Abstract
We describe MarkUs, a web server for analysis and comparison of the structural and functional properties of proteins. In contrast to a ‘structure in/function out’ approach to protein function annotation, the server is designed to be highly interactive and to allow flexibility in the examination of possible functions, suggested either automatically by various similarity measures or specified by a user directly. This is combined with tools that allow a user to assess independently whether or not a suggested function is consistent with the bioinformatic and biophysical properties of a given query structure, further allowing the user to generate testable hypotheses. The server is available at http://wiki.c2b2.columbia.edu/honiglab_public/index.php/Software:Mark-Us.
INTRODUCTION
Comparison to sequence and structural neighbors is one of the most powerful and widely-used methods to suggest a protein’s possible function(s). For a given query protein, a typical incarnation of this approach is to identify a single similar protein, where similarity can depend on a wide variety of properties: from sequence similarity as determined by a particular substitution matrix, to structural similarity as determined by root-mean-square deviation or other measures, to similarity in local features such as a particular configuration of active site residues. It is well known, however, that identification of a single neighbor that is optimal in terms of some measure of similarity will not necessarily provide a complete (or even accurate) description of a protein’s function. That is, a close sequence neighbor of an enzyme may have a slightly different substrate; or, conversely, a more remote homolog, even if less likely to accurately indicate a specific function, may provide more accurate information about general functional features, such as the overall location of a ligand binding site. In general, a given query protein may share the properties of a number of its neighbors with varying degrees of similarity.
Here we describe MarkUs, a server for the analysis and comparison of the structural and functional properties of proteins. Starting with a query protein structure in PDB format as input (either experimentally determined or a computational model), MarkUs calculates a number of bioinformatic and biophysical features of that protein itself. Figure 1 shows the overall flow of the annotation process and lists the specific features calculated by MarkUs which are stored in a relational database model implemented under MySQL. In addition, MarkUs identifies structural (and implicitly, sequence) neighbors of the query protein, using a measure of similarity [Protein Structural Distance (1)] that allows for the identification of both close and remote structural homologs. An underlying assumption in the design of the server is that useful information about a protein’s function may come from any of its structural neighbors (2) and that the annotation process will be facilitated by the ability to simultaneously browse and interrogate different sources of information about those neighbors, and to compare bioinformatic and biophysical features of the neighbors to those calculated for the query to confirm or refute a suggested function.
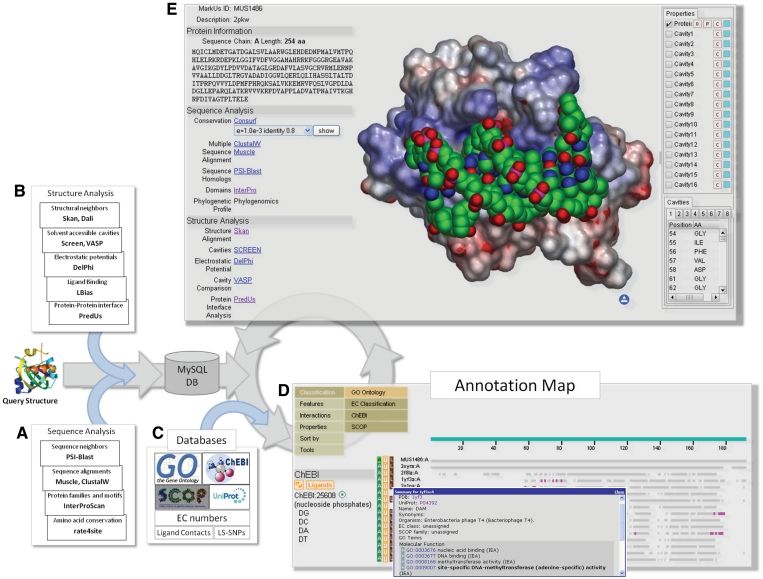
Figure 1.
