Abstract
High-throughput experimental technologies often identify dozens to hundreds of genes related to, or changed in, a biological or pathological process. From these genes one wants to identify biological pathways that may be involved and diseases that may be implicated. Here, we report a web server, KOBAS 2.0, which annotates an input set of genes with putative pathways and disease relationships based on mapping to genes with known annotations. It allows for both ID mapping and cross-species sequence similarity mapping. It then performs statistical tests to identify statistically significantly enriched pathways and diseases. KOBAS 2.0 incorporates knowledge across 1327 species from 5 pathway databases (KEGG PATHWAY, PID, BioCyc, Reactome and Panther) and 5 human disease databases (OMIM, KEGG DISEASE, FunDO, GAD and NHGRI GWAS Catalog). KOBAS 2.0 can be accessed at http://kobas.cbi.pku.edu.cn.
INTRODUCTION
High-throughput experimental technologies such as next generation sequencing, microarray profiling and proteomics profiling are widely used in current biological research and often identify dozens to hundreds of genes related to a biological or pathological process. Given such a set of genes, one wants to ask which metabolic and signaling pathways may be involved and which diseases may be implicated. As the number of genes is often large, it is desirable to have a computational tool to provide initial answers to these questions. However, ab initio prediction of pathways and diseases is challenging. One feasible approach is to use existing databases of known metabolic and signaling pathways and databases of known disease-associated genes as the starting point for annotation of a new set of genes.
We have previously reported a standalone software and a web server KOBAS 1.0 (1,2) that annotates an input set of genes or proteins by mapping to genes with known pathways in the KEGG PATHWAY database (3). KOBAS 1.0 was the first software to identify statistically significantly enriched pathways using a hypergeometric test. It has been successfully used in pathway analysis in plants, animals and bacteria [for instance, (4–6)].
During the past decade, many other functional enrichment analysis tools have become available. Most of them focus on identification of enriched functional categories based on Gene Ontology (GO) (7), such as FuncAssociate (8), Ontologizer (9), BiNGO (10), FatiGO (11), GOToolBox (11) and GFinder (12). Although tremendously useful, functional categories are not as informative and intuitive as metabolic and signaling pathways and human diseases. A growing number of tools have been developed for pathway and disease identification, including, but not limited to, MAPPFinder (13), EASE (14), DAVID (15,16), ArrayXPath (17), WebGestalt (18), FuncCluster (19), PageMan (20), GENECODIS (21,22), GeneTrail (23), g:Profiler (24), FunNet (25) and PaLS (26). Except for DAVID, all these tools integrate limited pathway and disease databases (for a comparison, see Supplementary Table S1). Furthermore, none of these tools support sequence similarity mapping, an important feature that allows the user to take advantage of data from other species. It is necessary and important to develop a web server tool which incorporates comprehensive pathway and disease databases and supports both ID mapping and sequence similarity mapping.
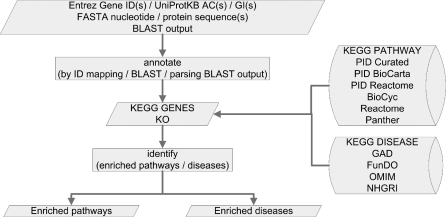
Here, we report a significantly expanded new version, KOBAS 2.0, which incorporates 5 pathway databases [KEGG PATHWAY, PID (27), BioCyc (28), Reactome (29,30) and Panther (31)] and 5 human disease databases [OMIM (http://www.ncbi.nlm.nih.gov/omim/), KEGG DISEASE (32), FunDO (33,34), GAD (35) and NHGRI GWAS Catalog (NHGRI) (36)]. Similar to version 1.0, KOBAS 2.0 supports not only ID mapping, but also sequence similarity mapping. KOBAS 2.0 consists of a standalone command line program written in Python which runs on most Linux systems as well as a user friendly web server developed using Java. Both command line program and web server are freely available at http://kobas.cbi.pku.edu.cn. KOBAS 2.0 flowchart is summarized in Figure 1 and detailed below.
Figure 1.
KOBAS 2.0 workflow. The types of input can be ID, FASTA sequence, or tabular BLAST output. KOBAS 2.0 has two programs ‘annotate’ and ‘identify’. The first program annotates input genes with pathways and diseases by ID mapping or sequence similarity mapping. The second program identifies statistically significantly enriched pathways and diseases.
