Abstract
Genome-wide expression profiles obtained with the use of DNA microarray technology provide abundance of experimental data on biological and molecular processes. Such amount of data need to be further analyzed and interpreted in order to obtain biological conclusions on the basis of experimental results. The analysis requires a lot of experience and is usually time-consuming process. Thus, frequently various annotation databases are used to improve the whole process of analysis. Here, we present RuleGO—the web-based application that allows the user to describe gene groups on the basis of logical rules that include Gene Ontology (GO) terms in their premises. Presented application allows obtaining rules that reflect coappearance of GO-terms describing genes supported by the rules. The ontology level and number of coappearing GO-terms is adjusted in automatic manner. The user limits the space of possible solutions only. The RuleGO application is freely available at http://rulego.polsl.pl/.
INTRODUCTION
Results of experiments with DNA microarrays are often summarized by lists of genes, called gene (molecular) signatures, that exhibit certain expression patterns across experimental conditions, e.g. are coexpressed, differentially expressed, overexpressed, etc. Study of biological facts behind gene compositions of gene signatures is often computationally supported by algorithms that aim at characterizing them by keywords provided by gene annotation databases. A special annotation database is the Gene Ontology (GO) database (1), which provides controlled and structured vocabulary of terms for genes and their products in the form of a directed acyclic graph (DAG). Due to its hierarchical structure, GO database represents the biological knowledge at different levels of specificity. Other databases used for annotations of gene signatures are e.g. KEGG Pathways (2), motifs from InterPro database (3) and keywords describing entries from UniProt database (4). Numerous programs and Internet services (5–8) have been developed for annotations of gene signatures. Such programs provide lists of annotation terms along with P-values of statistical tests to measure statistical significance of overrepresentation (enrichment) or underrepresentation (depletion) of terms in the analyzed gene signatures.
Recently, in the field of gene annotations, new ideas appeared, of using combinations of annotation terms (multiattribute annotations) rather than single annotation terms, to characterize gene signatures (9–11). The use of multiple instead of single annotation terms, potentially, offers advantages in researching gene signatures, such as: (i) combinations of annotation terms can define sets of genes with statistically significant deviations from totally random distribution, while single terms do not show statistically significant enrichment or depletion; (ii) sets of genes annotated by combinations of terms are smaller and therefore reflect more specific biological facts; (iii) combinations of annotation terms may lead to interesting biological interpretations, related e.g. to genetic pathways or their cross-talks. At present, there are already two Internet services: Annotation-Modules (9) and GeneCodis (10,11) allowing for characterization of gene signatures by combinations (associations) of annotation terms. Both these services are based on variants of the Apriori algorithm (12) mining association rules in databases.
Characterization of gene signatures by combinations of annotation terms leads to more difficult problems than those encountered when developing annotations by single terms. When constructing algorithms for searching through very large numbers of combinations of annotations terms, their designers must use heuristics to limit memory and time complexity. Therefore, results may become unpredictable, some important (interesting) multiattribute annotations may be overlooked and results of annotations obtained by using different algorithms may show significant differences. Due to very large number of associations, corrections for multiple testing become less reliable and more difficult to interpret. Finally, when designing annotation algorithms, one encounters the problem of accounting for the structural properties of GO graph. This important problem has already been addressed in the case of single-term annotations and methods have been proposed that allow decorrelating GO graph structure e.g. (13). However, for multiattribute annotations, existing tools do not provide the possibility of annotating gene sets by GO terms decorrelated with respect to the structure of GO graph.
In this article, we present a new web server, RuleGO, for multiattribute annotations of gene signatures. The annotation terms in our multiattribute rules are GO terms. The methodology used in the web server construction was presented in Refs. (14–16). Our algorithm is based on the extension of the Apriori algorithm, called Explore algorithm, published in (17), which introduces additional conditions that are used in the process of generating sets of multiattribute rules. The Explore algorithm allows searching for decision rules, which are combinations of annotation terms that differentiate the analyzed (signature) set of genes and the reference set of genes.
We take advantage of the possibilities given by the Explore algorithm to obtain decision rules satisfying quality criteria defined by the user. By using the searching method oriented toward induction of rules satisfying user preferences and by applying appropriate filtration methods, we can obtain sets of rules with higher statistical significance and, consequently, with better descriptive power than other rules induction methods based on simple generation of all combinations of annotation terms.
