Abstract
The Bioinformatics Links Directory continues its collaboration with Nucleic Acids Research to collaboratively publish and compile a freely accessible, online collection of tools, databases and resource materials for bioinformatics and molecular biology research. The July 2011 Web Server issue of Nucleic Acids Research adds an additional 78 web server tools and 14 updates to the directory at http://bioinformatics.ca/links_directory/.
To keep abreast of the growing needs of the bioinformatics community (both developers and users) and to facilitate communication of bioinformatics resources, tools and databases, the Bioinformatics Links Directory has undergone a major redevelopment. Existing and new links within the directory are now characterized as resources, tools or databases. With the release of the Nucleic Acids Research 2011 Web Server issue, there are now 144 resources, 480 databases and 1250 web server tools. Link owners now also have the opportunity to add useful content such as additional references, documentation, use-case examples and related announcements to their link page. With these and other new features, the Bioinformatics Links Directory is empowering the bioinformatics community to drive the continual development of this compendium of important bioinformatics resources, tools and databases to aid research.
COMMENTARY
The Bioinformatics Links Directory's ongoing partnership with Nucleic Acids Research herein adds the 2011 collection of web servers to its comprehensive public repository (1–6). The 2011 NAR Web Server issue describes the latest web servers and open access bioinformatic tools available online to guide and enable research in any number of life science domains. The complete listing of URLs cited in the 2011 Web Server issue can be accessed online at the Nucleic Acids Research web site, http://nar.oxfordjournals.org/, as well as at http://bioinformatics.ca/links_directory/narweb2011/.
The Bioinformatics Links Directory (http://bioinformatics.ca/links_directory/) is an open access and community-driven repository of bioinformatics links. Recent upgrades to the Bioinformatics Links Directory have reorganized all existing and new links into either: (i) Resource: a static resource whose intention is to convey bioinformatic information; (ii) Tool: a bioinformatic web server or downloadable software tool that can query, analyze, extract or modify input data; and (iii) Database: a biological data store that can be queried. The Bioinformatics Links Directory has also begun incorporating all of the bioinformatics databases released by the NAR Database issues (7,8), and is up to date with the 2010 and 2011 content. With this reclassification and added content, the Bioinformatics Links Directory now contains 144 resources, 480 databases and 1250 web server tools (Table 1).
Table 1.