MATERIALS AND METHODS
KOBAS 2.0 parses 10 pathway and disease databases and stores the data in a SQL relational database
Table 1 summarizes information about the pathway and disease databases that KOBAS 2.0 incorporates. Specifically, KEGG PATHWAY (3) and Reactome (29,30) are general pathway databases, whereas PID (27) and Panther (31) focus on signaling pathways and BioCyc (28) focuses on metabolic pathways. PID has only human data, whereas the others are multispecies databases. OMIM (http://www.ncbi.nlm.nih.gov/omim/) contains information on all known mendelian disorders and genes. KEGG DISEASE (32) collects knowledge on genetic and environmental factors of diseases. FunDO (33,34) is generated from GeneRIF using Disease Ontology Lite that is a condensed version of Disease Ontology. GAD (35) and NHGRI GWAS Catalog (36) both collect data from genetic association studies: GAD includes data from both candidate genes and GWAS studies, whereas NHGRI GWAS Catalog is a catalog of only GWAS studies.
Table 1.
Pathway and disease databases supported by KOBAS 2.0a
| Database name | Data content | File format | Number of species | Number of pathways or diseases in human | Number of genes mapped to KEGG GENES/all genes in human | URL |
|---|---|---|---|---|---|---|
| KEGG PATHWAY | Pathway | Text | 1327 | 220 | 5595/5595 | http://www.genome.jp/kegg/pathway.html |
| PID Curated | Pathway | XML | 1 | 192 | 2782/3315 | http://pid.nci.nih.gov/ |
| PID BioCarta | Pathway | XML | 1 | 254 | 1907/2391 | http://pid.nci.nih.gov/ |
| PID Reactome | Pathway | XML | 1 | 996 | 3783/4405 | http://pid.nci.nih.gov/ |
| BioCyc | Pathway | Text and Table | 6 | 277 | 1087/1120 | http://biocyc.org/ |
| Reactome | Pathway | Table | 22 | 68 | 4366/4534 | http://www.reactome.org/ReactomeGWT/entrypoint.html |
| Panther | Pathway | Table | 43 | 154 | 2170/2207 | http://www.pantherdb.org/ |
| OMIM | Disease | Table | 1 | 4990 | 3792/3792 | http://www.ncbi.nlm.nih.gov/omim |
| KEGG DISEASE | Disease | Text | 1 | 323 | 798/798 | http://www.genome.jp/kegg/disease/ |
| FunDO | Disease | Table | 1 | 561 | 3888/4029 | http://django.nubic.northwestern.edu/fundo/ |
| GAD | Disease | Table | 1 | 3770 | 3164/3238 | http://geneticassociationdb.nih.gov/ |
| NHGRI | Disease | Table | 1 | 369 | 1975/2191 | http://www.genome.gov/gwastudies/ |
aThe numbers in this table are summarized from KOBAS 2.0 backend database updated in November 23rd, 2010. And all the analyses using KOBAS 2.0 in this article are based on this data version.
KOBAS 2.0 downloaded the raw data files from each database. As shown in Table 1, the file formats include plain text, XML and table. We have written parsers for all the data files. For each pathway or disease database, we retrieve the gene-term mapping by parsing the raw data files. We retrieve the gene annotation and gene-ID relations from KEGG Genes and BioMart (37). To integrate across different databases, we mapped the genes in all databases to KEGG GENES and KEGG ORTHOLOGY (KO). The gene-pathway and gene-disease data is stored in our backend SQL relational database. The FASTA protein sequence files were preprocessed for BLAST. KOBAS 2.0 backend data is updated every 3 months.
KOBAS 2.0 annotates input genes with pathways and diseases and identifies enriched pathways and diseases
KOBAS 2.0 has two consecutive programs ‘annotate’ and ‘identify’, which is similar to KOBAS 1.0 (1,2). The first program ‘annotates’ each input gene with putative pathways and diseases by mapping the gene to genes in KEGG GENES or terms in KO which are linked to pathway and disease terms in backend databases. For ID mapping, input IDs are mapped directly to genes using the cross-links we parsed from KEGG GENES. Then, if necessary, IDs are mapped to KO terms. For sequence similarity mapping, each input sequence is BLASTed against all sequences in KEGG GENES. The default cutoffs are BLAST E-value <10−5 and rank ≤5. They mean that an input sequence is assigned KO term(s) of the first BLAST hit that (i) has known KO assignments; (ii) has BLAST E-value <10−5; and (iii) has less than five other hits with a lower E-value that do not have KO assignments (1). A new option in KOBAS 2.0 is that users can map against genes in user-specified species instead of all genes by BLASTing against only sequences of the user-specified species. In order to reduce possible false positives due to multidomain proteins, we added a new option to allow users to set a cutoff of BLAST subject coverage. Another new option allows users to restrict sequence mapping to only orthologs as defined by Ensembl Compara (38).
