Abstract
Mining genetic variation from personal genomes is a crucial step towards investigating the relationship between genotype and phenotype. However, compared to the detection of SNPs and small indels, characterizing large and particularly complex structural variation is much more difficult and less intuitive. In this article, we present a new scheme (inGAP-sv) to detect and visualize structural variation from paired-end mapping data. Under this scheme, abnormally mapped read pairs are clustered based on the location of a gap signature. Several important features, including local depth of coverage, mapping quality and associated tandem repeat, are used to evaluate the quality of predicted structural variation. Compared with other approaches, it can detect many more large insertions and complex variants with lower false discovery rate. Moreover, inGAP-sv, written in Java programming language, provides a user-friendly interface and can be performed in multiple operating systems. It can be freely accessed at http://ingap.sourceforge.net/.
INTRODUCTION
Structural variation (SV) generally refers to cytogenetically visible and submicroscopic variants, including insertions, deletions, inversions, translocations, duplications and copy number variants (1,2). Extensive studies have shown that structural variants are involved in various genetic disorders, including cancer (3). In 2005, Tuzun et al. (4) compared the human genome reference sequence with fosmid paired-end sequences from another human genome and identified a number of intermediate-sized structural variants. Since 2006, the advancement of next-generation sequencing technologies and paired-end mapping (PEM) approaches has greatly facilitated a high-throughput and comprehensive survey of SVsa in various organisms. In a pioneering study, Korbel et al. (5) sequenced over 5 billion base pairs from two human genomes using Roche 454 platform and identified many more SVs than initially hypothesized.
The huge amount of high-throughput sequencing data brings challenges to the development of bioinformatic analysis approaches. Current SV detection approaches, as reviewed by Medvedev et al. (6), can be classified into three categories based on what kind of signatures are used for SV discovery: PEM (7–13), depth of coverage (DOC) (14), and split read mapping (15,16). Each of these approaches has limits in terms of the type and size of SVs that they are able to detect, and there still lacks a one-stop solution for full range of structural variant detection. Instead, people use SVMerge (17) and programs alike to integrate multiple existing SV detection methods, which could complement each other and enhance capabilities for SV detection.
Due to the complex nature of structural variants, it is hard to find a simple rule to characterize all types of them. Breakdancer is a popular and sophisticated tool to detect large size SVs based on PEM data (7). Although it provides information like the type, size, the number of supporting read pairs and confidence score of each predicted SV, users still need more sufficient features to define a SV and its quality, e.g. the ratio of supporting read pairs to the local DOC, read mapping qualities, the presence of any combined SVs and tandem repeats, because all these components are critical to determine the type and quality of a SV. The same problem is also present in other PEM-based approaches. As shown in Supplementary Figure S1, different duplication types have distinct patterns on read depth, mapping distance and orientation, and mapping boundaries. A graphical visualization of these patterns together with other genomic properties (e.g. tandem repeat) can help users distinguish different types of SVs and filter false positives. Therefore, a combination of visual and automated approaches will help users seamlessly inspect and refine SV detection, classify complex forms of variants and minimize false discovery rate.
In addition, current PEM-based approaches are unable to detect large insertions when their size is larger than the insert size of the sequenced fragment, because of the lack of closely mapped read pairs which can span the entire insert region. Most recently, Medvedev (18) demonstrated that the integration of PEM and DOC analysis can detect large insertions and variants that do not create any discordant read pairs. Besides DOC signatures, single-end mapped reads on the flanking regions also provide valuable information to target the positions of large insertions, and should be taken into account.
Here, we present a new scheme, inGAP-sv, to detect and visualize large and complex SVs from PEM data. It utilizes both PEM and coverage of depth strategies to identify different types of SVs, including large indels, inversions, translocations, tandem duplications and segmental duplications. More importantly, it is possible to distinguish homozygous and heterozygous variants.
