Abstract
Mycobacterium avium complex (MAC) within macrophages undergoes a phenotype change that allows for more efficient entry into surrounding host cells. We hypothesized that, by developing an in vitro system resembling the intravacuolar environment, one could generate insights into the mycobacterial intracellular phenotype. MAC was incubated in “elemental mixtures” that reproduce metal concentrations and pH in the vacuoles at different time points and then used to infect fresh macrophages. Incubation of MAC with the mixture corresponding to the vacuole environment 24 h postinfection infected macrophages at a significantly higher rate than bacteria that were incubated in Middlebrook 7H9 broth. Uptake occurred by macropinocytosis, similar to the uptake of bacteria passed through macrophages. Genes reported to be upregulated in intracellular bacteria, such as Mav1365, Mav2409, Mav4487, and Mav0996, were upregulated in MAC incubated in the 24-h elemental mixture. Like MAC obtained from macrophages, the vacuoles of bacteria from the 24-h elemental mixture were more likely to contain lysosome-associated membrane protein 1 (LAMP-1). A stepwise reduction scheme of the 24-h elemental mixture indicated that incubation in physiologically relevant concentrations of potassium chloride, calcium chloride, and manganese chloride was sufficient to induce characteristics of the intracellular phenotype. It was demonstrated that bacteria harboring the intracellular phenotype induced early-onset macrophage death more efficiently than bacteria grown in broth. This new trace elemental mixture mimicking the condition of the vacuole at different time points has the potential to become an effective laboratory tool for the study of the MAC and Mycobacterium tuberculosis disease process, increasing the understanding of the interaction with macrophages.
INTRODUCTION
Pathogenic mycobacteria, such as Mycobacterium avium complex (MAC) and Mycobacterium tuberculosis, are a serious concern for immunocompromised individuals, such as AIDS patients. MAC is likely acquired via either the gastrointestinal or respiratory tract, frequently causing disseminated disease. In the host, the bacteria live and replicate in vacuoles within macrophages and exit phagosomes and eventually phagocytic cells, probably as a mechanism of dissemination (unpublished data). The mechanism(s) by which MAC moves from one macrophage to another has only begun to be elucidated.
Prior studies suggest that MAC senses and reacts to the environment within the macrophage and acquires a new, “intracellular phenotype” that appears to play a role in its ability to spread in the host (4). Specifically, MAC that has been inside host cells infects macrophages more efficiently than bacteria obtained from Middlebrook 7H9 broth and is more resistant to killing/inhibition of growth by macrophages stimulated with tumor necrosis factor alpha (TNF-α) (3). In addition, MAC from macrophages is more likely to use transferrin and β-1 integrin receptors and less likely to use complement receptor 3 than MAC from Middlebrook 7H9 broth (3) to gain access to phagocytic cells. Similarly, the vacuoles containing MAC passed in other macrophages have a lower pH and are more likely to contain lysosome-associated membrane protein 1 (LAMP-1) than vacuoles occupied by MAC from Middlebrook 7H9 broth (4). It was also observed that MAC enters the first macrophage by phagocytosis, while it sometimes uses macropinocytosis upon ingestion by subsequent macrophages (4). Studies in vivo support this observation (3).
Recent studies have indicated that MAC actively impacts the environment of the vacuole in macrophages by controlling the amount of single elements, such as iron (26, 45). Several other bacteria, such as Salmonella and Pseudomonas aeruginosa, are known to sense changes in metal concentrations, which lead to the regulation of the expression of a number of virulence determinants (2, 13). We therefore hypothesized that metals may be key factors in the environment of the macrophage that participate in the regulation of mycobacterial virulence genes. The elemental composition of the MAC vacuole was recently determined by hard X-ray microscopy at time points 1 h and 24 h after infection (44), while other work determined that the vacuolar pH is 5.8 to 6.1 (32). We used these parameters to model the condition of vacuoles that contain MAC in order to study the intracellular bacterial phenotype in a test tube. Mimicking the host environment has been attempted with other bacterial species, such as Pseudomonas (46) and Yersinia pestis (12, 37), and can become a powerful laboratory tool.
MATERIALS AND METHODS
Medium recipes.
Five hundred milliliters of Middlebrook 7H9 broth (Difco, Becton-Dickinson, Sparks, MD) was prepared as usual, and then the amounts of the chemicals or supplements listed in Table 1 were added to make a mixture that mimicked the vacuole 1 h after infection (1-h elemental mixture) and a mixture matching the vacuole 1 day after infection (24-h elemental mixture). All chemicals or supplements were from Sigma (St. Louis, MO), except for CaCl2, which was from Mallinckrodt (St. Louis, MO). The pH was adjusted to 6.6 for the 1-h mixture and 5.8 for the 24-h mixture. Both mixtures were then autoclaved and stored at 4°C until use.
Table 1.
Recipes for 1-h and 24-h elemental mixturesa
| Supplement | Amt (ml or μl) of supplement added to make: |
|
|---|---|---|
| 1-h elemental mixture | 24-h elemental mixture (mM concn) | |
| 1 M potassium chloride | 14.7 ml | 0.925 ml (1.79) |
| 1 M calcium chloride | 2 ml | 1.25 ml (2.42) |
| 1 M manganese chloride | 5.9 ml | 11.9 ml (23) |
| 1 M copper sulfate | 1.85 μl | 5.5 μl (0.01) |
| 1 M zinc chloride | 33 μl | 58.7 μl (0.11) |
| 0.25 M ferric pyrophosphate | 288 μl | 2 ml (0.97) |
| 1 M nickel chloride | 5 μl | 5 μl (0.01) |
The amounts of the elements listed were added to 500 ml of Middlebrook 7H9 broth to make a mixture that mimics the environment of the vacuole 1 h postinfection (1-h elemental mixture) or to make the mixture matching the vacuole 1 day after infection (24-h elemental mixture).
Infection assays.
Mycobacterium avium complex (MAC) strain 104 was obtained from blood from an AIDS patient, grown in Middlebrook 7H9 broth supplemented with oleic acid, albumin, dextrose, and catalase (OADC) (Hardy Diagnostics, Santa Maria, CA) for 5 days at 37°C with shaking, and then diluted to 3 × 108 bacteria/ml using McFarland standards. The bacteria were diluted 1:10 in Middlebrook 7H9 broth (lacking OADC) or in the indicated mixture and then incubated at 37°C with shaking for 1 h or 24 h. These samples were briefly vortexed, passed through a 24-gauge needle, and allowed to sediment for 5 min. Only the top half of the suspension was used as inoculum. The suspension was observed by microscopy and determined to consist of disperse bacteria. An aliquot was used to plate for CFU to determine the bacterial concentration in the inoculum. Another aliquot was used to infect Raw 264.7 (ATCC, Manassas, VA) macrophage monolayers. Bacterial viability of the inoculum was also observed by using the Live-Dead cell viability assay, as previously reported (3). Raw 264.7 cells were seeded overnight in Dulbecco's modified Eagle medium (DMEM) (Gibco, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS) (Gemini, Woodland, CA) in a 24-well plate (Costar, Corning, NY) and then infected at an multiplicity of infection (MOI) of ∼10 for 1 h. The macrophage monolayers were washed three times with Hanks' balanced salt solution (HBSS) (Gibco, Carlsbad, CA) to remove extracellular bacteria, which has been shown to remove 99.9% of the extracellular bacteria (5), lysed with sterile water, and then plated on Middlebrook 7H10 agar (Difco, Becton-Dickinson, Sparks, MD) with OADC to determine the number of CFU. This assay was repeated three times, with two wells per sample each time.
Infection control.