The second program ‘identifies’ statistically significantly enriched pathways and diseases by comparing results from the first program against the background (usually genes from the whole genome, or all probe sets on a microarray). Users can define their own background distribution in KOBAS 2.0 (for example, result from the first program to ‘annotate’ all probe sets on a microarray). If users do not upload a background file, KOBAS 2.0 uses the genes from whole genome as the default background distribution. Here, we consider only pathways and diseases for which there are at least two genes mapped in the input. Users can choose to perform statistical test using one of the following four methods: binomial test, chi-square test, Fisher's exact test and hypergeometric test, and perform FDR correction. The purpose of performing FDR correction is to reduce the Type-1 errors. When a large number of pathway and disease terms are considered, multiple hypotheses tests are performed, which leads to a high overall Type-1 error even for a relatively stringent P-value cutoff. KOBAS 1.0 supports the FDR correction method QVALUE (39). In KOBAS 2.0, we add two more popular FDR correction methods: Benjamini-Hochberg (40) and Benjamini-Yekutieli (41).
INPUT AND OUTPUT
Input
The input to ‘annotate’ can be a list of IDs, a FASTA sequence file or a tabular BLAST output. KOBAS 2.0 currently can accept three kinds of IDs: Entrez Gene ID, UniProtKB AC and GI. FASTA sequences can be protein or nucleotide sequences. Because BLAST is computationally intensive, the number of sequences that can be run on the online web server is limited to 500 per run. A new feature in KOBAS 2.0 is that, if users want to annotate more sequences online, they can run BLAST locally and upload the tabular BLAST output as the input to KOBAS 2.0. Or they can always run the standalone version of KOBAS 2.0 which has no limit. If users want to get the pathway and disease annotations of their genes, they only need to run ‘annotate’. If they want to find enriched pathways and diseases, they can feed the output of ‘annotate’ directly into ‘identify’ as input.
Output
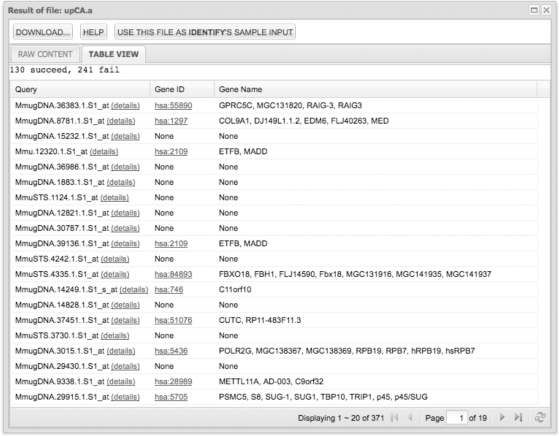
The example of the output of ‘annotate’ is shown in Figure 2. Each row corresponds to one input gene. The first column contains the input gene IDs. The second and third columns contain the mapped KEGG GENE IDs, hyperlinked to detailed descriptions in KEGG and the mapped KEGG GENE names. A user can click on ‘details’ next to the input gene ID to see details about the query and related pathways and diseases.
Figure 2.
Screenshot of the output of ‘annotate’. 371 upregulated probe sets in CA are assigned to KEGG human genes by sequence similarity mapping. Users can view the result in table format (by default) or raw format (which can be downloaded to local disks). Users can also directly use the result as the input of ‘identify’ to do further analysis.