METHODS
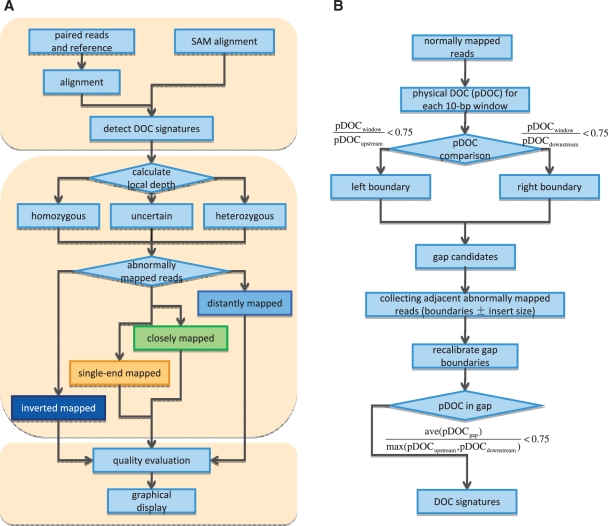
As shown in Figure 1, we employ a three-step strategy to discover structural variation: (i) scan the PE read alignment and detect all SV hotspots by counting the local mapping coverage; (ii) predict and classify SVs based on the combination of abnormally mapped read pairs; and (iii) evaluate variants by using mapping qualities and densities.
Figure 1.
The pipeline of SV detection in inGAP-sv. (A) A three-step strategy to detect SVs. (B) The workflow of DOC signature detection. Briefly, a DOC signature can be defined as a part of reference sequence covered by much fewer normally mapped reads than the local physical DOC. As shown in B, a gap is identified initially by pairing of its left and right boundaries. In a region with continuously descending pDOC values, the left boundary is set to be the location whose pDOC is smaller than three-quarters of its upstream local pDOC. The right boundary is determined based on the same rule. Incomplete gap signatures with only one side of boundaries will be ignored, which possibly result from sequencing coverage bias. Subsequently, inGAP-sv uses abnormally mapped read pairs adjacent to each gap to recalibrate boundaries. Finally, gaps with fine adjustment of boundaries are filtered out if its average pDOC exceeds three-fourth of the local pDOC value.
Mapping of high-throughput reads and statistical analysis of mapping coverage
inGAP-sv requires two files, a FASTA formatted reference sequence and a SAM alignment. The SAM format short read alignment can be generated using BWA (19) which has been integrated into the pipeline or other standalone mapping tools. After the SAM file is loaded into inGAP-sv, user-defined threshold of mapping quality (default value: 20) is applied to filter non-uniquely mapped reads. Read pairs with only one end mapped will be considered as single-end reads (SE reads). Repetitive reads are removed if they have identical matches on different loci of the genome.
inGAP-sv scans the entire short read alignment and identifies all gap signatures. The detailed workflow of this step is shown in Figure 1B. Briefly, a gap signature can be defined as a part of reference sequence covered by much fewer normally mapped reads than the local physical DOC (pDOC). Here normally mapped reads indicate those read pairs mapped with proper distance and orientation. The local pDOC refers to the physical coverage of the flanking 1-kb region of a gap. It should be noted that only normally mapped reads are taken into account in calculating the local pDOC. This is different from other DOC based approaches (14,18). For homozygous SVs, the pDOC values over the affected intervals are expected to be zero; whereas heterozygous variants may be represented by a reduced pDOC compared with their flanking regions. The identified DOC signatures may include extensive noise signals caused by sequencing bias and mapping errors (Supplementary Figure S2). In the following step, inGAP-sv will use PEM to remove false positive DOC signatures.
The distance between paired-end reads may vary owing to DNA library construction procedures before sequencing, but usually satisfies a normal distribution. inGAP-sv scans all normally mapped reads and obtains statistical information on average insert size (denoted by l) and standard variation (denoted by δ) of the insert fragments for input data automatically. A pair of reads are considered as closely (or distantly) mapped if their distance is shorter than l − 3δ (or longer than l + 3δ). This parameter is adjustable upon users’ own needs.