Raw 264.7 cells were treated with the elemental mixtures for 1 h and then infected with MAC from Middlebrook 7H9 broth at an MOI of 10. Raw 264.7 macrophage monolayers were incubated with the same concentration of 1-h mixture, 24-h mixture, and Middlebrook 7H9 broth as was used in the assay described above (1/10 the total volume) for 1 h, and one sample was incubated with DMEM as a control. The monolayers were washed three times with HBSS, and then fresh DMEM was added. Monolayers were then infected with MAC that was grown in Middlebrook 7H9 broth with OADC for 5 days at 37°C with shaking at an MOI of ∼10 for 1 h. Bacteria were harvested and plated onto Middlebrook 7H10 agar to determine the number of CFU. The assay was performed three times, with two wells per sample each time.
Real-time PCR.
MAC was grown in Middlebrook 7H9 broth supplemented with OADC for 5 days and then exposed to the elemental mixture for 24 h. RNA was extracted from the bacteria using phenol-chloroform according to previously published methods (10). The RNA was treated with DNase, and first-strand cDNA was synthesized using random hexamer primers and the SuperScript III cDNA synthesis kit according to the manufacturer's instructions (Invitrogen, Carlsbad, CA). Primers for genes previously described to be upregulated in macrophages (Table 2) were used for real-time PCR using SYBR green master mix (Bio-Rad, Hercules, CA) with a PCR protocol of 10 min at 95°C, followed by 40 cycles, with 1 cycle consisting of 1 min at 95°C, 1 min at 60°C, and 2 min at 72°C. The threshold cycle (CT) was determined as previously described, and fold change values were calculated using the ΔΔCT method (40). For real-time PCR with the 24-h elemental mixture, Mav4781 was used as a negative control (i.e., a gene that would not show any change in expression), since its expression does not appear to be influenced by the intracellular environment (10). Mav2036 was a positive control, since it has previously been shown to be upregulated in higher zinc concentrations (30). For all other real-time studies, Mav4357 was used as a negative control, since its expression level was not influenced by the 24-h elemental mixture. Data are from six real-time reactions, based on three biological samples.
Table 2.
Primers used for real-time PCR in elemental mixturea
| MAC strain 104 gene | M. tuberculosis H37Rv gene | Primer sequence | Reference |
|---|---|---|---|
| Mav1094 | Rv0981 | 5′CTGGAAATGCTGATCGCCAACC3′ | He et al. (18) |
| 5′TCACGGCGGCGTCTCCCGAAGCACGTA3′ | |||
| Mav3301 | Rv1477 | 5′ATGAGACGCACACGCTGG3′ | Gao et al. (15a) |
| 5′CTTGTTGACGCTTTCCTG3′ | |||
| Mav4487 | Rv1818c | 5′ATGTCGTTCGTTCAAGCGACTC3′ | Brennan et al. (8) |
| 5′GTTCATCAACTGCACGAACTGG3′ | |||
| Mav1365 | Rv1221 | 5′GATCTCCTCATACGACAG3′ | Manganelli et al. (28) |
| 5′GACCTTCATCCGGGTGTTC3′ | |||
| Mav5014 | Rv0170 | 5′CAGACTCAGCATCTTCTC3′ | Haile et al. (16a) |
| 5′GTCGACCAGCTGCACCGAAC3′ | |||
| Mav3270 | Rv1592 | 5′ATGGCAACCGAACTCCGAAAG3′ | Danelishvili et al. (11) |
| 5′GAAAACGAAACGACGGGTCGT3′ | |||
| Mav2420 | Rv3902 | 5′CACTTGGTGGAGGTGCACCCGACTC3′ | Danelishvili et al. (11) |
| 5′GGACGTGATGTTTCCGCTTATTC3′ | |||
| Mav1600 | 5′ATGTCCGACACCACAACAG3′ | Plum et al. (32a) | |
| 5′CTTCGACGAATTCGTTCCC3′ | |||
| Mav0996 | Rv0867 | 5′CACCACTTCCAGTGTCAG3′ | Hou et al. (19) |
| 5′GTGTTGATGCCCCAGTTG3′ | |||
| Mav0660 | Rv3496 | 5′GACTGTTTGACCTCGTCCCAC3′ | Hou et al. (19) |
| 5′GTGTATGTGCTGACGTCGCAAG3′ | |||
| Mav2409 | Rv2096c | 5′GTGGGCAGTCTTAGTCCGTG3′ | Hou et al. (19) |
| 5′CTTGAAGATCCGCATCGACTT3′ | |||
| Mav1424 | Rv1275 | 5′CTGACGGGCTGTTCCCGCTCG3′ | Danelishvili et al. (10) |
| 5′CTAGGAATTGGTCGCCAGCGT3′ | |||
| Mav0467 | Rv3663 | 5′ATGACCACCCCGCTGCTGTCG3′ | Danelishvili et al. (10) |
| 5′GCCACCTGGTCGACGAGGTCG3′ | |||
| Mav3275 | Rv1497 | 5′GCTGGAGCTGGCCGA3′ | Danelishvili et al. (10) |
| 5′CGGCACGTACAGGTTTCGGTC3′ | |||
| Mav2928 | Rv1787 | 5′ACGAATTCACGGTGTTTGACTTCGGAGCG3′ | Li et al. (26a) |
| 5′ACAAGCTTGTGGACTCTCGGTCGTGTTGAGG3′ | |||
| Mav4781 | Rv0359 | 5′CGGCTGGCTGGTGTCGCTGTG3′ | Danelishvili et al. (10) |
| 5′ACGCCGGAGAAGTCGAAC3′ | |||
| Mav2036 | Rv2359 | 5′CGACGGGGCAGGAGTCAG3′ | Milano et al. (30) |
| 5′ATGGTGTGGCTGACGTCGGAGAA3′ | |||
| Mav4357 | Rv3412 | 5′GACCACCTCCCACCGGGTTT3′ | Danelishvili et al. (10) |
| 5′TTGAGTGAGGCGTCGGCGTAG3′ |
Primers used for real-time PCR in the 24-h elemental mixture, along with the nearest M. tuberculosis homolog and reference in which the genes have been proven to be upregulated when Mycobacterium avium complex (MAC) is inside cells.
Macropinocytosis.
Raw 264.7 macrophages were infected as described above, except that after exposure to the 24-h elemental mixture or Middlebrook 7H9 broth, bacteria were stained by incubating ∼3 × 109 MAC with 100 μg/ml of 5 (and 6)-carboxytetramethylrhodamine succinimidyl ester (rhodamine) (Molecular Probes, Eugene, OR) for 1 h in the dark. The bacteria were subsequently diluted in HBSS before infection of macrophages in chamber slides (Nalge Nunc, Rochester, NY). The residual rhodamine was left in the presence of the bacteria during the infection to allow for visualization of the size of the compartment occupied by the bacteria. The slides were fixed in 2% paraformaldehyde (Sigma-Aldrich, St. Louis, MO) for 30 min and then visualized by using a Leica DM400B fluorescence microscope by counting 300 macrophages in each experiment.
Trafficking.