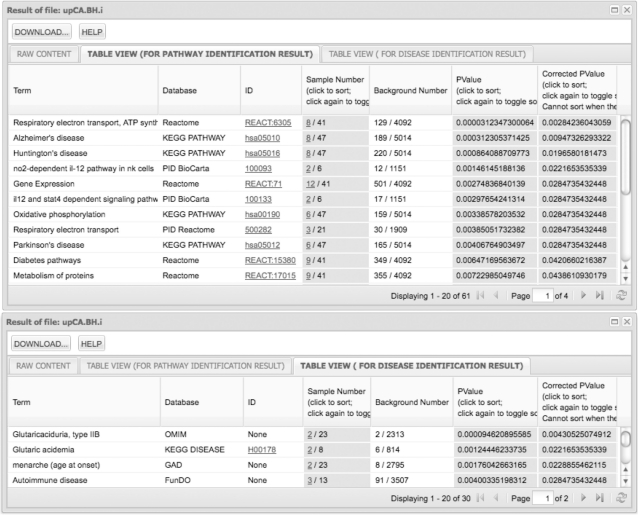
The examples of the output of ‘identify’ is shown in Figure 3. KOBAS 2.0 separates the results of pathways and diseases into two tables. In the pathway identification result, the first three columns show the pathway name, pathway database and pathway ID, hyperlinked to detailed description in the corresponding database. The fourth column lists two numbers of the input: the first one is the number of input genes mapped to the particular pathway and the second one is the total number of input genes mapped to any pathway in the pathway database. Users can click on the first number in the fourth column to see the list of input genes mapped to the particular pathway. The fifth column lists two numbers of the background: the first one is the number of background genes mapped to the particular pathway and the second one is the total number of background genes mapped to any pathway in the pathway database. The last two columns list the P-value and corrected P-value of the statistical test. In the disease identification result, the seven columns show the disease name, disease database, disease ID, numbers of the input, numbers of the background, P-value and corrected P-value similar to the pathway identification result. KOBAS 2.0 merges redundant pathway and disease terms from different databases.
Figure 3.
Screenshot of the output of ‘identify’. Statistically significantly enriched pathways and diseases of 371 upregulated probe sets in CA identified are sorted by increasing corrected P-value. Only those with corrected P ≤ 0.05 are shown. Similar to the output of ‘annotate’, users can view the result in table format (by default) or raw format (which can be downloaded to local disks).
BENEFIT OF CROSS-SPECIES SEQUENCE SIMILARITY MAPPING OVER ID MAPPING
Other existing pathway analysis tools accept only gene IDs as input and use only ID mapping to annotate their pathways. A benefit of KOBAS 2.0 is that it can use sequence similarity mapping to annotate input genes from species that are not yet well-represented in existing pathway databases. It can also map the genes from other species to human diseases to predict whether these genes may be good candidates to study any human diseases, an important question in the model organism research. To illustrate, we analyzed the microarray expression profiles in rhesus monkeys in two major hippocampal subdivisions critical for memory/cognitive function: cornu ammonis (CA) and dentate gyrus (DG) using data from Blalock et al. (42). We reanalyzed their raw data on six samples from CA and six samples from DG of young rhesus monkeys and identified 371 upregulated probe sets in CA using standard protocol [gcrma and limma through R and Bioconductor (43)]. We then used both DAVID (15,16) and KOBAS 2.0 to annotate these probe sets and identify enriched pathways and diseases by using the entire probe sets on the chip as background. DAVID can perform only ID mapping to rhesus genes in its two pathway databases (KEGG PATHWAY and Panther) and as a result, identified no statistically significantly enriched pathways or diseases (with default options and corrected P ≤ 0.05). On the other hand, KOBAS 2.0 supports sequence similarity mapping by BLAST to annotate the rhesus gene set and can thus take full advantage of the abundant data on human pathways and diseases. We used ‘annotate’ to map sequences of upregulated probe sets in CA as well as the entire probe sets to KEGG human genes with default cutoffs and then used ‘identify’ to perform hypergeometric test and Benjamini-Hochberg FDR correction to find significantly enriched pathways and diseases by using the two results of ‘annotate’ as input and background, respectively. Figure 3 shows significantly enriched pathways and diseases identified by KOBAS 2.0. The results are consistent with known functional differences between the two regions. For example, ‘respiratory electron transport, ATP synthesis by chemiosmotic coupling and heat production by uncoupling proteins’ pathway and ‘glutaricaciduria, type IIB’ and ‘Glutaric academia’ diseases are consistent with the known knowledge that the CA region showed greater expression than DG for genes associated with mitochondrial activity (42); while ‘no2-dependent il-12 pathway in nk cells’, ‘il12 and stat4 dependent signaling pathway in th1 development’ and ‘autoimmune disease’ are consistent with the known knowledge that CA region showed greater expression than DG for genes associated with inflammatory responses (42).
