. Struct. Mol. Biol. 2010;17:1169–1174. doi: 10.1038/nsmb.1921. [DOI] [PubMed] [Google Scholar]
- 20.Smith C, Heyne S, Richter AS, Will S, Backofen R. Freiburg RNA tools: a web server integrating intaRNA, expaRNA and locaRNA. Nucleic Acids Res. 2010;38:373–377. doi: 10.1093/nar/gkq316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gruber AR, Lorenz R, Bernhart SH, Neuböck R, Hofacker IL. The vienna RNA websuite. Nucleic Acids Res. 2008;36:70–74. doi: 10.1093/nar/gkn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC, Vogel J. Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol. Microbiol. 2009;74:139–158. doi: 10.1111/j.1365-2958.2009.06857.x. [DOI] [PubMed] [Google Scholar]
- 23.Pfeiffer V, Papenfort K, Lucchini S, Hinton JC, Vogel J. Coding sequence targeting by MicC RNA reveals bacterial mRNA silencing downstream of translational initiation. Nat. Struct. Mol. Biol. 2009;16:840–846. doi: 10.1038/nsmb.1631. [DOI] [PubMed] [Google Scholar]
- 24.Sledjeski DD, Gottesman S. Osmotic shock induction of capsule synthesis in escherichia coli K-12. J. Bacteriol. 1996;178:1204–1206. doi: 10.1128/jb.178.4.1204-1206.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 26.Bompfünwerer AF, Backofen R, Berhart SH, Hertel J, Hofacker IL, Stadler PF, Will S. Variations on RNA folding and alignment: Lessons from Benasque. J. Math. Biol. 2008;56:129–144. doi: 10.1007/s00285-007-0107-5. [DOI] [PubMed] [Google Scholar]
- 27.Berhart SH, Mückstein U, Hofacker IL. RNA Accessibility in cubic time. Algorithms. Mol Biol. 2011;6:3. doi: 10.1186/1748-7188-6-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Tafer H, Amman F, Eggenhofer F, Stadler PF, Hofacker IL. Fast Accessiblity-Based Prediction of RNA-RNA Interactions. Bioinformatics. 2011 doi: 10.1093/bioinformatics/btr281. (18 May, date last accessed) [DOI] [PubMed] [Google Scholar]