The MarkUs function annotation server. Starting with a query protein structure in PDB format a number of sequence and structure analysis methods are carried out. (A) Sequence based searches include a PSI-Blast (4) search against UniRef100 (3) and a sequence motif scan using InterProScan (6); a multiple sequence alignment of a subset of sequence neighbors clustered at 80% sequence identity or pre-calculated PFam (16) alignments are generated using Muscle (17). Muscle alignments are used for amino acid conservation analysis using rate4site (9). (B) Structural descriptors are generated using the programs DelPhi (10) to calculate electrostatic potentials; SCREEN to identify cavities (8); PredUs (11) to identify protein–protein interaction sites; Skan (18) and optionally DALI (19) to identify structural neighbors from a set of representative PDB (20) structures and SCOP (21) domains; LBias to identify potential ligands; and VASP (14) to identify proteins with similar cavity shapes. Results are stored in a MySQL database. (C) Annotation resources which can be visualized and queried in (D) the ‘Annotation Map’. This tool shows structural neighbors schematically aligned structurally to the query sequence. Currently the server integrates Gene Ontology (7), Enyzme Commission numbers (22), ChEBI (15), SCOP classification (21), UniProt features (13), LS-SNPs (23) and ligand contacts. (E) Properties of a particular structural neighbor (e.g. associated ligands, residue specific annotations) can be visualized in the context of query structure using AstexViewer (24) as molecular viewer. In this example, we show a DNA molecule (green, red and blue spheres) from a structural neighbor of a query protein, superimposed on the surface of the query structure which is colored by electrostatic potential (see main text).
This is enabled by a unique, interactive feature of MarkUs called the ‘annotation map’ (Figure 1D). The annotation map is a visual portal to a diverse set of biological databases and provides a set of tools that allow browsing, querying and filtering of the set of structural neighbors of the query based on different criteria. Via these tools, a user can ask both general and context-specific questions related to functional hypotheses that are generated either automatically in MarkUs or based on a user’s own assumptions (e.g. ‘What structural neighbors bind sugars?’, or ‘What are the active site residues in a functional subfamily of structural neighbors that bind specific sugars?’). Also via the annotation map, properties of a specific structural neighbor, with which a functional similarity is suspected, can be projected onto the query to allow a user to directly determine whether that functional hypothesis is consistent with the biochemical and biophysical features of the query itself.
A CASE STUDY
We describe the use of MarkUs by analyzing the NESG target StR221 (PDB code 2pkw) representing the protein yhiQ from Salmonella typhimurium. This example illustrates a subset of the functionalities of the server as well as the overall strategy of using MarkUs to explore structure-function space and hypothesize on the function of a protein structure. A help section describing all the tools is available on the web site. The data for the example described below can be found at http://luna.bioc.columbia.edu/honiglab/mark-us/cgi-bin/browse.pl?pdb_id=MUS1471. A tutorial for this example is also provided.
MarkUs identifies sequence neighbors by searching the UniRef100 database (3) using PSI-Blast (4) and scanning the InterPro database (5) using InterProScan (6). Sequences identified by these methods can be examined via an annotation map similar to that used for structural neighbors (Figure 1D, described in more detail below). For StR221, browsing the sequence neighbors does not provide an unambiguous functional hypothesis. InterProScan identifies StR221 as a member ‘DUF’ (domain of unknown function) and PSI-Blast results places it in a UPF (uncharacterized protein family). Gene Ontology terms (7) for sequence neighbors can also be browsed in the annotation map. In this case, some neighbors are annotated as having ‘methyltransferase activity’ inferred from electronic annotation (IEA) providing a first working hypothesis on the function of StR221.
A second important starting point for the annotation process is the identification of potential functional sites on the query structure itself. MarkUs uses SCREEN (8) to identify a set of cavities on the surface of the query protein. In the case of StR221, 16 cavities are identified and ranked by accessible surface area. The largest cavity on the surface of StR221 forms a tunnel extending deep into the interior of the globular protein structure. All cavities can be displayed individually in the molecular viewer and colored according to various pre-calculated properties.