Collecting abnormal mapping information and SV detection
Adjacent to each gap signature predicted from the first step, inGAP-sv collects information of abnormally mapped reads, as shown in Figure 2. Non-uniquely mapped reads (default as 20 for the BWA aligner) are ignored in this step. Single-end mapped, distantly/closely mapped and inverted mapped reads are grouped respectively according to their mapping orientation, and further divided into subgroups by using a center-based clustering algorithm, in which maximum distance among any pair within a subgroup should be shorter than the average insert size. Different types of SVs are predicted based on the combined patterns of abnormally mapped read pairs (Table 1). Deletions are detected by distantly mapped reads; small insertions, whose length are shorter than the insert size, are surrounded by single-end reads and have closely mapped reads; while large insertions are represented by flanking single-end reads with the absence of closely mapped read pairs; complex SVs including inversions, translocations and duplications also exhibit distinct patterns (Figure 2, Supplementary Figure S1 and Table 1). If the gap signature within a SV is fully spanned by continuously normally mapped read pairs, we then consider this SV to be heterozygous, otherwise to be homozygous.
Figure 2.
Illustrations of PEM patterns for different types of SVs. Grey links indicate normally mapped read pairs with proper read orientation and distance. Light blue links represent read pairs with proper read orientation but longer distance, which may indicate a deletion event in the query sequence. Green links represent read pairs with proper orientation but shorter distance, and thus indicate an insertion. Dark blue links show read pairs with abnormal orientation, in which paired ends are mapped to the wrong strand(s). Yellow lines indicate single-end mapped reads (SE reads), in which only one of the paired reads is mapped. Pink lines indicate a pair of reads mapped to different chromosomes. All gap signatures for different SVs are shown in blue oval circles. (A) For a small insertion (< the insert size), a fraction of paired reads (in green) that span the insertion is mapped too closely in the reference. Meantime, the insertion is surrounded by a set of single-end mapped reads (in yellow). (B) For a large insertion, no paired reads can span the insertion and only single-end mapped reads are present. (C) For a homozygous deletion, all the paired reads (in blue) are mapped farther than expected. (D) For a heterozygous deletion, normally mapped pairs (in grey) span the gap signature. (E) A translocation is represented by two sets of distantly mapped pairs and one set of inverted mapped pair (in dark blue). (F) An inversion causes the paired reads to change the orientation, and both ends will map to the same strand. (G) A segmental tandem duplication is represented by one set of distantly mapped reads and one set of inverted mapped reads.
Table 1.
Patterns for different types of SVs used by inGAP-sv
| SV Type | # Gap/Peak | Abnormal PEM Pattern |
|
|---|---|---|---|
| Single-end mapped | Paired-end mapped | ||
| Small insertion | 1/0 | SE+left, SE−right | PE+−close |
| Large insertion | 1/0 | SE+left, SE−right | none |
| Deletion | 1/0 | none | PE+−distant |
| Inversion | 2/0 | SE+left, SE−right | PE++, PE− − |
| Transposition | 2/0 | SE+left, SE−right | PE+−distant, PE−+ |
| Inverted transposition | 3/0 | SE+left, SE−right | PE+−distant, PE++, PE− − |
| Tandem duplication | 0/1 | SE+left, SE−right | PE−+ |
| Inverted tandem duplication | 1/1 | SE+left, SE−right | PE++, PE− − |
| Segmental duplication | 1/1 | SE+left, SE−right | PE+−distant, PE−+ |
| Inverted segmental duplication | 1/1 | SE+left, SE−right | PE++, PE− − |
SE+left refers to single-end mapped read on the plus strand of left flank region of a gap, while SE−right for that on the minus strand of right flank region. PE refers to paired-end mapped read, strand information of both ends are marked by +−(normal orientation) or ++/− −/−+ (inverted orientation), respectively.
SV evaluation by mapping qualities and densities
Quality of predicted SVs depends on its pattern and the ratio of observed supportive reads to expected supportive reads. The quality Q is calculated as  , where qsupport or qunsupport represents the mapping quality of an abnormally mapped read which may support or decline the existence of a SV, respectively. For a given SV, qsupport is the mapping quality of supported reads listed in Table 2, while the other reads are used to calculate qunsupport. For example, for homozygous deletions, all distantly mapped PE reads will be considered as supported reads, whereas normally, closely and single-end mapped reads will be considered as unsupported reads. avg(qglobal) is the average global mapping quality, while avg(qflanking) is the average local mapping quality 1 kb adjacent to a SV. Only normally mapped read contributes to the calculation of avg(qglobal) and avg(qflanking). Compared with homozygous SVs, heterozygous SVs tend to have 50% of normally mapped reads that can span the gap signature, so a factor δ (1 for homozygous SVs and 2 for heterozygous SVs) is set to recalibrate the qualities.