Our algorithm allows the user to chose among different quality indices of the decision rules: a rule quality measured by modified Yails measure (14), a rule length and a depth of the GO terms composing a rule. The RuleGO algorithm additionally incorporates a tool for controlling (limiting) the number of decision rules reported to the user, based on the appropriate rules filtration method.
We also address the problem of decorrelating GO graph when searching for multiattribute rules. The rules induction algorithm, which allows obtaining rules with non-redundant GO terms (14–16), is briefly described in the ‘Methods’ section. In the ‘RuleGO service’ and ‘Comparison to the existing tool’ sections, we present some possibilities offered by our web server and comparison to other tools.
METHODS
We denote by G1 and G2 two disjoint sets (groups) of gene symbols. G1 is the gene signature (primary) set and G2 is the reference set. Symbols of genes are denoted by letters g with appropriate indexes, e.g. Gi = {gi1, gi2, … ,giMi}, i = 1, 2. Here M1 and M2 are numbers of genes in gene groups G1 and G2.
GO graph is a directed acyclic graph (DAG), denoted GO = (A, ≤), where A denotes a set of all GO annotation terms and ≤ is a binary relation on A. GO terms are represented by letters a with appropriate indices. If there are two GO terms such that ak ≤ al, then GO term al is either equal to GO term ak or GO term al is a parent term to GO term ak. By a parent term, we understand each term al which is at the upper level of GO graph than term ak (i.e. term al is closer to the root of the graph than term ak) and there exists a path between both of the terms.
GO terms characterize (annotate) genes in the sense that each gene symbol g, has a set of GO terms associated to it.
A characterization of the gene signature G1 by GO terms describing genes composing this signature is given by a family of logical decision rules. Rules are denoted by letters r with appropriate indexes. The rule number i has the following form:
| (1) |
When specified to a particular gene, the rule ri in (1) has the following meaning: ‘if a gene is described by the GO terms that compose the rule ri, then it belongs to the group presented in the rule conclusion’.
The set (list) of decision rules, r1, r2, … , rN, which characterize the gene signature G1 is obtained by application of an appropriately designed algorithm for rules induction, described below.
Induction of multiattribute rules
The Explore algorithm (17) introduces structural modifications to the procedure for producing (generating) rules, such that only rules of certain properties are generated. Application of the idea of the Explore algorithm, in our web application RuleGO, allows inducing decision rules satisfying quality criteria defined by the user. Production of the set of decision rules follows by starting from a single GO term. The initial GO term belongs to the set of all GO terms, which annotate genes g11, g12, … , g1M1. Next, new GO terms are successively appended. Appending a new GO term generates a new rule. It is verified whether the generated rule satisfies certain criteria. If it does, then the rule is added to the output rules set. If it does not, it is still kept with a ‘temporary’ status. The algorithm verifies whether ‘temporary’ rules have potential for satisfying the quality criteria defined by the user by adding next GO terms. If, by using the appropriate condition, a temporary rule is verified to have no such potential, it is removed from further analysis. Otherwise, the process of appending new GO terms continues.
In order to address the problem of decorrelating the set of GO terms in the premise of the rule, we introduced additional modification to the above algorithm. The idea of the introduced modification is to take into account the hierarchy of genes assignment to GO terms by creating only such rules that include in a premise GO terms that do not lie on a common path on GO = (A, ≤) DAG. In other words, the algorithm will never produce a rule that includes in its premise GO terms that are in ≤ (parent–child) relation.
The algorithm determines all possible logical rules in which statistical significance level is equal to (or less than) a threshold defined by the user. For assessment of statistical significance of a rule, we consider the following null hypothesis: ‘assignment of genes described by the rule to the signature group indicated by the rule is equivalent to a random assignment of the genes to the group’. To verify the hypothesis, the one-side (right side) hypergeometric test is used, because we search for combinations of GO terms which are overrepresented in analyzed gene signature. To adjust for multiple hypothesis testing, a false discovery rate (FDR) coefficient, given in Ref. (18), for the P-value is also computed. To compute FDR coefficient, we sort the obtained P-values in the ascending order (starting with the most significant P-value). Assuming that we have obtained n multiattribute rules (we count all rules generated during analysis, not only statistically significant ones), and denoting p(k) as the k-th smallest P-value, we estimate the FDR corrected P-value as:
| (2) |
Multiattribute rules quality measure
Experimental analysis of various data sets shows that the number of created rules is very large and usually varies from several to a few dozen thousand rules. Interpretation of such set is impossible, therefore the procedure of evaluation and filtration of the determined rules was prepared. Three criteria are taken into account during the rule r evaluation: Length(r) is the number of GO terms occurring in the rule premise (we assume that the more terms the better, because longer rules give us more complete biological description of genes), Depth(r) is the normalized sum of levels in GO graph to which terms appearing in the rule premise are assigned (the lower level the better, since we deal then with the more precise knowledge) and q(r) is the rule quality based on the modified Yails measure. The q(r) measure reflects the compromise between rule accuracy and generality (according to Knowledge Discovery requirements, discovered patterns should be accurate and general). A modification of the q(r) measure, proposed in (14), allows obtaining more general rules (describing more genes) without decreasing the accuracy.