We also compared KOBAS 2.0 with popular GO enrichment analysis tools, FuncAssociate 2.0 (8), Ontologizer 2.0 (9), BiNGO (10) and EASE (14) using the same data set. Because these other tools can only take IDs as input, we first mapped the rhesus probe sets to human genes using sequence similarity. Then we ran the four GO enrichment analysis tools, the results of which are shown in Supplementary Table S2. The list of enriched pathways identified by KOBAS 2.0 is more specific and informative than the lists of functional categories identified by the GO enrichment analysis tools, and offers more insights into the biological processes.
CONCLUSIONS
KOBAS 2.0 has an expanded reservoir of underlying pathway databases and statistical tests, and the addition of disease databases. In future research, we aim to improve the graphical representation of the output pathways. We will continue to update KOBAS 2.0 with new pathway and disease data.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
‘National Outstanding Young Investigator’ from Natural Science Foundation of China (31025014); Johnson and Johnson (scholarship); China Ministry of Science and Technology 863 Hi-Tech Research and Development Programs (2007AA02Z165) and 973 Basic Research Program (2011CBA01102, 2007CB946904). Funding for open access charge: 973 Basic Research Program (2011CBA01102).
Conflict of interest statement. None declared.
REFERENCES
- 1.Mao X, Cai T, Olyarchuk JG, Wei L. Automated genome annotation and pathway identification using the KEGG Orthology (KO) as a controlled vocabulary. Bioinformatics. 2005;21:3787–3793. doi: 10.1093/bioinformatics/bti430. [DOI] [PubMed] [Google Scholar]
- 2.Wu J, Mao X, Cai T, Luo J, Wei L. KOBAS server: a web-based platform for automated annotation and pathway identification. Nucleic Acids Res. 2006;34:W720–W724. doi: 10.1093/nar/gkl167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Shi YH, Zhu SW, Mao XZ, Feng JX, Qin YM, Zhang L, Cheng J, Wei LP, Wang ZY, Zhu YX. Transcriptome profiling, molecular biological, and physiological studies reveal a major role for ethylene in cotton fiber cell elongation. Plant Cell. 2006;18:651–664. doi: 10.1105/tpc.105.040303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Huang J, Chen T, Liu X, Jiang J, Li J, Li D, Liu XS, Li W, Kang J, Pei G. More synergetic cooperation of Yamanaka factors in induced pluripotent stem cells than in embryonic stem cells. Cell Res. 2009;19:1127–1138. doi: 10.1038/cr.2009.106. [DOI] [PubMed] [Google Scholar]
- 6.Sridhar J, Rafi ZA. Functional annotations in bacterial genomes based on small RNA signatures. Bioinformation. 2008;2:284–295. doi: 10.6026/97320630002284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Berriz GF, Beaver JE, Cenik C, Tasan M, Roth FP. Next generation software for functional trend analysis. Bioinformatics. 2009;25:3043–3044. doi: 10.1093/bioinformatics/btp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bauer S, Grossmann S, Vingron M, Robinson PN. Ontologizer 2.0–a multifunctional tool for GO term enrichment analysis and data exploration. Bioinformatics. 2008;24:1650–1651. doi: 10.1093/bioinformatics/btn250. [DOI] [PubMed] [Google Scholar]
- 10.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 11.Al-Shahrour F, Diaz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 12.Masseroli M, Martucci D, Pinciroli F. GFINDer: Genome Function INtegrated Discoverer through dynamic annotation, statistical analysis, and mining. Nucleic Acids Res. 2004;32:W293–W300. doi: 10.1093/nar/gkh432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salomonis N, Hanspers K, Zambon AC, Vranizan K, Lawlor SC, Dahlquist KD, Doniger SW, Stuart J, Conklin BR, Pico AR. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hosack DA, Dennis G, Jr, Sherman BT, Lane HC, Lempicki RA. Identifying biological themes within lists of genes with EASE. Genome Biol. 2003;4:R70. doi: 10.1186/gb-2003-4-10-r70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huang da W, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Huang da W, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat. Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 17.Chung HJ, Park CH, Han MR, Lee S, Ohn JH, Kim J, Kim JH. ArrayXPath II: mapping and visualizing microarray gene-expression data with biomedical ontologies and integrated biological pathway resources using Scalable Vector Graphics. Nucleic Acids Res. 2005;33:W621–W626. doi: 10.1093/nar/gki450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Kirov S, Snoddy J. WebGestalt: an integrated system for exploring gene sets in various biological contexts. Nucleic Acids Res. 2005;33:W741–W748. doi: 10.1093/nar/gki475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Henegar C, Cancello R, Rome S, Vidal H, Clement K, Zucker JD. Clustering biological annotations and gene expression data to identify putatively co-regulated biological processes. J. Bioinform. Comput. Biol. 2006;4:833–852. doi: 10.1142/s0219720006002181. [DOI] [PubMed] [Google Scholar]
- 20.Usadel B, Nagel A, Steinhauser D, Gibon Y, Blasing OE, Redestig H, Sreenivasulu N, Krall L, Hannah MA, Poree F, et al. PageMan: an interactive ontology tool to generate, display, and annotate overview graphs for profiling experiments. BMC Bioinformatics. 2006;7:535. doi: 10.1186/1471-2105-7-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carmona-Saez P, Chagoyen M, Tirado F, Carazo JM, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo JM, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37:W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Backes C, Keller A, Kuentzer J, Kneissl B, Comtesse N, Elnakady YA, Muller R, Meese E, Lenhof HP. GeneTrail–advanced gene set enrichment analysis. Nucleic Acids Res. 2007;35:W186–W192. doi: 10.1093/nar/gkm323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reimand J, Kull M, Peterson H, Hansen J, Vilo J. g:Profiler–a web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007;35:W193–W200. doi: 10.1093/nar/gkm226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Prifti E, Zucker JD, Clement K, Henegar C. FunNet: an integrative tool for exploring transcriptional interactions. Bioinformatics. 2008;24:2636–2638. doi: 10.1093/bioinformatics/btn492. [DOI] [PubMed] [Google Scholar]
- 26.Alibes A, Canada A, Diaz-Uriarte R. PaLS: filtering common literature, biological terms and pathway information. Nucleic Acids Res. 2008;36:W364–W367. doi: 10.1093/nar/gkn251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schaefer CF, Anthony K, Krupa S, Buchoff J, Day M, Hannay T, Buetow KH. PID: the Pathway Interaction Database. Nucleic Acids Res. 2009;37:D674–D679. doi: 10.1093/nar/gkn653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahren D, Tsoka S, Darzentas N, Kunin V, Lopez-Bigas N. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Croft D, O'Kelly G, Wu G, Haw R, Gillespie M, Matthews L, Caudy M, Garapati P, Gopinath G, Jassal B, et al. Reactome: a database of reactions, pathways and biological processes. Nucleic Acids Res. 2011;39:D691–D697. doi: 10.1093/nar/gkq1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Osborne JD, Flatow J, Holko M, Lin SM, Kibbe WA, Zhu LJ, Danila MI, Feng G, Chisholm RL. Annotating the human genome with Disease Ontology. BMC Genomics. 2009;10(Suppl 1):S6. doi: 10.1186/1471-2164-10-S1-S6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Du P, Feng G, Flatow J, Song J, Holko M, Kibbe WA, Lin SM. From disease ontology to disease-ontology lite: statistical methods to adapt a general-purpose ontology for the test of gene-ontology associations. Bioinformatics. 2009;25:i63–i68. doi: 10.1093/bioinformatics/btp193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nat. Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 36.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38:D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Storey JD. A direct approach to false discovery rates. J. R. Statist. Soc. B. 2002;64:479–498. [Google Scholar]
- 40.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Statist. Soc. B. 1995;57:289–300. [Google Scholar]
- 41.Benjamini Y, Yekutieli D. The control of the false discovery rate in multiple testing under dependency. Ann. Statist. 2001;29:1165–1188. [Google Scholar]
- 42.Blalock EM, Grondin R, Chen KC, Thibault O, Thibault V, Pandya JD, Dowling A, Zhang Z, Sullivan P, Porter NM, et al. Aging-related gene expression in hippocampus proper compared with dentate gyrus is selectively associated with metabolic syndrome variables in rhesus monkeys. J. Neurosci. 2010;30:6058–6071. doi: 10.1523/JNEUROSCI.3956-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]