MAC was incubated in Middlebrook 7H9 broth or in the 24-h elemental mixture for 24 h, and then a 1-mg/ml 5 (and 6)-carboxyfluorescein, succinimidyl ester (Molecular Probes, Eugene, OR) solution in HBSS was added to the MAC for 1 h in the dark. Excess dye was removed by washing the cells three times with centrifugation using HBSS. The stained bacteria were used to infect Raw 264.7 cells on 2-well chamber slides at an MOI of 10 for 1 h, and extracellular MAC was killed for 2 h with 200 μg/ml amikacin (Sigma) and then washed away three times with HBSS. The cells were fixed with 2% paraformaldehyde in HBSS for 60 min in the dark at room temperature, washed with HBSS, and then permeabilized with 0.1% Triton X-100 (J. T. Baker, Phillipsburg, NJ) at room temperature for 5 min. Next, they were blocked for 1 h with 10% bovine serum albumin (Sigma-Aldrich, St. Louis, MO) in HBSS, washed with HBSS, exposed to the appropriate primary antibody for 1 h, washed again, and treated with Texas Red-conjugated secondary antibody. These cells were analyzed by fluorescence microscopy for colocalization of the stained bacteria with the vacuole marker, with about 40 stained bacteria per treatment group analyzed during each repetition. For primary antibodies, we used rabbit anti-Rab5a (Santa Cruz Biotechnology, Santa Cruz, CA), rabbit anti-Rab7 (Santa Cruz Biotechnology, Santa Cruz, CA), goat anti-early endosomal antigen 1 (anti-EEA-1) (Santa Cruz Biotechnology, Santa Cruz, CA), and mouse anti-LAMP-1 (BD Biosciences, San Jose, CA). The secondary antibodies were all Texas Red conjugates: Texas Red-conjugated donkey anti-rabbit IgG (Amersham Biosciences, NJ), Texas Red-conjugated mouse anti-goat IgG (Santa Cruz Biotechnology, Santa Cruz, CA), and Texas Red-conjugated sheep anti-mouse IgG (Amersham Biosciences, NJ).
Live-Dead cell viability assay.
Raw 264.7 cells were seeded on 2-well chamber slides and allowed to attach overnight. MAC was incubated in Middlebrook 7H9 broth without OADC or the 24-h elemental mixture for 24 h and then used to infect Raw 264.7 cells at an MOI of 1,000 or 100 for bacteria from Middlebrook 7H9 broth or at an MOI of 100 or 10 for bacteria from the elemental mixture to attain roughly the same number of intracellular bacteria. The large number of bacteria was chosen to ensure that a great number of cells in the monolayer were infected. After a 1-h infection, the monolayers were washed three times with HBSS for removal of extracellular bacteria, and the cell culture medium was replenished. Two hours later, the Raw 264.7 cells were analyzed by Live-Dead cell viability assay kit (Molecular Probes, Eugene, OR) using fluorescence microscopy, according to the manufacturer's instructions. Three hundred cells were counted in each sample per repetition. For bacteria that had been passed through Raw 264.7 cells, Raw 264.7 cell monolayers in 6-well plates were infected at an MOI of 100 for 1 h, the monolayers were washed, the medium was replenished, and 24 h later, the monolayers were lysed for 10 min in 0.25% SDS at room temperature. The cellular debris was centrifuged for 5 min at 500 × g, the supernatant was collected, and then bacteria were pelleted by centrifugation at 12,000 × g for 1 min. Pelleted bacteria were resuspended in HBSS and used to infect fresh monolayers of Raw 264.7 cells seeded in chamber slides at an MOI of 50. Infection with MAC from Middlebrook 7H9 broth was performed using an MOI of 500. For a control, uninfected Raw 264.7 cells were also lysed and centrifuged twice as described above, and the resuspended lysate was used to inoculate fresh macrophage monolayers.
TUNEL assay.
The terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) assay was carried out similarly to the Live-Dead assay, except that 8-well chamber slides were used instead of 2-well chamber slides, and the fluorescein in situ cell death detection kit (Roche, Indianapolis, IN) was used for analysis.
Statistical analysis.
The two-sided Student t test was performed comparing the experimental data to the negative-control values from three biological repetitions using Microsoft Excel software, with a level of significance cutoff value of P value < 0.05 or P value < 0.10 as indicated in the figures.
RESULTS
Bacterial growth.
To determine whether incubation in the presence of the elemental mixture was associated with different growth or clumping of Mycobacterium avium complex (MAC), bacteria were incubated in Middlebrook 7H9 broth supplemented with oleic acid, albumin, dextrose, and catalase (OADC), or elemental mixture for up to 2 days. Table 3 shows that no difference was seen between growth in Middlebrook 7H9 broth or in the elemental mixture.
Table 3.
Growth of MAC under different conditions
| Conditions | Inoculum (time zero) | No. of MAC CFU/ml (mean ± SD) |
|
|---|---|---|---|
| 24 h | 48 h | ||
| Middlebrook 7H9 broth + OADC | 1 × 105 ± 0.6 × 105 | 6.8 × 105 ± 0.5 × 105 | 1.7 × 106 ± 0.4 × 106a |
| 24-h elemental mixture | 1 × 105 ± 0.2 × 105 | 2.8 × 105 ± 0.4 × 105 | 7.3 × 105 ± 0.3 × 105a |
| Raw 264.7 macrophages | 2 × 105 ± 0.4 × 105 | 4.3 × 105 ± 0.5 × 105 | 8.9 × 105 ± 0.3 × 105a |
P < 0.05 compared with the number of bacteria at time zero.
Uptake by Raw 264.7 macrophages.
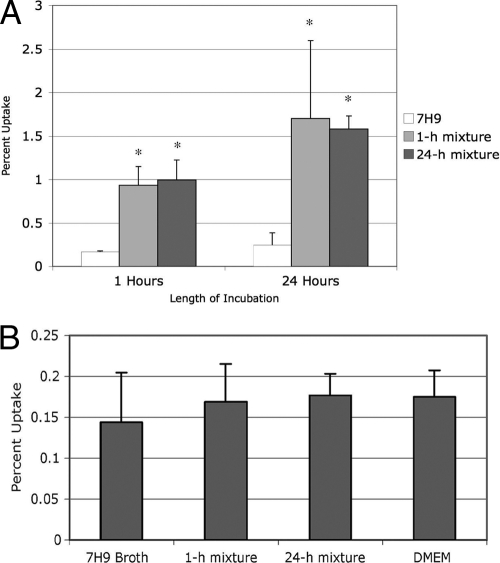
We exposed MAC for either 1 h or 24 h to two solutions that mimicked metal concentrations and pH in the macrophage vacuole 1 h postinfection and 24 h postinfection, which were named the 1-h and 24-h elemental mixture, respectively. We washed the monolayers, which removed 99.8% ± 0.1% of the extracellular bacteria. MAC exposed to an elemental mixture was taken up by Raw 264.7 macrophages approximately 8-fold more efficiently than bacteria that had been grown in Middlebrook 7H9 broth (Fig. 1 A), similar to bacteria that have previously been passed in macrophages (3). To ensure that the difference in uptake by macrophages was not an effect of increased activation of the macrophages secondary to the exposure to metals absorbed on the bacteria, macrophage monolayers were exposed to the 1-h elemental mixture, 24-h elemental mixture, Middlebrook 7H9 broth, or DMEM, washed, and infected with MAC. All bacterial preparations were taken up by macrophages with equal efficiency (Fig. 1B), suggesting that the bacteria exposed to the elemental mixtures undergo a phenotypic change that results in more efficient macrophage uptake. Approximately 78% ± 10% of the macrophages were infected with four to seven bacteria.
Fig. 1.
Uptake of Mycobacterium avium complex (MAC) incubated in the elemental mixture by macrophages. (A) MAC was incubated in Middlebrook 7H9 broth, the 1-h elemental mixture, and the 24-h elemental mixture for 1 h and 24 h, respectively, and then used to infect Raw 264.7 macrophages. The values (indicated by the bars) are the ratios of the number of bacteria inside cells divided by inocula from three repetitions. Bacteria that were incubated in either elemental mixture infected macrophages more efficiently than bacteria grown in Middlebrook 7H9 broth. Values that were significantly different (P < 0.05) from the control value (Middlebrook 7H9 broth) are indicated by an asterisk. (B) Raw 264.7 macrophages were incubated in the indicated solution for 1 h, washed, and then infected with MAC. Incubation of macrophages prior to infection had no effect on the rate of bacterial uptake. The assay was repeated three times.