A compound quality measure that enables the user to evaluate the rule quality from the point of view of the aspects presented above is the product of all component measures:
| (3) |
Rules filtration
The filtration algorithm that uses rules ranking obtained on the basis of the measure defined by the Equation (3) is executed in a loop. Beginning from the best rule in the ranking, all rules covering the same set of genes or its subset are candidates to be removed from the result rules set. However, before removing any rule, its similarity to the reference rule is verified. If a rule is similar to the reference rule in more than a threshold defined by the user, it is removed from the set of determined rules, otherwise it remains in the output rules set. The similarity of two rules ri and rj is determined by the following formula:
| (4) |
where: #GOterms(ri, rj) is a number of unique GO-terms occurring in the rule ri and not occurring in the rule rj. The GO-term a from the rule ri is recognized as the unique if it does not occur directly in the rule rj and there is no path in GO graph that includes both term a and any term b from rule rj premise; #GOterms(ri), #GOterms(rj) are the numbers of GO-terms in the rules ri and rj premises respectively.
RULEGO SERVICE
Input data and algorithm parameters
The user initializes an experiment by choosing an organism and sending two disjoint gene groups or one group of genes to the service. In the latter case, the second group is created automatically as the group of the remaining genes from the genome of the considered organism (rest of the genome). The service allows using various popular gene identifiers such as Gene ID, gene symbol, Ensembl, etc. The list of supported gene formats is provided to the users of service.
Then, the user defines the set of parameters which are used by rules generation and filtration algorithms. The form for defining parameters configuration is divided into sections concerning selection of statistical significance threshold, Gene Ontology annotations, rules generation options and rules filtration parameters. Figure 1 presents all parameters that can be defined by the user of RuleGO service.
Figure 1.
RuleGO parameters configuration form.
The first, Statistical test, section allows the user to determine statistical features which should characterize discovered rules. To compute the P-value of determined rules, the hypergeometric test for over-representation is used. Only the rules with the P-value less or equal to the threshold defined are generated.
In the section ‘Gene Ontology’, the user can define the GO aspects which should be used for genes annotations. ‘Hierarchical annotations’ parameter determines whether hierarchy of GO graph should be considered during annotation process. If the option ‘Hierarchical annotations’ is selected, hierarchical dependencies among GO terms are analyzed according to the ‘true path rule’, which means that genes annotations are propagated to upper levels of GO graph. If the ‘Hierarchical annotations’ option is off, then all GO terms (between selected values of minimal and maximal ontology levels) are annotated directly from GO database.
‘Ontology level’ parameter allows setting the minimal and maximal levels of GO terms annotating genes, and thus defines the level of detail of the obtained description.
‘Algorithm options’ section allows the user to provide parameters for rules generation algorithm and thus, to limit the searched space. Minimal number of genes described by a GO term allows selecting only these GO terms, which describe more (or equal) number of genes than a threshold defined by the user. This parameter removes GO terms from analysis describing too few genes. For example, if we search for rules which describe at least three genes, there is no sense in including GO terms into the analysis annotating less than three genes from the signature set. ‘Maximal number of GO terms’ parameter is used to limit the number of GO terms which can be placed in a rule premise.
It is worth noticing, that increasing the value of ‘Maximal number of GO terms’ parameter results in the generation of more specific rules (described by lower number of genes). However, too specific rules may not satisfy other limitations applied by the algorithm parameters (i.e. statistical significance, minimal number of genes described by the rule) and thus, increasing the value of this parameter above a certain threshold does not result in generation of any new combinations of GO terms satisfying defined criteria.
The next, ‘minimal support’ parameter is used to determine the minimal number of genes that each of determined rules should describe. Usually, we would like to obtain rules that are general, that is, which describe at least several genes from the analyzed group.