For example, MarkUs uses the program rate4site (9) to calculate residue conservation and the individual cavities can be colored according to this property. For StR221, it can be seen that the top-ranked cavity is also well-conserved, with 38 residues lining the tunnel having a rate4site score below −1. Specific conserved residues can be identified by clicking in the molecular viewer, and the entire set can be listed by following the ‘ConSurf’ link to the raw rate4site results. Among the most strongly conserved residues are a set of eight basic amino acids (R241, R219, K218, K184, K185, K179, K238) that are near the opening of the top-ranked cavity. MarkUs also uses the program DelPhi (10) to calculate the electrostatic potential of the query structure. Coloring the molecular surface based on the DelPhi potential shows that the conserved basic residues are part of a large positive surface patch that surrounds the opening to the putative functional tunnel. Potential sites for protein-protein interactions are calculated by the program PredUs (11), which, when mapped to the molecular surface also form a distinct patch in close proximity to the opening of the top-ranked SCREEN cavity. Taken together, these attributes calculated by MarkUs are a strong indicator of a functional nature of this cavity and suggest properties of potential ligands/substrates.
An important feature of MarkUs is the ability to explore and, to a certain extent, manually validate a functional hypothesis based on an examination of structural neighbors via the annotation map (Figure 1D). In particular, as discussed above, sequence information suggests ‘methyltransferase’ as a possible function for StR221. The annotation map allows a user to explore structure space to identify proteins with properties consistent with a proposed function and to ascertain whether they are conserved in the query protein. For example, various ranking operators can be applied to the set of structural neighbors. Based on a score called the SAS, which reflects a combination of RMSD and alignment length (12), the two closest neighbors share unique structural features with StR221 (two large insertions, one of which is close the suggested functional tunnel). Unfortunately, these proteins (structural genomics targets SfR275 and NgR48 from NESG, PDB codes 2oyr and 2r6z) are currently unannotated. However, the server allows a user to examine ligands in the structural neighbors via the annotation map, and in this case it can be seen that the neighbor SfrR275 was co-crystallized with S-adenosyl-homocysteine (SAH), a known cofactor of methyltransferases. The annotation map allows a user to project ligands from a structural neighbor into the structure of the query itself and in this case, the SAH fits into the query structure with very minor clashes. Moreover, as can be seen in the molecular viewer, the sugar moiety in the S-adenosyl homocysteine overlaps with a solvent molecule (TRIS, a sugar/carbohydrate analog) co-crystalized with the query structure.
Another important feature of the annotation map is the ability to extend this type of analysis further. The annotation map allows a user to filter the set of structural neighbors based on their functional properties. Given the evidence for the methyltransferase activity, we would like to examine all possible neighbors, regardless of their degree of sequence or structural similarity, that have functions consistent with this hypothesis and compare them to the target protein in order to attempt to identify a more specific suggested function. Searching the annotation map for the GO term ‘methyltransferase activity’ reveals protein RsmC from Thermus thermophilus (PDB code 3dmg) to be the closest neighbor annotated with a more specific activity [‘rRNA (adenine-N6,N6-)-dimethyltransferase activity’]. RsmC is also in complex with the coenzyme S-adenosyl-l-homocysteine but no substrate analog is bound that could be used to infer functional consistency with the target structure.
To go further, we attempt to confirm this more specific hypothesis by further restricting the set of structural neighbors to RNA methyltransferases, resulting in a set of 39 proteins (22 ‘rRNA methyltransferase activity’, 10 ‘tRNA methyltransferase activity’, one ‘rRNA methyltransferase activity’, one ‘RNA trimethylguanosine synthase activity’). For reliable transfer of annotation, it is of course important that the specific residues mediating the function be conserved. Conservation of important residues can also be evaluated within the annotation map, which highlights individual residues associated with a function, based on UniProt sequence ‘features’ (13). For the sub-group of tRNA methyltransferases two different active site residues can be identified: an active site cysteine (for PDB code 3bt7) and an active site aspartate (for PDB codes 3ckk and 2vdv). These positions correspond to very well conserved residues in the target (M173 and P220, respectively), but are not identical, arguing against a tRNA methyltransferase hypothesis.