Summary of the number of resources, databases and web server tools listed in each subcategory of the Bioinformatics Links Directory, July 2011
| Resources | Databases | Tools | Total | |
|---|---|---|---|---|
| Computer related | ||||
| Bio-* Programming Tools | 6 | 0 | 12 | 18 |
| C/C++ | 1 | 0 | 1 | 2 |
| Databases | 0 | 1 | 5 | 6 |
| Java | 3 | 0 | 1 | 4 |
| Linux/Unix | 5 | 1 | 4 | 10 |
| PERL | 4 | 0 | 1 | 5 |
| PHP | 0 | 0 | 1 | 1 |
| Statistics | 2 | 0 | 7 | 9 |
| Web Development | 2 | 0 | 0 | 2 |
| Web Services | 3 | 0 | 16 | 19 |
| Workflows | 1 | 0 | 5 | 6 |
| DNA | ||||
| Annotations | 1 | 7 | 65 | 73 |
| Databases | 0 | 56 | 0 | 56 |
| DNA and Genomic Analysis | 5 | 1 | 22 | 28 |
| Gene Prediction | 1 | 1 | 35 | 37 |
| Mapping and Assembly | 3 | 2 | 16 | 21 |
| Phylogeny Reconstruction | 4 | 4 | 45 | 53 |
| Sequence Polymorphisms | 1 | 8 | 39 | 48 |
| Sequence Retrieval and Submission | 1 | 6 | 23 | 30 |
| Structure and Sequence Feature Detection | 3 | 10 | 150 | 163 |
| Tools For the Bench | 2 | 7 | 67 | 76 |
| Education | ||||
| Bioinformatics-related News Sources | 9 | 0 | 0 | 9 |
| Community | 17 | 0 | 2 | 19 |
| Courses, Programs and Workshops | 3 | 0 | 0 | 3 |
| Directories and Portals | 12 | 0 | 6 | 18 |
| General | 13 | 1 | 0 | 14 |
| Tutorials and Directed Learning Resources | 10 | 0 | 0 | 10 |
| Expression | ||||
| cDNA, EST, SAGE | 1 | 10 | 38 | 49 |
| Databases | 0 | 21 | 0 | 21 |
| Gene Regulation | 3 | 16 | 127 | 146 |
| Gene Set Analysis | 0 | 1 | 29 | 30 |
| Networks | 0 | 2 | 15 | 17 |
| Protein Expression | 1 | 3 | 18 | 22 |
| Splicing | 0 | 3 | 17 | 20 |
| Transcript Expression & Microarrays | 5 | 10 | 107 | 122 |
| Human Genome | ||||
| Annotations | 2 | 5 | 39 | 46 |
| Databases | 0 | 31 | 1 | 32 |
| Ethics | 5 | 1 | 0 | 6 |
| Genomics | 3 | 2 | 15 | 20 |
| Health and Disease | 1 | 7 | 23 | 31 |
| Other Resources | 9 | 4 | 17 | 30 |
| Sequence Polymorphisms | 2 | 4 | 43 | 49 |
| Literature | ||||
| Databases | 0 | 7 | 0 | 7 |
| Goldmines | 1 | 4 | 0 | 5 |
| Open Access Resources | 1 | 3 | 0 | 4 |
| Search Tools | 0 | 6 | 8 | 14 |
| Text Mining and Semantics | 0 | 4 | 30 | 34 |
| Model Organisms | ||||
| Databases | 0 | 95 | 0 | 95 |
| Fish | 0 | 5 | 6 | 11 |
| Fly | 2 | 8 | 13 | 23 |
| General Resources | 3 | 12 | 16 | 31 |
| Microbes | 3 | 12 | 49 | 64 |
| Mouse and Rat | 4 | 17 | 21 | 42 |
| Other Organisms | 3 | 6 | 13 | 22 |
| Other Vertebrates | 1 | 3 | 7 | 11 |
| Plants | 2 | 10 | 18 | 30 |
| Worm | 1 | 4 | 6 | 11 |
| Yeast | 2 | 9 | 10 | 21 |
| Other Molecules | ||||
| Carbohydrates | 0 | 0 | 7 | 7 |
| Compounds | 0 | 1 | 14 | 15 |
| Databases | 0 | 26 | 0 | 26 |
| Metabolites | 0 | 1 | 12 | 13 |
| Small Molecules | 0 | 2 | 13 | 15 |
| Protein | ||||
| 2D Structure Prediction | 1 | 0 | 65 | 66 |
| 3D Structural Features | 1 | 2 | 103 | 106 |
| 3D Structure Comparison | 1 | 6 | 68 | 75 |