Both previously mentioned parameters (‘minimal support’ and ‘maximal number of GO terms’) can have a big influence on the time of rules generation and it is important to notice that by increasing ‘maximal number of GO terms’ value and decreasing ‘minimal support’ value, one can significantly multiply the number of combinations that need to be analyzed and thus extend the computation time. The analysis of how different settings of this both parameters can influence the computation time and the number of obtained rules are available as Supplementary Data F01.
The last parameter from this section limits the number of generated rules. As rules are generated in order to describe the specified group of genes and they are further presented to the user, the number of generated rules should not be very big, according to the limitations of the human perception. Following the user decision, only n best rules are generated, where n is the value of ‘Maximal number of generated rules’ parameter.
‘Rules filtration’ section includes parameters that are required by rules filtration algorithm. First three parameters are used to compute the compound rule quality measure defined by the Equation (3). These parameters allow the user to select which aspects of the rule quality are most important for the specific application. If the ‘Rules similarity’ options is selected, a set of generated rules is filtered according to the method described in ‘Rules filtration’ section. ‘Minimal similarity threshold’ parameter is used to define the similarity threshold [Equation (4)].
Generated rules are sorted according to one of the selected criteria: the compound quality measure [Equation (3)] or by the P-value computed using the hypergeometric test. Different rankings of rules allow the user to analyze different aspects of the obtained list of rules.
Output
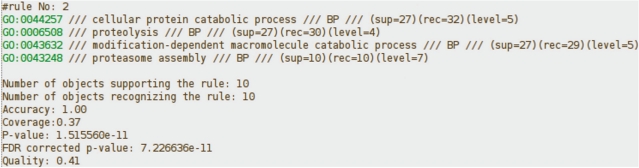
Results of analysis are presented in the form of a list of multiattribute rules. We present exemplary output rule in Figure 2. The resulting rule is a set of GO terms which describe all genes described by the rule (a list of these genes is presented below the rule). For each GO term, we present its symbol and description. We also provide information of how many genes are described by this (single) GO term in the signature group (sup parameter), how many genes are described in both (signature and reference) groups of genes (rec parameter) and the level of this GO term on GO graph (we assume that the root of GO graph is on the level 0).
Figure 2.
Example of multiattribute rule with quality evaluation parameters.
For each rule, we provide a set of values that allow evaluating different aspects of the rule quality. We present the number of genes that are described by this rule in the primary set G1 (number of genes supporting the rule) and in both sets, G1 and G2 (number of genes recognizing the rule). For each rule, we also compute its accuracy (ratio of the number of genes supporting the rule to the number of genes recognizing the rule) and coverage (ratio of the number of genes supporting the rule to the number of genes in primary set). These parameters allow deciding whether the rule is specific to genes from the signature set (accuracy parameter) and/or it is general (coverage parameter). For each rule, we also provide its P-value and FDR adjusted P-value. We also present the value of the compound quality measure computed according to the formula [Equation (3)] and parameters selected by the user.
Usually, a single rule describes only a subset of genes from the analyzed group. To obtain the description of the whole group of genes, we need to analyze the list of all rules induced. Thus, it is important to know how many genes from the group are described by the rules generated by our algorithm. This information is presented on the top of the results page, by the parameter ‘percent covered’, which provides the information about percentage of genes from the signature group described (covered) by the generated rules.
COMPARISON TO THE EXISTING TOOL
In this section, we present a comparison of the results of the analysis performed with the use of the RuleGO service with the existing tool, GeneCodis (10,11). GeneCodis is the tool that uses rule discovery algorithm based on Apriori method, which finds significant combinations of annotations. The RuleGO service is based on same idea of searching all possible significant combinations; however, our algorithm does not generate rules that include in their premises GO terms that are in parent–child relation. In addition, we provide advanced methods of rules filtration and quality evaluation that allow users to select the most interesting rules, according to the user preferences.
We used the GeneCodis interface available form the Babelomics service (19) due to the fact that it allows us to control more parameters concerning GO annotations than the original GeneCodis web site. We analyzed the set of 224 genes, which we call peroxisome gene set. The analyzed peroxisome gene set was obtained in the Smith et al. (20), in the DNA microarray experiment concerning coexpression of peroxisome genes in yeast. The peroxisome gene set was also analyzed (annotated) in Ref. (10).
For analysis of the peroxisome gene set with the use of the GeneCodis tool, we used the following parameters: GO biological process (levels from 4 to 19); allowed range of term annotations among 1 to 1000 (from genome); minimum number of genes: 3; each term parent within levels has been included. Using the above settings, we generated GeneCodis rules describing the peroxisome set of genes. The complete set of obtained rules is available as Supplementary Table R01.