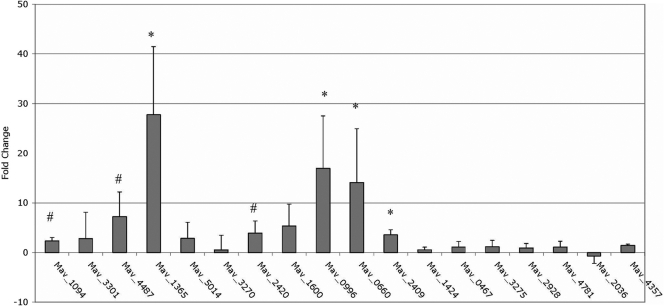
Gene expression by real-time PCR.
Real-time PCR analysis of several bacterial genes that have previously been reported in at least two publications (10, 19, 23, 34) to be upregulated during macrophage infection showed upregulation upon exposure to the 24-h elemental mixture for a 24-h period (Fig. 2). These genes were Mav1094, the homolog to mprA, which is part of a 2-component regulatory system protein that induces sigE expression in Mycobacterium tuberculosis (18), Mav4487, a PE-PGRS family protein with potential role in the interaction with macrophages (8), Mav1365, the homolog to sigE, an alternative sigma factor required for M. tuberculosis virulence in mice (28), Mav2420, a hypothetical protein within a pathogenicity island (11), Mav0996, a gene associated with the resuscitation of nongrowing cells (19), Mav0660, the homolog to mce4D and a member of a family of proteins potentially involved in mammalian cell entry (19), and Mav2409, the homolog to tatA, a gene of the twin-arginine translocase system, which is used to transport folded proteins across the mycobacterial membrane (19, 29).
Fig. 2.
Real-time PCR of MAC strain 104 exposed to the 24-h elemental mixture. Total bacterial RNA was extracted from MAC strain 104 exposed to the 24-h elemental mixture for 24 h and bacteria exposed to Middlebrook 7H9 broth as a control. cDNA was made from the RNA and used for real-time PCR to study expression of genes previously shown to be upregulated by intracellular bacteria or bacteria entering macrophages. The values are means plus standard deviations (error bars) from three repetitions. The real-time PCR results shown in the graph take the medium control into consideration and report the difference from the control value. Values that were significantly different from the control value are indicated as follows: #, P < 0.10; *, P < 0.05.
Macropinocytosis.
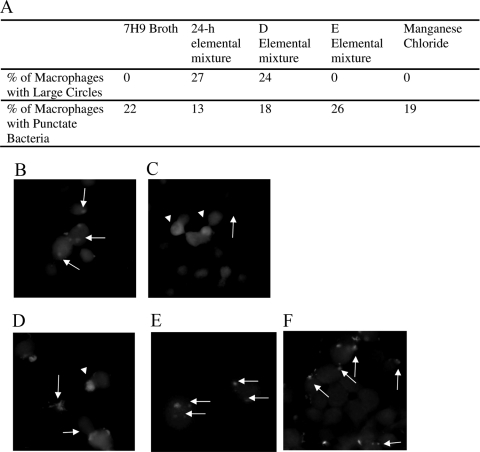
To visualize the compartment macrophages use to take up MAC, rhodamine-labeled MAC was used to infect macrophages in the presence of residual unincorporated rhodamine. MAC that had been exposed to the 24-h elemental mixture showed large fluorescent circles and punctate fluorescent bacteria. In contrast, MAC from Middlebrook 7H9 broth showed only punctate fluorescent bacteria. This suggests that some MAC from the 24-h elemental mixture was taken up in large compartments via macropinocytosis, while bacteria from Middlebrook 7H9 broth entered by phagocytosis, similar to previously described data (using several different approaches) regarding MAC from macrophages versus MAC from broth (Fig. 3 A, first 3 columns, and Fig. 3B and C) (4).
Fig. 3.
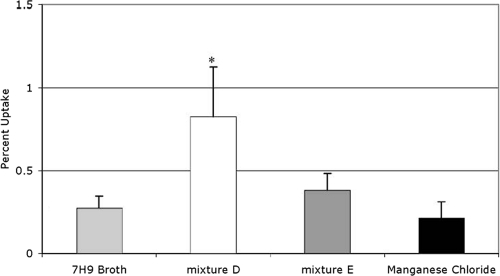
Uptake of MAC by phagocytosis and macropinocytosis. (A) Percentage of total MAC strain 104-infected macrophages containing punctate bacteria or large circles with bacteria inside following bacterial incubation in the indicated solution. (B) Raw 264.7 macrophages were infected by rhodamine-labeled MAC incubated in Middlebrook 7H9 broth prior to infection in the presence of rhodamine. (C) Bacteria incubated in the 24-h elemental mixture before rhodamine staining. (D to F) Bacteria incubated in elemental mixture D (D), elemental mixture E (E), and manganese chloride (F). The experiment was repeated three times. The white arrows point to evidence of bacterial entry by phagocytosis; the white arrowheads point to evidence of macropinocytosis.
Minimal requirements for induction of the intracellular phenotype.
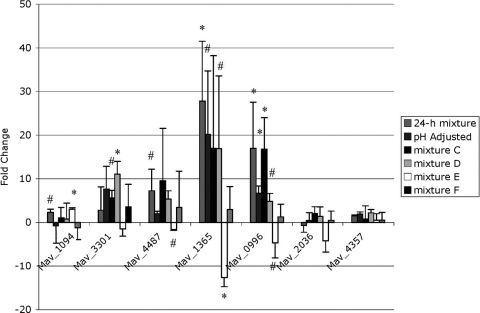
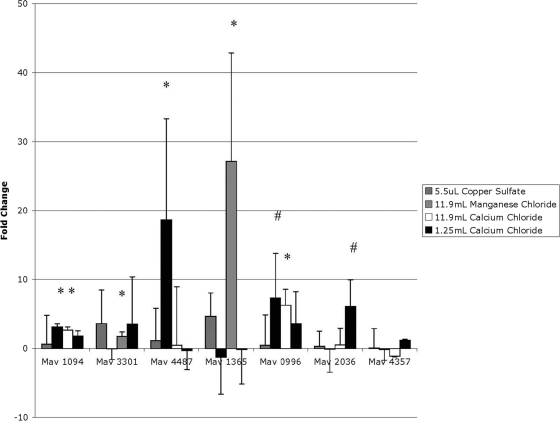
To determine which component(s) of the 24-h elemental mixture induce the MAC intracellular phenotype, we made several versions of our elemental mixture, labeled mixture A through mixture F, each of which was missing one component more than the previous version (Table 4). Mixture A contained all metal components except nickel chloride, while mixture B lacked nickel chloride and ferric pyrophosphate, and so on until mixture F contained only potassium chloride. We then extracted RNA from bacteria incubated in each of these elemental mixtures and carried out real-time PCR to determine which mixture no longer induced gene upregulation. Several of the genes were upregulated similarly to the 24-h elemental mixture until elemental mixture E, which contained only potassium chloride and calcium chloride (Fig. 4). Bacteria incubated in elemental mixture E were not taken up by macrophages any more efficiently than bacteria incubated in Middlebrook 7H9 broth, but bacteria incubated in elemental mixture D, which contained potassium chloride, calcium chloride, and manganese chloride, had a more efficient uptake rate (Fig. 5). Likewise, microscopy images of bacteria from elemental mixture D indicated entry in macrophages by macropinocytosis and phagocytosis, while microscopy of bacteria from elemental mixture E only suggested entry by phagocytosis (Fig. 3D and E). The difference between inducing mixture D and noninducing mixture E was the addition of manganese chloride into mixture D. We therefore hypothesized that manganese chloride was responsible for the induction of the intracellular phenotype. MAC was then exposed to Middlebrook 7H9 broth containing the same physiologically relevant concentration of manganese chloride as the 24-h elemental mixture for 24 h, and real-time PCR and fluorescence microscopy analyses were carried out as described above. For a control, we exposed MAC to Middlebrook 7H9 broth containing the same concentration of copper sulfate as the 24-h elemental mixture. To determine whether the genes were upregulated in response to manganese or chloride, we added calcium chloride at the concentration that manganese chloride was in the 24-h elemental mixture to Middlebrook 7H9 broth in another sample. As expected, exposure to the copper sulfate did not induce changes in gene expression (Fig. 6). MAC exposed to the physiological concentration of manganese chloride showed upregulation of the protein that binds the SigE promoter, Mav1094, PE-PGRS family protein Mav4487, and growth-promoting protein Mav0996, but not the hypothetical invasion protein Mav3301 or alternative sigma factor SigE Mav1365. To our surprise, MAC exposed to the calcium chloride control showed upregulation in not only the mprA homolog Mav1094 and resuscitation gene Mav0996 but also in the hypothetical proteins Mav3301 and sigE Mav1365, suggesting that Mav3301 and Mav1365 respond to high concentrations of calcium. The fluorescence microscopy for bacteria incubated in manganese chloride showed no evidence of macropinocytosis (Fig. 3F). To determine what role the calcium chloride had in the regulation of these genes, we also exposed MAC to a sample containing Middlebrook 7H9 broth with the physiologically relevant amount of calcium chloride. Only one gene that has previously been reported to respond to changes in zinc, Mav2036 (30) appeared to be upregulated under these conditions (Fig. 6). These data suggest that chloride, manganese, calcium, and potassium together are necessary for inducing the intracellular phenotype. For a control, we determined the growth of MAC in vitro in the elemental mixture (Table 5).