, where qsupport or qunsupport represents the mapping quality of an abnormally mapped read which may support or decline the existence of a SV, respectively. For a given SV, qsupport is the mapping quality of supported reads listed in Table 2, while the other reads are used to calculate qunsupport. For example, for homozygous deletions, all distantly mapped PE reads will be considered as supported reads, whereas normally, closely and single-end mapped reads will be considered as unsupported reads. avg(qglobal) is the average global mapping quality, while avg(qflanking) is the average local mapping quality 1 kb adjacent to a SV. Only normally mapped read contributes to the calculation of avg(qglobal) and avg(qflanking). Compared with homozygous SVs, heterozygous SVs tend to have 50% of normally mapped reads that can span the gap signature, so a factor δ (1 for homozygous SVs and 2 for heterozygous SVs) is set to recalibrate the qualities.
Table 2.
Quality scoring for different types of SVs
| SV Type | SV quality |
|---|---|
| Small insertion |  |
| Large insertion |  |
| Deletion |  |
| Inversion |  |
| Translocation |  |
| Inverted translocation |  |
| Tandem duplication |  |
| Inverted tandem duplication |  |
| Segmental duplication |  |
| Inverted segmental duplication |  |
 . A maximum value 100 is assigned to Qs if it is higher than 100.
. A maximum value 100 is assigned to Qs if it is higher than 100.
Data sets
To evaluate the performance of our method, we used inGAP (20) to simulate both haploid and diploid paired-end data sets from a 10-Mb region in chr20: 1 000 001–11 000 000. The read length was 50 bp and the average insert size was 300 bp with a standard deviation ranging from 5 to 20%. Three million paired-end reads were simulated, with an average DOC as 30X. For both data sets, we simulated 0.1% of SNPs and 0.01% of large indels ranging from 50 bp to 1 Kb. Randomized mutations and 1-bp indels were incorporated to mimic sequencing errors.
In order to test the sensitivity of gap search by pDOC under different depths of sequence data, we downloaded the whole-genome paired-end sequencing data (SRX000600, ∼38X depth), NA18507, sequenced by Bentley et al. (21). This data set consists of 35-bp paired-end reads sequenced on an Illumina GA platform, with a mean insert size of 200 bp. Four subsets were sampled, representing 5-, 10-, 20- and 30-fold of genomic coverage, respectively.
Most recently, Mills et al. reported a map of unbalanced SVs based on 185 human resequencing data sets (22). In the article, they applied various SV detection algorithms to DNA sequence data from NA12878, and also provided a list of reported SV coordinates, which can be downloaded at http://www.nature.com/nature/journal/v470/n7332/extref/nature09708-s5.zip. Here we applied inGAPsv to the same sequence data from NA12878 (ERX000080, ∼8X), and compared the identified SVs with reported SV calls by VariationHunter, Breakdancer, PEMer, Spanner, Cortex and Pindel. We used the same gold standard data sets (GS1) in Mills's study, which were collected from previous published data (23–25). Through the visualization of inGAP-sv, we noticed that some deletions in the gold standard data set were well-supported by normally mapped read pairs, and thus might be false positives (Supplementary Figure S3). We filtered GS1 to generate a clean set (GS2). If a deletion in GS1 cannot be detected by any of above SV detection algorithm, it will be ruled out in GS2. Deletions predicted by inGAP-sv were compared with GS1 and GS2 respectively to calculate sensitivity values.
RESULTS
A scheme to visualize PEM and deduce SVs
inGAP-sv provides users a visualized interface to further inspect predicted SVs, as all detailed information is shown graphically. For paired-end libraries, normally mapped reads have their left end map to the positive strand (colored in blue bars, as shown in Figure 2) and right end to the negative strand (in red). Non-uniquely mapped reads are shown in grey. We use a link to connect a pair of forward and reverse reads and various types of links are shown in different colors. Combination of these links is used to represent different types of SVs shown in Figure 2.