We compared the results obtained by using the GeneCodis tool to the set (sets) of rules obtained by using the RuleGO service. We defined algorithm parameters used in the RuleGO service, such that they corresponded to those used in the GeneCodis service, namely: Ontology level: min 3, max 18; minimal number of genes described by GO term: 3; minimal support: 3; hierarchical annotations: yes. For RuleGO analysis, we also reduced the maximal number of GO terms in a rule premise to 5 and we set significance level value to 0.05. The similarity threshold used in filtration process was set to the default value 0.5. In the RuleGO service, apart from parameters described above, we also have filtration and ranking options that allow presenting the obtained rules to the user on the basis of different criteria. We have analyzed several possible combinations of these options as described in the subsections below. Complete sets of obtained rules are available as Supplementary Tables R2–R10.
One of the important motivations for using multiattribute rules, given by combinations of GO annotation terms, is that they can define sets of genes with statistically significant deviations from totally random distribution, despite that single terms do not show statistically significant enrichment or depletion. Analysis of our Supplementary Tables R01–R10 shows that both services, GeneCodis and RuleGO, indeed return many rules that include in premises GO terms which separately do not have the power to differentiate between the primary and reference sets. However, if the same GO terms are analyzed together, they compose statistically significant multiattribute rules. One example, returned by RuleGO service, is (see Supplementary Table R04):
The P-value of the above rule is 0.047, which satisfies the established criterion for statistical significance. This rule describes all genes from peroxisome signature set, which are involved in transcription elongation from RNA polymerase II promoter process. These genes are as follows: EAF3, RSC30, HIR1. The rule is composed of five GO terms and for each term we have provided, on the list above, the number of supported and recognized genes. One can see that none of the GO terms that compose the rule premise shows statistical significance, including the GO term ‘transcription elongation from RNA polymerase II promoter’, whose P-value is 0.19. Only the combination of the above statistically insignificant GO terms can give the significant rule, indicating genes from perixosome signature that are involved in transcription elongation process. Analogous examples can be seen as a result of the use of the GeneCodis service, and were also reported in Carmona-Saez et al. (10).
Quality indices of sets of multiattribute rules
The first aspect of the comparison of the rule sets obtained with the use of GeneCodis and RuleGO services concerns indices that describe the quality of the obtained set of rules. The quality indices, which we consider here, are as follows:
mean P-value of rules;
number of rules;
coverage.
The mean P-value index concerns averaging over P-values without FDR correction. The last index, coverage, is defined as the ratio of the number of those genes, which support at least one of the generated rules, to the number of genes from analyzed peroxisome gene set described by at least one GO term. In the peroxisome gene set, the number of such genes that each has at least one GO term associated to it, is equal to 171.
For the generated set of GeneCodis rules, we have obtained the following values for these three indices:
mean P-value: 0.0083;
number of rules: 73;
coverage: 69%.
The same quality indices were also computed for characterization of peroxisomal gene signature by multiattribute rules obtained by using our RuleGO service. We generated nine different sets of rules using all possible settings of rankings and filtration options. The results of analysis are presented in Table 1.
Table 1.
Indices describing RuleGO rule sets obtained for different filtration and ranking settings
| Filtration | Ranking method |
Compound quality measure |
Mean P-value | Rules number | Coverage (%) | ||
|---|---|---|---|---|---|---|---|
| m.Yails | Length | Depth | |||||
| NO | * | * | * | * | 0.0052 | 7813 | 85 |
| YES | P-value | * | * | * | 0.00063 | 41 | 69 |
| Q measure | YES | YES | YES | 0.0063 | 79 | 85 | |
| YES | YES | NO | 0.0062 | 84 | 85 | ||
| YES | NO | YES | 0.0073 | 53 | 85 | ||
| YES | NO | NO | 0.005 | 60 | 85 | ||
| NO | YES | YES | 0.01 | 83 | 85 | ||
| NO | YES | NO | 0.011 | 77 | 85 | ||
| NO | NO | YES | 0.0082 | 57 | 85 | ||
Bold values denote rule set with the best mean P-value.
Two rows (groups of rows) of Table 1 are labeled ‘filtration NO' and ‘filtration YES'. In the ‘filtration NO' row, all rules obtained in the search process are reported. As it can be seen in ‘rules number' column, 7813 rules satisfying the criterion P ≤ 0.05 were obtained. Among these 7813 rules, many are repeating in the sense that they define the same set of genes. In the file returned by the RuleGO service, all rules defining the same set of (supporting) genes are grouped together. If we limit the set of obtained rules to rules supported by different sets of genes, then we obtain 293 rules (this value is further shown in Table 2).