Table 4.
Reduction scheme used to determine which elements trigger the MAC intracellular phenotypea
| pH or supplement | 24-h elemental mixture | 24-h elemental mixture with adjusted pH | Elemental mixture |
|||||
|---|---|---|---|---|---|---|---|---|
| A | B | C | D | E | F | |||
| pH | 5.8 | 6.9 | 6.9 | 6.9 | 6.9 | 6.9 | 6.9 | 6.9 |
| Amt (ml or μl) of supplement added | ||||||||
| 1 M KCl (ml) | 0.925 | 0.925 | 0.925 | 0.925 | 0.925 | 0.925 | 0.925 | 0.925 |
| 1 M CaCl2 (ml) | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | 1.25 | |
| 1 M Mn2Cl (ml) | 11.9 | 11.9 | 11.9 | 11.9 | 11.9 | 11.9 | ||
| 1 M CuSo4 (μl) | 5.5 | 5.5 | 5.5 | 5.5 | 5.5 | |||
| 1 M ZnCl2 (μl) | 58.7 | 58.7 | 58.7 | 58.7 | ||||
| 0.25 M FePO4 (ml) | 2 | 2 | 2 | |||||
| 1 M NiCl2 (μl) | 5 | 5 | ||||||
Several versions of the 24-h elemental mixture were made, each one missing one component more than the previous one.
Fig. 4.
Real-time PCR of MAC strain 104 in the subtracted 24-h elemental mixtures. Total bacterial RNA was extracted from bacteria exposed to the indicated mixture for 24 h and bacteria exposed to Middlebrook 7H9 broth as a reference. cDNA was made from the RNA and used for real-time PCR. Values represent the difference from the background control. The values are the means plus standard deviations from three repetitions. Values that were significantly different from the control value are indicated as follows: #, P < 0.10; *, P < 0.05.
Fig. 5.
Uptake of MAC strain 104 by macrophages after incubation in subtraction elemental mixtures. MAC was incubated in the indicated mixture for 24 h and then used to infect Raw 264.7 macrophage monolayers. Percent uptake was determined by dividing the number of bacteria that were inside macrophages after a 1-h infection by the number of bacteria that were added to the monolayer and then multiplying by 100. The experiment was repeated three times. Values that were significantly different (P < 0.05) from the control value (Middlebrook 7H9 broth) are indicated by an asterisk.
Fig. 6.
Real-time PCR of MAC strain 104 incubated in single metals. MAC was exposed to Middlebrook 7H9 broth containing the indicated volume of a 1 M concentration of the indicated metal per 500 ml of Middlebrook 7H9 broth for 24 h, and then RNA was extracted and used for cDNA synthesis and real-time PCR and compared to MAC exposed to Middlebrook 7H9 broth. Values represent the difference from the background control. The values are means plus standard deviations (error bars) from three repetitions. Values that were significantly different from the control value are indicated as follows: #, P < 0.10; *, P < 0.05.
Table 5.
MAC growth in elemental mixtures in vitroa
| Conditions | Initial inoculum (mean ± SD) | No. of CFU 4 days postinfection (mean ± SD) |
|---|---|---|
| Middlebrook 7H9 broth | 3 × 105 ± 0.5 × 105 | 7.3 × 107 ± 0.8 × 107 |
| 24-h elemental mixture | 3 × 105 ± 0.5 × 105 | 3.5 × 107 ± 0.4 × 107 |
M. avium complex strain 104 bacteria were grown at 37°C in 15-ml plastic tubes.
Association with vacuole markers.
We tested which phagosome markers were present on macrophage vacuoles occupied by bacteria incubated in the 24-h elemental mixture. Using fluorescence microscopy, fluorescein isothiocyanate (FITC)-stained bacteria, and Texas Red-conjugated antibodies to phagosome marker proteins, we determined the percent colocalization between the stained bacteria and vacuolar proteins. Rab5, Rab7, and early endosomal antigen 1 (EEA-1) colocalized with the bacteria at rates similar to those of the group incubated in Middlebrook 7H9 broth and the group exposed to the 24-h elemental mixture (Table 6). On the other hand, lysosome-associated membrane protein 1 (LAMP-1) colocalized with the bacteria exposed to Middlebrook 7H9 broth 30% of the time, while 60% of the stained bacteria from the 24-h elemental mixture colocalized with LAMP-1. This trend with Rab5 and LAMP-1 is consistent with bacteria that have been passed through macrophages prior to macrophage infection, further supporting induction of the intracellular phenotype in our 24-h elemental mixture (4).
Table 6.
Colocalization of MAC and phagosome markersa
| MAC growth condition | % colocalization (mean ± SD) of MAC with the following protein: |
|||
|---|---|---|---|---|
| Rab5 | Rab7 | EEA-1 | LAMP-1 | |
| Middlebrook 7H9 broth | 63 ± 16 | 22 ± 7 | 46 ± 14 | 30 ± 13 |
| 24-h elemental mixture | 71 ± 16 | 25 ± 7 | 46 ± 24 | 60 ± 9b |
Raw 264.7 cells were infected with rhodamine-labeled MAC that had previously been incubated in either Middlebrook 7H9 broth or 24-h elemental mixture for 24 h. The cells were then washed, treated with amikacin to kill extracellular bacteria, fixed, permeablized, and appropriately stained for analysis with a fluorescence microscope. The assay was repeated three times, with about 40 stained bacteria analyzed per sample per time.
This value was significantly different (P < 0.05) from the value for MAC incubated in Middlebrook 7H9 broth.
Cell death.