To better understand the structure of complex SVs, we simulated indels, inversions, translocations and a variety of duplications in the 10-Mb region of chr20 to generate a pseudo sequence. Then we simulated paired-end reads from this pseudo sequence and mapped them back to the original 10-Mb sequence. As shown in Figure 2, a small insertion is represented by green links and adjacent yellow lines (i.e. SE reads), while a large insertion only consists of yellow lines. As for complex SVs, they can be characterized by a combination of several forms of colored links and gap signatures. For example, translocation involves two deletion patterns (light blue links), one inversely orientated pattern (dark blue links) and multiple SE reads. Inversion has two inversely oriented patterns, multiple SE reads and two gap signatures. Moreover, the visualization scheme used in this study can also distinguish different duplication forms, such as tandem and segmental duplications (Supplementary Figure S1).
A simulation study
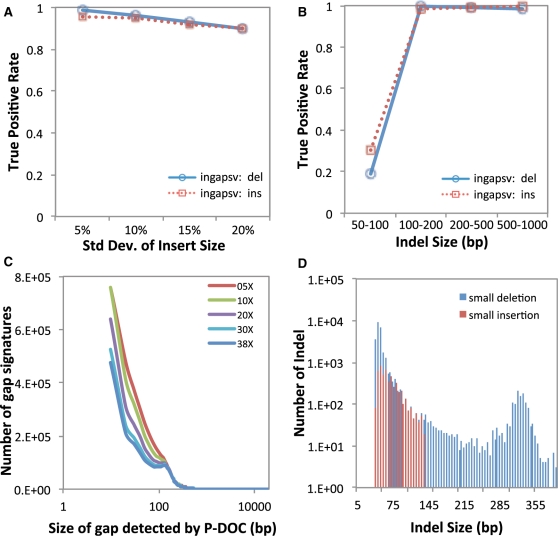
We firstly tested how standard deviation of insert size could affect the performance of SV detection. As shown in Figure 3A, along with the increase of insert size variance, the sensitivity (i.e. true positive rate) of indel detection using inGAP-sv slightly decreased, and down to 90% when variance reached to 20% of insert size. Most of paired-end sequencing libraries for either NA18507 or NA12878 are constructed with variance range given in Figure 3A. inGAP-sv failed to detect some small indels <50 bp (Figure 3B) but performed well on detection of larger indels.
Figure 3.
Performance of indel detection by inGAP-sv on both simulated (A and B) and real data sets (C). (A) True positive rate of indel detection at different levels of standard deviation of insert size. When the insert size variance increases, the true positive rate of deletion detection slightly decreases; (B) Plot of true positive rate of indel size. inGAP-sv fails to detect very small indels (<50 bp), while works well on the detection of large indels. (C) Size of gap signatures detected by inGAP-sv under different sequence depth from NA18507; (D) Distribution of indel size predicted by inGAP-sv with 38X data from NA18507. Blue bars indicate the number of small deletions, while red bars indicate small insertions. Moreover, inGAP-sv detected 729 deletions >400 bp and 524 insertions larger than the insert size, which are not shown in the graph.
We secondly simulated different depth of sequences by combining different runs from NA18507 (see ‘Methods’ section for details) to investigate the sensitivity of gap searching by pDOC. As shown in Figure 3C, the total number of gaps detected is not that sensitive to the sequence coverage. However, with more sequence data, inGAP-sv can detect fewer small gaps, indicating that higher coverage of sequence data can help reduce pDOC bias. A length distribution of identified SVs using the 38X data from NA18507 was given in Figure 3D, in which the size of deletions ranges from 40 bp to 78 kb. Since the standard deviation of insert size for NA18507 is 13 bp, deletions <39 bp cannot be detected due to the recognition strategy of abnormally mapped reads (see ‘Methods’ section). However, these short deletions could be recognized by setting different parameters, yet sacrificing the specificity of SV prediction.
Case studies on real data
We applied inGAP-sv to the paired-end sequencing data of NA12878 to detect SVs and assessed its sensitivity using the gold standard data sets (GS1 and GS2). The calculation time of inGAP-sv is linearly proportional to the number of mapped reads and the length of the reference sequence. In this study, with 8X data from NA12878 on a 2 Gb memory desktop, it took 13 min to run inGAP-sv on chr1 and 5 min on chr12. As shown in Supplementary Table S1, inGAP-sv could detect 58 and 80% of deletions in GS1 and GS2, respectively. This sensitivity ratio is higher than those reported in Mills's study (Figure 4A).