The closest ‘rRNA methyltransferase’ neighbor with a bound substrate is TGS1 (trimethylguanosine synthase 1, PDB code 3gdh), co-crystallized with the minimal substrate m7GTP and the reaction product S-adenosyl-l-homocysteine (AdoHcy). Individual structural neighbors can also be superimposed on the query structure and viewed in the molecular viewer. In this case, comparing TGS1 in the context of the target reveals that TGS1 has a significantly smaller positive surface patch surrounding the putative active site. The active site cavity in TGS1 is also smaller, as indicated by a volumetric comparison of the putative functional cavity in the target using the program VASP (14) (volumetric comparison can be accessed for each structural neighbor by following the ‘VASP’ link on the main results page).
The above analysis was carried out by using specific known annotations to filter structural neighbors. The annotation map also provides the functionality to use purely geometric properties to filter the list of structural neighbors and by this to identify proteins sharing similar ligand binding sites, as determined by the program LBias (manuscript in preparation). Sorting the set of structural neighbors based on their LBias score in the annotation map identifies a protein from Listeria monocytogenes (PDB code 2f8l) as having the best similarity in binding interactions between the protein and the co-factor molecule (excluding unannotated close sequence neighbors). This protein is annotated with the GO term ‘DNA binding’. As described above, we can again filter the list of neighbors based on the general term ‘DNA binding’ to attempt to identify more specific functions.
Among the filtered set of proteins are a set of five neighbors which are annotated as ‘site-specific DNA–methyltransferase (adenine-specific)’, which are of particular interest because of the larger positive surface patches in our target, compared to other structural neighbors. That is, the large electropositive patch is consistent with the idea that our target binds nucleic acid oligomers as opposed to isolated nucleic acids. MarkUs provides the tools to explore this further by allowing a user to filter the set of structural neighbors based on the types of ligand they bind, using the ChEBI ontology (15). Filtering based on the ChEBI class ‘nucleoside phosphates’ identifies four proteins that are in complex with DNA oligomers, two of which are also annotated as ‘site-specific DNA-methyltransferase (adenine-specific)’. Again, placing the DNA oligomer from the structural neighbor in the context of the query structure using the annotation map shows that they interact with the query at the large electropositive patch (Figure 1E). Furthermore, the electropositive patches are of similar size in both the target and this subset of structural neighbors. It is also noteworthy that one of these structures, the DNA adenine methylase DAM from Enterobacteria phage T4, partly possesses residues corresponding to the unique insertions of the target, described above.
DISCUSSION
Ideally, function annotation would be an entirely automated process. While MarkUs provides predictive tools that attempt to recognize specific sequence and geometric similarities between a query structure and its structural neighbors to transfer an annotation, the server is more specifically designed to provide a set of interactive tools to carry out the additional steps of examining a particular hypothesis further, to determine whether a function is consistent with the properties of the query structure itself and to identify function-determining properties that will guide experimental validation. In the above analysis, close sequence neighbors suggested only a general membership in a highly functionally diverse set of proteins. Structural neighbors yielded the more specific methyltransferase hypothesis, but specific substrates such as isolated nucleic acids were rejected based on an analysis of the structural determinants of those functions. Finally, a purely geometric analysis of ligand binding site similarity suggested a function that is consistent with biophysical and geometric features of the target. Of course, this requires experimental validation, but it is evident that the integration of these tools combined with the interactive nature of the server allows a user to go further in generating and exploring hypotheses than any single tool on its own.
FUNDING
National Institutes of Health (grant numbers GM030518, CA121852, GM094597). Funding of open access charge: Howard Hughes Medical Institute.
Conflict of interest statement. None declared.