| 3D Structure Prediction | 4 | 1 | 85 | 90 |
| 3D Structure Retrieval/Viewing | 4 | 7 | 47 | 58 |
| Annotation and Function | 0 | 5 | 57 | 62 |
| Biochemical Features | 2 | 2 | 43 | 47 |
| Databases | 0 | 116 | 1 | 117 |
| Do-it-all Tools for Protein | 3 | 0 | 12 | 15 |
| Domains and Motifs | 0 | 14 | 115 | 129 |
| Interactions, Pathways, Enzymes | 3 | 21 | 113 | 137 |
| Localization and Targeting | 0 | 3 | 39 | 42 |
| Molecular Dynamics and Docking | 1 | 0 | 44 | 45 |
| Phylogeny Reconstruction | 4 | 5 | 47 | 56 |
| Presentation and Format | 1 | 0 | 13 | 14 |
| Protein Expression | 1 | 2 | 7 | 10 |
| Proteomics | 3 | 4 | 32 | 39 |
| Sequence Comparison | 0 | 0 | 18 | 18 |
| Sequence Data | 1 | 6 | 3 | 10 |
| Sequence Features | 2 | 2 | 46 | 50 |
| Sequence Retrieval | 1 | 6 | 21 | 28 |
| RNA | ||||
| Databases | 0 | 24 | 1 | 25 |
| Functional RNAs | 0 | 5 | 39 | 44 |
| General Resources | 4 | 3 | 4 | 11 |
| Motifs | 0 | 1 | 26 | 27 |
| Sequence Retrieval | 0 | 2 | 7 | 9 |
| Structure Prediction, Visualization and Design | 0 | 3 | 65 | 68 |
| Sequence Comparison | ||||
| Alignment Editing and Visualization | 0 | 0 | 25 | 25 |
| Analysis of Aligned Sequences | 0 | 0 | 65 | 65 |
| Comparative Genomics | 2 | 7 | 40 | 49 |
| Multiple Sequence Alignments | 1 | 0 | 67 | 68 |
| Other Alignment Tools | 0 | 0 | 13 | 13 |
| Pairwise Sequence Alignments | 1 | 0 | 34 | 35 |
| Similarity Searching | 1 | 2 | 49 | 52 |
| Total Resources, Databases and Tools in Bioinformatics Links Directory | 144 | 480 | 1250 | 1874 |
Within each link type, links remain organized by biological subject such as ‘DNA’, ‘RNA’, ‘Protein’, ‘Expression’ and ‘Sequence Comparison’, etc., with subcategories of common tasks relevant to each subject listed. Table 1 summarizes the links available within each of these categories and subcategories according to the Bioinformatics Link Directory reclassification schema.
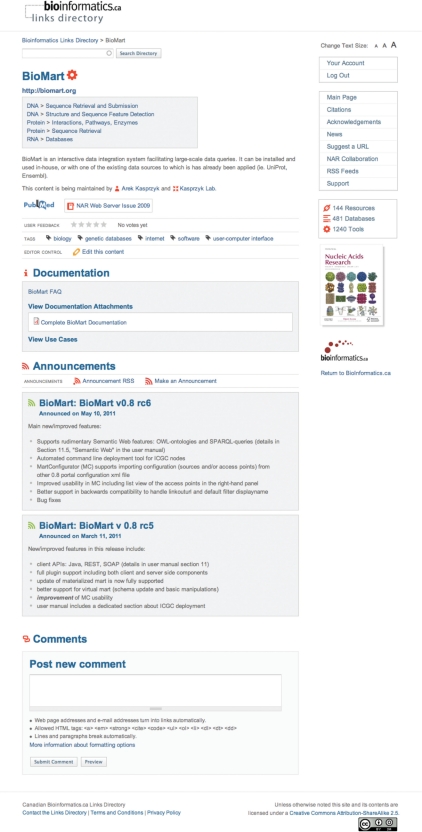
Since most links within the Bioinformatics Links Directory have an associated publication, keyword tags obtained from the link's PubMed MeSH terms have been added (Figure 1). Keyword tags may also be added independently of PubMed MeSH terms. Tags describe the contents of a link and can describe the contents in a more versatile manner than the directory structure, so that subtle context about a link's functionality can be gleaned. Tags facilitate the linkage of similar tools beyond the Directory structure so that users can quickly locate bioinformatic links with similar function or which handle similar datatypes, etc.
Figure 1.
Example management page for a link in the Bioinformatics Links Directory. Link owners can add keyword tags, PubMed citations, documentation, use-case examples and announcements to their link page. Users can post comments and feedback about a link for public review.