Figure 4.
Performance comparison between inGAP-sv and other tools (Breakdancer, VariationHunter and Spanner). (A) The Gold standard SV set (GS2) is used to assess the detection sensitivity of the four methods for an individual NA12878. inGAP-sv can call 80% of deletions in GS2, which is slightly higher than the other three tools. Most importantly, the number of deletions (shown in blue) identified by inGAP-sv and another tool simultaneously is significantly higher than that without inGAP-sv (shown in red). The detailed list of identified deletions for each tool is shown in Supplementary Table S1. (B) A Venn diagram shows the comparison of the deletion calls made by the four tools on chr20 of NA12878.
As to chr20, inGAP-sv could identify 140 insertions, 435 deletions, and most of these indels were heterozygous. After filtering out low quality (Q < 5, 14% of the total) and short indels (<50 bp, 70% of the rest), there were 69 insertions and 73 deletions remaining. This number is slightly higher than those reported by other approaches (e.g. VariationHunter, 70; Breakdancer, 60; Spanner, 51). Moreover, inGAP-sv also found that five deletions reported in the Mills study were <50 bp. Notably, the number of deletions identified by inGAP-sv and another approach simultaneously is significantly higher than that without inGAP-sv (Wilcoxon rank test, P = 0.03) (Figure 4B). This indicates that the SVs detected by inGAP-sv are more likely to be supported by other independent tools.
As shown in Figure 4B, each approach also reported a small percentage of algorithm-specific deletions unrecognized by other approaches. We manually checked them based on the inGAP-sv visualization. We found that among the 13 inGAP-sv specific deletions, three have size >100 bp, and the remaining 10 have size ranging from 50 to 60 bp. We speculate that these short deletions could have been identified by other tools, but might have their size been underestimated (<50 bp) and thus were filtered out. It should be mentioned that although both VariationHunter and Breakdancer reported a significant fraction of deletions (25.7 and 30%, respectively), after manual inspection a majority of them seem to be false positives (16 out of 18 by VariationHunter, 14 out of 18 by Breakdancer, e.g. Supplementary Figure S4 and Supplementary Table S2). Detailed comparison can be downloaded from http://sourceforge.net/projects/ingap/files/ingap/datasets/NA12878_chr20.tgz.
For complex SVs, inGAP-sv identified one tandem duplication (chr20: 2 164 990–2 165 120) and two segmental duplications (chr20: 2 308 470–2 308 690, chr20: 18 189 590–18 190 140). In contrast, PEMer reported two of them and Pindel identified one, and all of them were mis-classified as deletions. inGAP-sv also identified a number of very low-quality SVs (Q < 5), which were generally false positives or located in tandem repeat regions. Users can manually check the reliability of the predicted SVs through the graphic interface. inGAP-sv also provides a function to design primers that can amplify SV sequences for further experimental verification.
As another demonstration of the algorithm, inGAP-sv had been applied to an Arabidopsis thaliana genome re-sequencing project (unpublished data). Using the PEM data, inGAP-sv identified 815 insertions and 1000 deletions. We then compared these indels to the Monsanto A. thaliana assembly, and found that 78% of the deletions could be covered by the Monsanto assembly and 99% of them were correct. As for insertions, 71% could be covered and 96% of them were correct.
DISCUSSIONS
In this study, we employed a new strategy to deduce and visualize SVs from PEM data. We used DOC information to detect SV hotspot regions and then clustered all the surrounding abnormally mapped reads to classify the SV type. The context information was used to determine their qualities. These improvements make it possible to (i) identify large insertions and complex forms of SVs; (ii) reduce false discovery rate in tandem repeat regions; and (iii) distinguish homozygous and heterozygous SVs. Moreover, inGAP-sv is a one-stop SV detector, in which users can identify, visualize, annotate and manually edit SVs.