REFERENCES
- 1.Yang AS, Honig B. An integrated approach to the analysis and modeling of protein sequences and structures. I. Protein structural alignment and a quantitative measure for protein structural distance. J. Mol. Biol. 2000;301:665–678. doi: 10.1006/jmbi.2000.3973. [DOI] [PubMed] [Google Scholar]
- 2.Petrey D, Fischer M, Honig B. Structural relationships among proteins with different global topologies and their implications for function annotation strategies. Proc. Natl Acad. Sci. USA. 2009;106:17377–17382. doi: 10.1073/pnas.0907971106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suzek BE, Huang H, McGarvey P, Mazumder R, Wu CH. UniRef: comprehensive and non-redundant UniProt reference clusters. Bioinformatics. 2007;23:1282–1288. doi: 10.1093/bioinformatics/btm098. [DOI] [PubMed] [Google Scholar]
- 4.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 7.The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nayal M, Honig B. On the nature of cavities on protein surfaces: application to the identification of drug-binding sites. Proteins. 2006;63:892–906. doi: 10.1002/prot.20897. [DOI] [PubMed] [Google Scholar]
- 9.Pupko T, Bell RE, Mayrose I, Glaser F, Ben-Tal N. Rate4Site: an algorithmic tool for the identification of functional regions in proteins by surface mapping of evolutionary determinants within their homologues. Bioinformatics. 2002;18(Suppl. 1):S71–S77. doi: 10.1093/bioinformatics/18.suppl_1.s71. [DOI] [PubMed] [Google Scholar]
- 10.Rocchia W, Sridharan S, Nicholls A, Alexov E, Chiabrera A, Honig B. Rapid grid-based construction of the molecular surface and the use of induced surface charge to calculate reaction field energies: applications to the molecular systems and geometric objects. J. Comput. Chem. 2002;23:128–137. doi: 10.1002/jcc.1161. [DOI] [PubMed] [Google Scholar]
- 11.Zhang QC, Petrey D, Norel R, Honig BH. Protein interface conservation across structure space. Proc. Natl Acad. Sci. USA. 107:10896–10901. doi: 10.1073/pnas.1005894107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolodny R, Koehl P, Levitt M. Comprehensive evaluation of protein structure alignment methods: scoring by geometric measures. J. Mol. Biol. 2005;346:1173–1188. doi: 10.1016/j.jmb.2004.12.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen BY, Honig B. VASP: a volumetric analysis of surface properties yields insights into protein-ligand binding specificity. PLoS Comput. Biol. 2010;6:e1000881. doi: 10.1371/journal.pcbi.1000881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Degtyarenko K, de Matos P, Ennis M, Hastings J, Zbinden M, McNaught A, Alcantara R, Darsow M, Guedj M, Ashburner M. ChEBI: a database and ontology for chemical entities of biological interest. Nucleic Acids Res. 2008;36:D344–D350. doi: 10.1093/nar/gkm791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Finn RD, Mistry J, Schuster-Bockler B, Griffiths-Jones S, Hollich V, Lassmann T, Moxon S, Marshall M, Khanna A, Durbin R, et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–D251. doi: 10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Petrey D, Honig B. GRASP2: visualization, surface properties, and electrostatics of macromolecular structures and sequences. Methods Enzymol. 2003;374:492–509. doi: 10.1016/S0076-6879(03)74021-X. [DOI] [PubMed] [Google Scholar]
- 19.Holm L, Sander C. Protein structure comparison by alignment of distance matrices. J. Mol. Biol. 1993;233:123–138. doi: 10.1006/jmbi.1993.1489. [DOI] [PubMed] [Google Scholar]
- 20.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ryan M, Diekhans M, Lien S, Liu Y, Karchin R. LS-SNP/PDB: annotated non-synonymous SNPs mapped to Protein Data Bank structures. Bioinformatics. 2009;25:1431–1432. doi: 10.1093/bioinformatics/btp242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. J. Computer-aided Mol. des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]