Apoptosis and necrosis are associated with spread of infections. Since MAC infection of the “first macrophage” only exits the cells upon apoptosis, we investigated whether MAC exhibiting the intracellular phenotype induce apoptosis in macrophages more efficiently than bacteria that have been grown in Middlebrook 7H9 broth. MAC was exposed either to Middlebrook 7H9 broth or the 24-h elemental mixture for 24 h and then used to infect Raw 264.7 cells. Two hours later, we determined that Raw 264.7 cells infected by bacteria exposed to the 24-h elemental mixture had an apoptosis rate of 19.5%, in comparison with 8.6% of the cells infected by bacteria from Middlebrook 7H9 broth (Table 7). Similarly, 19.8% of cells infected by bacteria from the 24-h elemental mixture gave a positive terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) signal, while 11% of cells from bacteria exposed to Middlebrook 7H9 broth indicated apoptotic death (Table 7). We also performed control tests in which the Raw 264.7 cells were treated with Middlebrook 7H9 broth and 24-h elemental mixture without bacteria, and no significant difference in cell death by the Live-Dead cell viability assay or TUNEL assay was observed.
Table 7.
Macrophage death after infection by MAC exhibiting the intracellular phenotypea
| Inoculum | % dead macrophages (MOI of 1,000/MOI of 100) | % cells stained by TUNEL assay (MOI of 1,000/MOI of 100) |
|---|---|---|
| Middlebrook 7H9 broth (control) | 4.02 ± 1.6/4.3 ± 0.6 | 12.3 ± 5.0/13.5 ± 2 |
| 24-h elemental mixture (control) | 5.04 ± 2.2/4.8 ± 1.9 | 15.8 ± 2.1/12.6 ± 3 |
| MAC from Middlebrook 7H9 broth | 8.58 ± 4.4/8.2 ± 2.5 | 11.0 ± 2.1/10.6 ± 2.7 |
| MAC from 24-h elemental mixture | 19.52 ± 4.9b/20.1 ± 3.7b | 19.8 ± 6.8/19.4 ± 4.9b |
Two hours postinfection, Raw 264.7 cells infected by MAC from the 24-h elemental mixture undergo cell death and apoptosis (measured by the TUNEL assay) more frequently than Raw 264.7 cells infected by MAC from Middlebrook 7H9 broth (control). The assay was repeated three times.
These values were significantly different (P < 0.05) from the value for MAC incubated in Middlebrook 7H9 broth.
To confirm that the increase in percent macrophage death in the elemental mixture was consistent with the intracellular phenotype, Raw 264.7 cell monolayers were infected with bacteria that were recovered from macrophages, and percent cell death was compared to macrophages infected by bacteria from Middlebrook 7H9 broth, as described previously (3). In these assays, only 3.4% of macrophages infected by bacteria from Middlebrook 7H9 broth stained dead, while 8.5% of macrophages infected by bacteria from other macrophages were dead (Table 8). We found no difference in percent death between our control groups: uninfected macrophages, macrophages inoculated with Middlebrook 7H9 broth, and macrophages inoculated by macrophage lysate. These data suggest that bacteria of the intracellular phenotype induce apoptosis of the host macrophage more efficiently than bacteria from Middlebrook 7H9 broth.
Table 8.
Macrophage death after infection by MAC from different sources and conditionsa
| Macrophage | Avg inoculumb | % dead macrophagesc |
|---|---|---|
| Uninfected macrophages | N/A | 2.1 ± 0.9 |
| Macrophages incubated in Middlebrook 7H9 broth | N/A | 1.9 ± 0.7 |
| Macrophages incubated with Raw 264.7 lysate | N/A | 1.7 ± 0.5 |
| Macrophages inoculated with MAC from the following: | ||
| Middlebrook 7H9 broth | 1.40E + 10 | 3.4 ± 0.8 |
| Raw 264.7 cells | 8.10E + 08 | 8.5 ± 3.0d |
Bacteria from Middlebrook 7H9 broth or collected from macrophages 24 h postinfection were used to infect fresh macrophage monolayers. Two hours later, the Live-Dead cell viability assay was performed to determine percent macrophage death.
N/A, not available.
Values represent means ± standard deviations from three repetitions.
This value was significantly different (P < 0.05) from the value for MAC incubated in Middlebrook 7H9 broth.
DISCUSSION
Consistent with previous reports regarding Mycobacterium avium complex (MAC) from macrophages, we observed that MAC from 24-h elemental mixture was taken up by macrophages 8 times more efficiently than MAC from Middlebrook 7H9 broth and used macropinocytosis in addition to phagocytosis as an entry mechanism (3, 4). While we observed upregulation of several genes that have previously been reported to be upregulated in macrophages using 24-h elemental mixture, other mycobacterial genes were not upregulated. This could be due to differences in technique, i.e., real-time PCR versus promoter-green fluorescent protein (GFP) system, or due to differences in the time point of expression inside macrophages, but is more likely because our mimicry system contains only metals and pH changes, and these genes may require proteins, lipids, or other factors not included in the mixture that are specific to the macrophage.
Despite several differences in metal concentration between the 1-h elemental mixture and the 24-h elemental mixture, incubation of MAC in either mixture resulted in increased uptake by macrophages. We did not examine the 1-h elemental mixture in detail in this study, so we are unsure whether other traits of the MAC intracellular phenotype were induced with the 1-h elemental mixture. Considering that elemental mixture D, which contained potassium chloride, calcium chloride, and manganese chloride at the concentrations relevant to the vacuole 24 h postinfection, was able to induce the MAC intracellular phenotype, it seems valuable to compare the concentrations of these components in the 1-h elemental mixture to the 24-h elemental mixture. The 1-h elemental mixture contains higher concentrations of potassium chloride (29.4 mM versus 1.9 mM) and calcium chloride (4 mM versus 2.5 mM) but a lower concentration of manganese chloride (11.8 mM versus 23.8 mM) than the 24-h elemental mixture. Further study in determining which components of the 1-h elemental mixture are necessary to induce the intracellular phenotype will help narrow down the required concentrations for inducing the MAC intracellular phenotype. It may be that both solution concentrations are within the threshold.
Mav2036 did not appear to be upregulated in response to physiological concentrations of zinc, but it was upregulated in M. tuberculosis at similar zinc concentrations (30). This gene has not been studied in MAC, and the difference in gene regulation could be one of several differences between the two Mycobacterium species. Alternately, the addition of several other metals to the elemental mixture may have altered the effect of zinc on Mav2036.
Mav1094, the homolog to M. tuberculosis mprA, was upregulated under some conditions, such as during incubation in elemental mixture E, in which Mav1365, the homolog to M. tuberculosis sigE was not upregulated. In M. tuberculosis, sigE is upregulated in macrophages and is essential for virulence (23, 27, 28). MprA binds to the promoter region of sigE to lead to increased expression of sigE (18). In MAC, the promoter region of sigE contains an untested putative binding motif for MprA, but our data suggest an additional step of regulation between overexpression of mprA and overexpression of sigE, since upregulation of the MprA homolog showed no upregulation of the SigE homolog. Alternatively, MAC may use a different or additional method of sigE regulation than what is observed in M. tuberculosis.