A graphic visualization is irreplaceable in SV detection and verification because of the complex structure of certain types of SVs and the repetitive nature of eukaryotic genomes. Paired-end reads mapped onto the reference genome are linked with colored lines and can be visualized intuitively. When right click a specific read or SV, auxiliary information will be shown, such as ID, location, read length and orientation, mapping quality, mate position and insert size. Notably, tandem repeat information of the reference sequence is also computed and displayed below the alignment panel, which is helpful to filter false positives. In addition, inGAP-sv provides extensive flexibility to change the appearance of the displayed short read alignment and colored links. Such a color or format setting function is necessary, because users usually need a better display when exploring SVs from hundreds of fold coverage of short read alignments.
Exploration of the local physical DOC is a critical step in the pipeline of inGAP-sv. We use the weighted pDOC instead of the sequencing DOC to detect gap signatures, in which only high-quality mapped read pairs (Q ≥ 20) are considered. This step can filter falsely mapped reads in repetitive regions and increase the accuracy of SV prediction. In addition, pDOC provides higher coverage than sequencing DOC, and thus provides higher confidence for gap prediction. inGAP-sv employs the gap signature to determine the SV breakpoint and to cluster different types of colored links (e.g. blue links, dark blue links, etc). This strategy is particularly useful to define complex SVs. If nested links share the same gap signature, they may belong to one SV. In this way, inGAP-sv has successfully classified various types of duplications as shown in Supplementary Figure S1. Another advantage of exploring the gap signature is that it can distinguish the actual link types from other noises. For example, wrongly-placed reads, due to sequencing errors or genome complexities (e.g. tandem repeats) may lead to the formation of colored links. But these artifacts do not have to form a gap signature. Applications of inGAP-sv on real data sets further confirm that the integration of the gap signature in SV prediction can significantly reduce the false discovery rate.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Fudan University (the ‘985’ and ‘211’ Programs of the Ministry of Education of China); Rijk Zwaan (Netherlands), and CAS grants (to F.Z.) (Y064021BJ1, 0869011BJ5).
Conflict of interest statement. None declared.
REFERENCES
- 1.Feuk L, Carson AR, Scherer SW. Structural variation in the human genome. Nat. Rev. Genet. 2006;7:85–97. doi: 10.1038/nrg1767. [DOI] [PubMed] [Google Scholar]
- 2.Sharp AJ, Cheng Z, Eichler EE. Structural variation of the human genome. Annu. Rev. Genomics Hum. Genet. 2006;7:407–442. doi: 10.1146/annurev.genom.7.080505.115618. [DOI] [PubMed] [Google Scholar]
- 3.Stankiewicz P, Lupski JR. Structural variation in the human genome and its role in disease. Annu. Rev. Med. 2010;61:437–455. doi: 10.1146/annurev-med-100708-204735. [DOI] [PubMed] [Google Scholar]
- 4.Tuzun E, Sharp AJ, Bailey JA, Kaul R, Morrison VA, Pertz LM, Haugen E, Hayden H, Albertson D, Pinkel D, et al. Fine-scale structural variation of the human genome. Nat. Genet. 2005;37:727–732. doi: 10.1038/ng1562. [DOI] [PubMed] [Google Scholar]
- 5.Korbel JO, Urban AE, Affourtit JP, Godwin B, Grubert F, Simons JF, Kim PM, Palejev D, Carriero NJ, Du L, et al. Paired-end mapping reveals extensive structural variation in the human genome. Science. 2007;318:420–426. doi: 10.1126/science.1149504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Medvedev P, Stanciu M, Brudno M. Computational methods for discovering structural variation with next-generation sequencing. Nat. Methods. 2009;6:S13–S20. doi: 10.1038/nmeth.1374. [DOI] [PubMed] [Google Scholar]