Table 2.
Overlapping sets of genes supporting RuleGO and GeneCodis rules, unique sets of genes in the RuleGO service
| Filtration | Ranking method |
Compound quality measure |
Overlapping gene sets | Unique gene sets | |||
|---|---|---|---|---|---|---|---|
| Yails | Length | Depth | |||||
| NO | * | * | * | * | 25 | 293 | |
| YES | P-value | * | * | * | 9 | 37 | |
| Q measure | YES | YES | YES | 16 | 70 | ||
| YES | YES | NO | 12 | 72 | |||
| YES | NO | YES | 11 | 51 | |||
| YES | NO | NO | 11 | 52 | |||
| NO | YES | YES | 13 | 72 | |||
| NO | YES | NO | 10 | 68 | |||
| NO | NO | YES | 14 | 57 | |||
The ‘filtration YES’ group of rows is further stratified into two subgroups, ‘ranking method P-value’ and ‘ranking method Q measure’. In the ‘ranking method P-value' row, we reported results of the greedy search through the obtained set of rules, based on the P-values. The search was terminated when the coverage equal to 69% (equal to the coverage obtained by using the GeneCodis service) was reached. The number of rules in this row is much lower than the number reported by the GeneCodis service and the average P-value is more than 10 times lower than the corresponding average P-value obtained by using the GeneCodis service.
In the ‘ranking method Q measure’ group of rows, we reported results of the greedy searches through the obtained set of rules, based on (compound) Q measure with different options, defined by YES or NO entries in the appropriate row–column crossings. As can be seen in Table 1, in most cases, the RuleGO rule sets are characterized by better average P-values and, importantly, by over 15% better coverages. The latter allows us to better describe the analyzed group of genes.
GeneCodis service reports 73 rules from the set of several thousands rules satisfying the defined criteria. Thus, clearly a filtration operation is applied, oriented toward selection of most significant and most important rules. However, (i) this operation is not optimized with respect to one of the several possible criteria and (ii) the user of the service cannot influence the process of final selection of output rules. In case of RuleGO service, we provide a set of filtration parameters allowing the user to select the most interesting aspects of rule quality, depending on the experiment purposes.
The above results also show that RuleGO rules generation method allows obtaining rules that describe more genes from analyzed signature group. Applied ranking method influences the number of output rules and their mean P-value, however, the filtration method always guarantees obtaining the best possible coverage.
Overlapping gene sets
For different sets of multiattribute rules, we have obtained different sets of genes supporting single rules. To further compare GeneCodis and RuleGO, results, we also analyzed overlap among the genes supporting single rules. The overlap is measured by the number of identical gene sets supporting rules generated by both services. Such identical gene sets are called overlapping gene sets.
The results are presented in Table 2. The row structure in Table 2 repeats that of Table 1. The column ‘overlapping gene sets’ shows numbers of overlapping gene sets obtained by GeneCodis and RuleGO services. Contemplation of entries in this column in Table 2 shows that the use of the two services leads to two rather different sets of genes, following from obtained rules, with little overlap.
The last column in Table 2 gives us information on the repeating structure of gene sets obtained by the RuleGO service, for different options. Such information is not provided for the GeneCodis rules due to the fact that there are no repeating gene sets among GeneCodis rules. On the contrary, results returned by our RuleGO service can have the structure with repeating gene sets supporting different rules. The reason for leaving repeating gene sets in the output of the service, is that they are (may be) defined by rules with the structure different enough for suspecting that they may provide different (new) information. Entries in the last column in Table 2 provide numbers of unique gene sets in the output of the RuleGO service. The analysis of the last column of Table 2 shows that in most cases we have obtained lower number of unique rules, which cover more genes from analyzed group than GeneCodis rules. Due to the fact, that results are further presented to an expert who is able to analyze only limited number of rules, the selection of the most significant and interesting rules (according to the user preferences) is one of the most important parts of rules generation process.
Decorrelation with respect to GO graph
Decision rules returned by the RuleGO service are decorrelated with respect to the GO graph structure, i.e. no rule can contain two GO terms lying on the same ontology path. We also analyzed the structure of GeneCodis rules with the aim to find out whether these rules can include, in their premises, GO terms lying on the same ontology path. After such analysis, we obtained 48 rules (out of 73) including in their premises GO terms lying on the same ontology path. Due to the fact that all GO annotations must follow the true path rule, which means that if a gene is annotated by a single GO term it is also annotated by all its parent terms, and in our opinion such rules provide redundant information.