The means by which manganese chloride, calcium chloride, and potassium chloride together were able to induce the intracellular phenotype of MAC is unknown. Manganese contributes to virulence of other bacteria, such as Salmonella enterica serovar Typhimurium, and Bacillus anthracis via the use of Nramp and ABC transporters (7, 24). In Pseudomonas aeruginosa and Yersinia, type III secretion system genes are upregulated, and its effectors are transported in response to low concentrations of calcium, similar to what happens when in contact with host cells (13, 25, 38, 41). Similarly, in Salmonella, potassium induces expression of the Salmonella pathogenicity island 1 (SPI1) type III secretion system and increases host cell invasion (39). While MAC has no type III secretion system, the M. tuberculosis potassium/proton antiporter kefB (homolog = Mav_4198) was found to contribute to virulence in mice (34, 36). M. avium also contain natural resistance-associated macrophage protein (Nramp) homologs and ABC homologs, both of which are known to utilize manganese, often preferentially over other divalent metals (22, 24). In M. tuberculosis, mramp does not appear to be required for virulence (6), but our study suggests an important role for manganese in MAC virulence that may be through the nramp homolog or some other gene(s). Likewise, the mycobacterium-specific family of PE-PGRS proteins, some of which play a role in virulence, contain calcium-binding repeats (2). Further study would potentially shed light unto the molecular details of how these genes and others respond to specific metals and are used by MAC for virulence.
The observation that bacteria sense their environment and respond, potentially in a way to enhance virulence, is not new (1, 15). In other bacteria, such as Escherichia coli, Pseudomonas aeruginosa, and M. tuberculosis, sensor kinases of 2-component regulatory systems detect environmental changes, which lead to phosphorylation of the response regulator, and this induces a conformational change that permits binding of the response regulator to promoters of the DNA, resulting in changes in gene expression and phenotype (16, 21, 33). In MAC, the mtrAB system has already been shown to contribute to virulence, but its environmental cues have not been determined (9). On the basis of sequence similarity, other potential two-component regulatory systems are present in MAC, but the environmental stimuli and role in virulence have not been elucidated. The new incubation media presented in this study could become a useful laboratory tool to further decipher the sensing mechanisms of the intracellular phenotype of MAC.
It is important to consider, though, that other stimuli inside the vacuole, such as superoxide anion or NO, can trigger a similar response. In fact, while it is possible that noxious conditions may induce a comparable response, data on the response of M. tuberculosis to oxidative stress (O2− and NO) do not suggest it (42, 43), and with the exception of a few genes, the majority of the genes regulated by the “intravacuolar supernatant” are not regulated under oxidative stress (35).
It is interesting to note that MAC incubated with the elemental mixture is found in intracellular vacuoles that contain LAMP-1 more often than vacuoles of MAC growing in culture. It confirms the phenotype observed previously (4) that bacteria with the intracellular phenotype are seen in a phagosome that differs in some aspects from the phagosomes of MAC grown in medium. Perhaps the intracellular phenotype uses the environment to trigger expression or to suppress genes involved in the ability of the bacteria to leave the macrophages and spread to surrounding macrophages or other cells.
Our observation that MAC obtained from the 24-h elemental mixture and macrophages is associated with a higher percentage of macrophage death brings forth the question of whether the bacteria or macrophage is responsible for inducing cell death/apoptosis. It has previously been assumed that apoptosis of the macrophage is induced by the host as a defense mechanism (14). Other pathogens, such as Yersinia paratuberculosis and Salmonella Typhimurium, induce apoptosis to prevent uptake and to escape the host cell, respectively (20, 31). One could surmise, however, that bacteria induce apoptosis, especially after passage through macrophages, as a method of macrophage escape, which would aid with the process of dissemination. Alternatively, MAC within the host may have a preference for a subpopulation of macrophages, and therefore, MAC escapes macrophages until it enters a macrophage with the desired characteristics, similar to the association of Listeria monocytogenes with a subpopulation of macrophages (17). It is also possible that an effect of the method of bacterial entry into the macrophage, i.e., macropinocytosis, may induce apoptosis, resulting in our observed increase in percent cell death. More research with this aspect could lead to important discoveries of how MAC causes disseminated disease.
ACKNOWLEDGMENTS
This work was supported by the NIH grants AI43199 and AI47010.
We thank Denny Weber for assistance with the manuscript.
Footnotes
Published ahead of print on 28 March 2011.
REFERENCES
- 1. Altier C. 2005. Genetic and environmental control of salmonella invasion. J. Microbiol. 43(1):85–92 [PubMed] [Google Scholar]
- 2. Bachhawat N., Singh B. 2007. Mycobacterial PE_PGRS proteins contain calcium-binding motifs with parallel beta-roll folds. Genomics Proteomics Bioinformatics 5:236–241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bermudez L. E., Parker A., Goodman J. R. 1997. Growth within macrophages increases the efficiency of Mycobacterium avium in invading other macrophages by a complement receptor-independent pathway. Infect. Immun. 65:1916–1925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bermudez L. E., Petrofsky M., Sangari F. 2004. Intracellular phenotype of Mycobacterium avium enters macrophages primarily by a macropinocytosis-like mechanism and survives in a compartment that differs from that with extracellular phenotype. Cell Biol. Int. 28:411–419 [DOI] [PubMed] [Google Scholar]
- 5. Bermudez L. E., Young L. S. 1988. Tumor necrosis factor, alone or in combination with IL-2, but not IFN-gamma, is associated with macrophage killing of Mycobacterium avium complex. J. Immunol. 140:3006–3013 [PubMed] [Google Scholar]
- 6. Boechat N., et al. 2002. Disruption of the gene homologous to mammalian Nramp1 in Mycobacterium tuberculosis does not affect virulence in mice. Infect. Immun. 70:4124–4131 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Boyer E., Bergevin I., Malo D., Gros P., Cellier M. F. 2002. Acquisition of Mn(II) in addition to Fe(II) is required for full virulence of Salmonella enterica serovar Typhimurium. Infect. Immun. 70:6032–6042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Brennan M. J., et al. 2001. Evidence that mycobacterial PE_PGRS proteins are cell surface constituents that influence interactions with other cells. Infect. Immun. 69:7326–7333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cangelosi G. A., et al. 2006. The two-component regulatory system mtrAB is required for morphotypic multidrug resistance in Mycobacterium avium. Antimicrob. Agents Chemother. 50:461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Danelishvili L., Poort M. J., Bermudez L. E. 2004. Identification of Mycobacterium avium genes up-regulated in cultured macrophages and in mice. FEMS Microbiol. Lett. 239:41–49 [DOI] [PubMed] [Google Scholar]
- 11. Danelishvili L., et al. 2007. Identification of Mycobacterium avium pathogenicity island important for macrophage and amoeba infection. Proc. Natl. Acad. Sci. U. S. A. 104:11038–11043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feodorova V. A., Devdariani Z. L. 2001. Expression of acid-stable proteins and modified lipopolysaccharide of Yersinia pestis in acidic growth medium. J. Med. Microbiol. 50:979–985 [DOI] [PubMed] [Google Scholar]
- 13. Frank D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 26:621–629 [DOI] [PubMed] [Google Scholar]
- 14. Fratazzi C., et al. 1999. Macrophage apoptosis in mycobacterial infections. J. Leukoc. Biol. 66:763–764 [DOI] [PubMed] [Google Scholar]