- 7.Chen K, Wallis JW, McLellan MD, Larson DE, Kalicki JM, Pohl CS, McGrath SD, Wendl MC, Zhang Q, Locke DP, et al. BreakDancer: an algorithm for high-resolution mapping of genomic structural variation. Nat. Methods. 2009;6:677–681. doi: 10.1038/nmeth.1363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Korbel JO, Abyzov A, Mu XJ, Carriero N, Cayting P, Zhang Z, Snyder M, Gerstein MB. PEMer: a computational framework with simulation-based error models for inferring genomic structural variants from massive paired-end sequencing data. Genome Biol. 2009;10:R23. doi: 10.1186/gb-2009-10-2-r23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee S, Hormozdiari F, Alkan C, Brudno M. MoDIL: detecting small indels from clone-end sequencing with mixtures of distributions. Nat. Methods. 2009;6:473–474. doi: 10.1038/nmeth.f.256. [DOI] [PubMed] [Google Scholar]
- 10.Hormozdiari F, Alkan C, Eichler EE, Sahinalp SC. Combinatorial algorithms for structural variation detection in high-throughput sequenced genomes. Genome Res. 2009;19:1270–1278. doi: 10.1101/gr.088633.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hajirasouliha I, Hormozdiari F, Alkan C, Kidd JM, Birol I, Eichler EE, Sahinalp SC. Detection and characterization of novel sequence insertions using paired-end next-generation sequencing. Bioinformatics. 2010;26:1277–1283. doi: 10.1093/bioinformatics/btq152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zeitouni B, Boeva V, Janoueix-Lerosey I, Loeillet S, Legoix-ne P, Nicolas A, Delattre O, Barillot E. SVDetect: a tool to identify genomic structural variations from paired-end and mate-pair sequencing data. Bioinformatics. 2010;26:1895–1896. doi: 10.1093/bioinformatics/btq293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sindi S, Helman E, Bashir A, Raphael BJ. A geometric approach for classification and comparison of structural variants. Bioinformatics. 2009;25:i222–i230. doi: 10.1093/bioinformatics/btp208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Campbell PJ, Stephens PJ, Pleasance ED, O'Meara S, Li H, Santarius T, Stebbings LA, Leroy C, Edkins S, Hardy C, et al. Identification of somatically acquired rearrangements in cancer using genome-wide massively parallel paired-end sequencing. Nat. Genet. 2008;40:722–729. doi: 10.1038/ng.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: a pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Abel HJ, Duncavage EJ, Becker N, Armstrong JR, Magrini VJ, Pfeifer JD. SLOPE: a quick and accurate method for locating non-SNP structural variation from targeted next-generation sequence data. Bioinformatics. 2010;26:2684–2688. doi: 10.1093/bioinformatics/btq528. [DOI] [PubMed] [Google Scholar]
- 17.Wong K, Keane TM, Stalker J, Adams DJ. Enhanced structural variant and breakpoint detection using SVMerge by integration of multiple detection methods and local assembly. Genome Biol. 2010;11:R128. doi: 10.1186/gb-2010-11-12-r128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Medvedev P, Fiume M, Dzamba M, Smith T, Brudno M. Detecting copy number variation with mated short reads. Genome Res. 2010;20:1613–1622. doi: 10.1101/gr.106344.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qi J, Zhao F, Buboltz A, Schuster SC. inGAP: an integrated next-generation genome analysis pipeline. Bioinformatics. 2010;26:127–129. doi: 10.1093/bioinformatics/btp615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, Bignell HR, et al. Accurate whole human genome sequencing using reversible terminator chemistry. Nature. 2008;456:53–59. doi: 10.1038/nature07517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mills RE, Walter K, Stewart C, Handsaker RE, Chen K, Alkan C, Abyzov A, Yoon SC, Ye K, Cheetham RK, et al. Mapping copy number variation by population-scale genome sequencing. Nature. 2011;470:59–65. doi: 10.1038/nature09708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Conrad DF, Pinto D, Redon R, Feuk L, Gokcumen O, Zhang Y, Aerts J, Andrews TD, Barnes C, Campbell P, et al. Origins and functional impact of copy number variation in the human genome. Nature. 2010;464:704–712. doi: 10.1038/nature08516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McCarroll SA, Kuruvilla FG, Korn JM, Cawley S, Nemesh J, Wysoker A, Shapero MH, de Bakker PI, Maller JB, Kirby A, et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 25.Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N, Graves T, Hansen N, Teague B, Alkan C, Antonacci F, et al. Mapping and sequencing of structural variation from eight human genomes. Nature. 2008;453:56–64. doi: 10.1038/nature06862. [DOI] [PMC free article] [PubMed] [Google Scholar]