Below we present comparative analysis of two similar rules generated using GeneCodis and RuleGO methods, respectively. Both the rules are supported by the same set of three genes: NED2, RKI1, MDH3. The rule obtained in the GeneCodis service is as follows:
The analysis of the structure of GO graph for biological process ontology revealed that there are following relations among three of GO terms composing the above rule: monosaccharide metabolic process ≤ hexose metabolic process ≤ glucose metabolic process.
The rule, generated using the RuleGO service (see Supplementary Table R04), corresponding to the same gene set is as follows:
Analysis of the structure of the GO graph shows that both rules provide very similar functional description. ‘Glucose metabolic process’ is common term for both rules. ‘Carbohydrate catabolic process’ term from RuleGO rule is immediate child of ‘carbohydrate metabolic process’ term from GeneCodis rule, while RuleGO ‘nicotinamide nucleotide metabolic process’ term is second-level child of GeneCodis ‘nucleotide metabolic process’ term. It is worth noticing, that both terms from RuleGO rule are child-terms of corresponding GeneCodis terms, and from three GeneCodis terms lying on common path, the term representing the lowest level was selected by RuleGO algorithm. This indicates that our algorithm allows obtaining rules that provide more specific description of analyzed genes.
CONCLUSIONS
Using different methods for induction of multiattribute rules can lead to substantial differences in their outcomes, as shown by our comparisons. Our web-based application for induction of multiattribute rules can outperform the existing tool in several aspects of the quality including the coverage of the analyzed signature gene set by multiattribute rules. The novelty of our service is in providing to its users the possibility of rules quality evaluation and filtration, and in creating rules that do not include in their premises terms lying on the same path in GO graph. The presented set of various parameters allows the user to create different rankings of the generated rules and evaluate different features of the obtained rules, according to specific requirements.
The RuleGO service enables the user to obtain gene group descriptions by means of multiattribute logical decision rules. Obtained rules reflect co-appearance of GO-terms describing genes supported by the rules. The ontology level and the number of co-appearing GO-terms is adjusted in automatic manner. The RuleGO provides a tool that allows selecting the most interesting combinations of GO-terms from all possible significant combinations, which can save an expert time and improve the whole process of analysis.
The RuleGO service provides multiattribute rules which do not include in their premise GO terms lying on the common path. The presented algorithm allows avoiding generation of rules that provide redundant information.
Our method guarantees that all statistically significant rules are determined. However, the experimental analysis shows that even if we generate only statistically significant rules, we still can observe the very large number of output rules. In such case, we cannot expect that a human expert will be able to review all the generated rules. For that reason, RuleGO provides a set of methods for evaluation of rules quality that allows limiting the number of output rules and selecting only the most interesting ones. However, presented filtration method does not guarantee that during the filtration process some of the interesting rules will not be removed. The manner of rules removing depends on rules ranking, which is fixed by applied compound quality measure.
The parameters available to the RuleGO users are set to default values based on our experience and analyses performed on various data sets. In most cases, the user should be able to generate a description of a signature group using the default values. However, if the obtained list of rules does not satisfy requirements (e.g. the number of obtained rules is too large or to small; the rules describe too few genes from a signature group), we recommend comparing results of different designs of analysis with different values of parameters.
One limitation of our method is its computational complexity (we look for all possible statistically significant rules), which may cause long wait for experiment results in extreme cases. After completing the computations, the results are stored at our web site and the user is notified by e-mail.
The need of deciding about different sets of parameters before one can obtain a satisfactory description of an analyzed gene group can be regarded as another disadvantage of the presented method. However, with some experience in using these parameters, altering values of the algorithm parameters can lead to the possibility of analyzing different aspects of the ontological structures of the studied gene signatures.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
European Social Fund (A.G.); EC FP6 Grant (GENEPI lowRT to A.P.) in part. Funding for the open access charge: European Social Fund.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank anonymous referees for their helpful comments. We would also like to thank our colleague, Łukasz Chróst for his technical help.