Entries in the Bioinformatics Links Directory also contain a short description of the link's function, as well as the accompanying PubMed citation and web server URL (Figure 1), all searchable and hyperlinked. As a community-driven repository, anyone in the research or bioinformatics community may suggest a link through links@bioinformatics.ca or through the ‘Suggest a URL’ page on the Bioinformatics Links Directory. To keep pace with the growing needs of both the developers and users of the Bioinformatics Links Directory, and to facilitate link communication, the recent Directory upgrades also provide a number of features to empower the link owners. Previously, links were suggested by users, and entered and maintained by the Directory curators. Link owners now have the capacity to edit their own link's short description, to add PubMed citations and keyword tags, and more importantly to upload supporting user documentation and use-case examples, and make link announcements (Figure 1). In this manner, link owners have the opportunity to educate users of their link and enhance their link's usability with the addition of supporting read-me documentation that contains useful tips on how to navigate within and use a given tool, as well as the addition of brief tutorials or use-case examples to show potential users how to perform simple to advanced analyses with the tool or how to use a tool for a given research problem. The announcement feature further facilitates communication about a link by allowing the link owner to post version updates, new feature or other link announcements for the wider user community to see. If a user desires, communication of a particular link's announcements may also be communicated directly through subscription to that link's RSS feed.
In addition to being able to search and locate a list of potential tools applicable to a given problem, users of the Bioinformatics Links Directory now also have the option to review a tool's feedback. Registered users may post comments and rate a tool's usefulness for other users to review (Figure 1). Such ratings and commentary allow users to rapidly narrow down which tool will be most applicable to their research problem.
In summary, the Bioinformatics Links Directory has been enhanced with more resources, tools and databases from Nucleic Acids Research publications, and by a major upgrade to provide more features for communicating on bioinformatics links. With these new additions, it is hoped that the Bioinformatics Links Directory will continue to empower the community and grow the use of bioinformatics in research.
FUNDING
This work was conducted with the support of the Ontario Institute for Cancer Research through funding provided by the Government of Ontario. The open access publication charge for this paper has been waived by Oxford University Press in recognition of their work on behalf of the journal.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The Bioinformatics Links Directory is a community resource built on a commitment to the spirit of open access under the Attribution-Share Alike 2.5 Canada (CC BY-SA 2.5) license. The authors wish to acknowledge the efforts of Nucleic Acids Research and the researchers and developers worldwide who invest considerable effort into ensuring that their research is freely accessible to all. In particular, the authors would like to acknowledge all of the contributors to the Bioinformatics Links Directory for their valuable input and suggestions for improvements to the directory; these individuals are listed on the Acknowledgements page at http://bioinformatics.ca/links_directory/acknowledgements/.
REFERENCES
- 1.Brazas MD, Fox JA, Brown T, McMillan S, Ouellette BF. Keeping pace with the data: 2008 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2008;36:W2–W4. doi: 10.1093/nar/gkn399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Brazas MD, Yamada JT, Ouellette BF. Evolution in bioinformatic resources: 2009 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2009;37:W3–W5. doi: 10.1093/nar/gkp531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brazas MD, Yamada JT, Ouellette BF. Providing web servers and training in Bioinformatics: 2010 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2010;38:W3–W6. doi: 10.1093/nar/gkq553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fox JA, Butland SL, McMillan S, Campbell G, Ouellette BF. The Bioinformatics Links Directory: a compilation of molecular biology web servers. Nucleic Acids Res. 2005;33:W3–W24. doi: 10.1093/nar/gki594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fox JA, McMillan S, Ouellette BF. A compilation of molecular biology web servers: 2006 update on the Bioinformatics Links Directory. Nucleic Acids Res. 2006;34:W3–W5. doi: 10.1093/nar/gkl379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fox JA, McMillan S, Ouellette BF. Conducting research on the web: 2007 update for the bioinformatics links directory. Nucleic Acids Res. 2007;35:W3–W5. doi: 10.1093/nar/gkm459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cochrane GR, Galperin MY. The 2010 Nucleic Acids Research Database Issue and online Database Collection: a community of data resources. Nucleic Acids Res. 2010;38:D1–D4. doi: 10.1093/nar/gkp1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Galperin MY, Cochrane GR. The 2011 Nucleic Acids Research Database Issue and the online Molecular Biology Database Collection. Nucleic Acids Res. 2011;39:D1–D6. doi: 10.1093/nar/gkq1243. [DOI] [PMC free article] [PubMed] [Google Scholar]