- 15. Gancz H., Jones K. R., Merrell D. S. 2008. Sodium chloride affects Helicobacter pylori growth and gene expression. J. Bacteriol. 190:4100–4105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15a. Gao L. Y., Guo S., McLaughlin B., Morisaki H., Engel J. N., Brown E. J. 2004. A mycobacterial virulence gene cluster extending RD1 is required for cytolysis, bacterial spreading and ESAT-6 secretion. Mol. Microbiol. 53:1677–1693 [DOI] [PubMed] [Google Scholar]
- 16. Gooderham W. J., Hancock R. E. 2009. Regulation of virulence and antibiotic resistance by two-component regulatory systems in Pseudomonas aeruginosa. FEMS Microbiol. Rev. 33:279–294 [DOI] [PubMed] [Google Scholar]
- 16a. Haile Y., Caugant D. A., Bjune G., Wiker H. G. 2002. Mycobacterium tuberculosis mammalian cell entry operon (mce) homologs in Mycobacterium other than tuberculosis (MOTT). FEMS Immunol. Med. Microbiol. 33:125–132 [DOI] [PubMed] [Google Scholar]
- 17. Harrington-Fowler L., Wilder M. S. 1982. Fate of Listeria monocytogenes in murine peritoneal macrophage subpopulations. Infect. Immun. 35:124–132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. He H., Hovey R., Kane J., Singh V., Zahrt T. C. 2006. MprAB is a stress-responsive two-component system that directly regulates expression of sigma factors SigB and SigE in Mycobacterium tuberculosis. J. Bacteriol. 188:2134–2143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hou J. Y., Graham J. E., Clark-Curtiss J. E. 2002. Mycobacterium avium genes expressed during growth in human macrophages detected by selective capture of transcribed sequences (SCOTS). Infect. Immun. 70:3714–3726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ibarra J. A., Steele-Mortimer O. 2009. Salmonella-the ultimate insider. Salmonella virulence factors that modulate intracellular survival. Cell. Microbiol. 11:1579–1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Igo M. M., Ninfa A. J., Silhavy T. J. 1989. A bacterial environmental sensor that functions as a protein kinase and stimulates transcriptional activation. Genes Dev. 3:598–605 [DOI] [PubMed] [Google Scholar]
- 22. Jabado N., et al. 2000. Natural resistance to intracellular infections: natural resistance-associated macrophage protein 1 (Nramp1) functions as a pH-dependent manganese transporter at the phagosomal membrane. J. Exp. Med. 192:1237–1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Jensen-Cain D. M., Quinn F. D. 2001. Differential expression of sigE by Mycobacterium tuberculosis during intracellular growth. Microb. Pathog. 30:271–278 [DOI] [PubMed] [Google Scholar]
- 24. Kehres D. G., Zaharik M. L., Finlay B. B., Maguire M. E. 2000. The NRAMP proteins of Salmonella typhimurium and Escherichia coli are selective manganese transporters involved in the response to reactive oxygen. Mol. Microbiol. 36:1085–1100 [DOI] [PubMed] [Google Scholar]
- 25. Kim J., et al. 2005. Factors triggering type III secretion in Pseudomonas aeruginosa. Microbiology 151:3575–3587 [DOI] [PubMed] [Google Scholar]
- 26. Kuhn D. E., Lafuse W. P., Zwilling B. S. 2001. Iron transport into Mycobacterium avium-containing phagosomes from an Nramp1(Gly169)-transfected RAW264.7 macrophage cell line. J. Leukoc. Biol. 69:43–49 [PubMed] [Google Scholar]
- 26a. Li Y., Miltner E., Wu M., Petrofsky M., Bermudez L. E. 2005. A Mycobacterium avium PPE gene is associated with the ability of the bacterium to grow in macrophages and virulence in mice. Cell Microbiol. 7:539–548 [DOI] [PubMed] [Google Scholar]
- 27. Manganelli R., et al. 2004. The extracytoplasmic function sigma factor sigma(E) is essential for Mycobacterium tuberculosis virulence in mice. Infect. Immun. 72:3038–3041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Manganelli R., Voskuil M. I., Schoolnik G. K., Smith I. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423–437 [DOI] [PubMed] [Google Scholar]
- 29. McDonough J. A., Hacker K. E., Flores A. R., Pavelka M. S., Jr., Braunstein M. 2005. The twin-arginine translocation pathway of Mycobacterium smegmatis is functional and required for the export of mycobacterial beta-lactamases. J. Bacteriol. 187:7667–7679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Milano A., Branzoni M., Canneva F., Profumo A., Riccardi G. 2004. The Mycobacterium tuberculosis Rv2358-furB operon is induced by zinc. Res. Microbiol. 155:192–200 [DOI] [PubMed] [Google Scholar]
- 31. Monack D. M., Mecsas J., Ghori N., Falkow S. 1997. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc. Natl. Acad. Sci. U. S. A. 94:10385–10390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Oh Y. K., Straubinger R. M. 1996. Intracellular fate of Mycobacterium avium: use of dual-label spectrofluorometry to investigate the influence of bacterial viability and opsonization on phagosomal pH and phagosome-lysosome interaction. Infect. Immun. 64:319–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32a. Plum G., Brenden M., Clark-Curtiss J. E., Pulverer G. 1997. Cloning, sequencing, and expression of the mig gene of Mycobacterium avium, which codes for a secreted macrophage-induced protein. Infect. Immun. 65:4548–4557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Saini D. K., Malhotra V., Tyagi J. S. 2004. Cross talk between DevS sensor kinase homologue, Rv2027c, and DevR response regulator of Mycobacterium tuberculosis. FEBS Lett. 565:75–80 [DOI] [PubMed] [Google Scholar]
- 34. Sassetti C. M., Rubin E. J. 2003. Genetic requirements for mycobacterial survival during infection. Proc. Natl. Acad. Sci. U. S. A. 100:12989–12994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schnappinger D., et al. 2003. Transcriptional adaptation of Mycobacterium tuberculosis within macrophages: insights into the phagosomal environment. J. Exp. Med. 198:693–704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Stewart G. R., Patel J., Robertson B. D., Rae A., Young D. B. 2005. Mycobacterial mutants with defective control of phagosomal acidification. PLoS Pathog. 1:269–278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Straley S. C., Brubaker R. R. 1981. Cytoplasmic and membrane proteins of yersiniae cultivated under conditions simulating mammalian intracellular environment. Proc. Natl. Acad. Sci. U. S. A. 78:1224–1228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Straley S. C., Plano G. V., Skrzypek E., Haddix P. L., Fields K. A. 1993. Regulation by Ca2+ in the Yersinia low-Ca2+ response. Mol. Microbiol. 8:1005–1010 [DOI] [PubMed] [Google Scholar]
- 39. Su J., Gong H., Lai J., Main A., Lu S. 2009. The potassium transporter Trk and external potassium modulate Salmonella enterica protein secretion and virulence. Infect. Immun. 77:667–675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Tenant R., Bermudez L. E. 2006. Mycobacterium avium genes upregulated upon infection of Acanthamoeba castellanii demonstrate a common response to the intracellular environment. Curr. Microbiol. 52:128–133 [DOI] [PubMed] [Google Scholar]
- 41. Vallis A. J., Yahr T. L., Barbieri J. T., Frank D. W. 1999. Regulation of ExoS production and secretion by Pseudomonas aeruginosa in response to tissue culture conditions. Infect. Immun. 67:914–920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Voskuil M. I., et al. 2003. Inhibition of respiration by nitric oxide induces a Mycobacterium tuberculosis dormancy program. J. Exp. Med. 198:705–713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Voskuil M. I., Visconti K. C., Schoolnik G. K. 2004. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis (Edinb.) 84:218–227 [DOI] [PubMed] [Google Scholar]
- 44. Wagner D., et al. 2005. Elemental analysis of Mycobacterium avium-, Mycobacterium tuberculosis-, and Mycobacterium smegmatis-containing phagosomes indicates pathogen-induced microenvironments within the host cell's endosomal system. J. Immunol. 174:1491–1500 [DOI] [PubMed] [Google Scholar]
- 45. Wagner D., et al. 2005. Changes of the phagosomal elemental concentrations by Mycobacterium tuberculosis Mramp. Microbiology 151:323–332 [DOI] [PubMed] [Google Scholar]
- 46. Wolfgang M. C., Lee V. T., Gilmore M. E., Lory S. 2003. Coordinate regulation of bacterial virulence genes by a novel adenylate cyclase-dependent signaling pathway. Dev. Cell 4:253–263 [DOI] [PubMed] [Google Scholar]