REFERENCES
- 1.Ashburner M, Ball C, Blake J, Botstein D, Butler H, Cherry J, Davis A, Dolinski K, Dwight S, Eppig J, et al. Gene Ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Minoru K, Michihiro A, Susumu G, Masahiro H, Mika H, Masumi I, Toshiaki K, Shuichi K, Shujiro O, Toshiaki T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huner S. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Al-Shahrour F, Daz-Uriarte R, Dopazo J. FatiGO: a web tool for finding significant associations of Gene Ontology terms with groups of genes. Bioinformatics. 2004;20:578–580. doi: 10.1093/bioinformatics/btg455. [DOI] [PubMed] [Google Scholar]
- 6.Dennis G, Sherman B, Hosack D, Yang J, Gao W, Lane H, Lempicki R. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:R60. [PubMed] [Google Scholar]
- 7.Maere S, Heymans K, Kuiper M. BiNGO: a Cytoscape plugin to assess overrepresentation of Gene Ontology categories in biological networks. Bioinformatics. 2005;21:3448–3449. doi: 10.1093/bioinformatics/bti551. [DOI] [PubMed] [Google Scholar]
- 8.Zhang B, Schmoyer D, Kirov S, Snoddy J. GOTree Machine (GOTM): a web-based platform for interpreting sets of interesting genes using Gene Ontology hierarchies. BMC Bioinformatics. 2004;5:16. doi: 10.1186/1471-2105-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hackenberg M, Matthiesen R. Annotation-Modules: a tool for finding significant combinations of multisource annotations for gene lists. Bioinformatics. 2008;24:1386–1393. doi: 10.1093/bioinformatics/btn178. [DOI] [PubMed] [Google Scholar]
- 10.Carmona-Saez P, Chagoyen M, Tirado F, Carazo J, Pascual-Montano A. GENECODIS: a web-based tool for finding significant concurrent annotations in gene lists. Genome Biol. 2007;8:R3. doi: 10.1186/gb-2007-8-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nogales-Cadenas R, Carmona-Saez P, Vazquez M, Vicente C, Yang X, Tirado F, Carazo J, Pascual-Montano A. GeneCodis: interpreting gene lists through enrichment analysis and integration of diverse biological information. Nucleic Acids Res. 2009;37(Suppl. 2):W317–W322. doi: 10.1093/nar/gkp416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Agrawal R, Srikant R. Fast algorithms for mining association rules. In: Bocca J, Jarke M, Zaniolo C, editors. VLDB'94, Proceedings of 20th International Conference on Very Large Data Bases. Chile: Morgan Kaufmann, Santiago de Chile; 1994. pp. 487–499. [Google Scholar]
- 13.Alexa A, Rahnenführer J, Lengauer T. Improved scoring of functional groups from gene expression data by decorrelating GO graph structure. Bioinformatics. 2006;22:1600–1607. doi: 10.1093/bioinformatics/btl140. [DOI] [PubMed] [Google Scholar]
- 14.Gruca A. 2009. Characterization of gene groups using decision rules (in Polish). Ph.D. Thesis. Silesian University of Technology Gliwice, Poland. [Google Scholar]
- 15.Sikora M, Gruca A. Quality improvement of rules based gene groups descriptions using information about GO terms importance occurring in premises of determined rules. Int. J. Appl. Math. Comput. Sci. 2010;20:555–570. [Google Scholar]
- 16.Sikora M, Gruca A. Induction and selection of the most interesting Gene Ontology based multiattribute rules for descriptions of gene groups. Pattern Recognit. Lett. 2011;32:258–269. [Google Scholar]
- 17.Stefanowski J, Vanderpooten D. Induction of decision rules in classification and discovery-oriented perspectives. Int. J. Intell. Syst. 2001;16:13–27. [Google Scholar]
- 18.Benjamini Y, Hochberg T. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. Roy. Stat. Soc. Ser. B. 1995;57:289–300. [Google Scholar]
- 19.Medina I, Carbonell J, Pulido L, Madeira S, Goetz S, Conesa A, Tarraga J, Pascual-Montano A, Nogales-Cadenas R, Santoyo J, et al. Babelomics: an integrative platform for the analysis of transcriptomics, proteomics and genomic data with advanced functional profiling. Nucleic Acids Res. 2010;38(Suppl. 2):W210–W213. doi: 10.1093/nar/gkq388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith J, Marelli M, Christmas R, Vizeacoumar F, Dilworth D, Ideker T, Galitski T, Dimitrov K, Rachubinski R, Aitchison J. Transcriptome profiling to identify genes involved in peroxisome assembly and function. J. Cell Biol. 2002;158:259–271. doi: 10.1083/jcb.200204059. [DOI] [PMC free article] [PubMed] [Google Scholar]