Abstract
Uncomplicated urinary tract infections (UTI) are caused most commonly by uropathogenic Escherichia coli (UPEC). Whole-genome screening approaches, including transcriptomic, proteomic, and signature-tagged mutagenesis, have shown that UPEC highly expresses or requires genes for translational machinery, capsule, lipopolysaccharide, type 1 fimbriae, and iron acquisition systems during UTI. To identify additional genes expressed by UPEC during UTI, an immunoscreening approach termed in vivo-induced antigen technology (IVIAT) was employed to identify antigens produced during experimental infection that are not produced during in vitro culture. An inducible protein expression library, constructed from genomic DNA isolated from UPEC strain CFT073, was screened using exhaustively adsorbed pooled sera from 20 chronically infected female CBA/J mice. Using this approach, we identified 93 antigens induced by UPEC in vivo. A representative subset of these genes was tested by quantitative PCR for expression by CFT073 in vivo and during growth in human urine or LB medium in vitro; proWX, narJI, lolA, lolD, tosA (upxA), c2432, katG, ydhX, kpsS, and yddQ were poorly expressed in vitro but highly expressed in vivo. Of these, tosA, a gene encoding a predicted repeat-in-toxin family member, was expressed exclusively during UTI. Deletion of tosA in UPEC strain CFT073 resulted in significant attenuation in bladder and kidney infections during ascending UTI. By screening for in vivo-induced antigens, we identified a novel UPEC virulence factor and additional proteins that could be useful as potential vaccine targets.
INTRODUCTION
The Gram-negative bacterium Escherichia coli is responsible for 80% of community-acquired urinary tract infections (UTI) (38). Uncomplicated UTI create a significant burden on human health, with more than 50% of adult women suffering a UTI during their lifetime (16). An equally large burden is placed on our health care system, with estimates of direct and indirect costs of $1.6 billion per year in the United States alone (15). While antibiotics have proven to be an effective treatment in the past, a worldwide increase in antibiotic resistance in uropathogenic E. coli (UPEC) isolates (1, 6, 8, 14) has created an urgent need for alternative treatment and prevention strategies to combat this important and widespread human pathogen.
In addition to the core genetic backbone of this species, UPEC contains a unique repertoire of genetic material distinct from what is present in commensal strains or other E. coli pathotypes (39). This set of accessory genes provides the mechanisms for colonizing the host urinary tract and eliciting the signs and symptoms of an uncomplicated UTI (28). The host urinary tract environment challenges bacteria with an immune response that the bacteria must overcome to persist in the host (33). Recently, our group correlated antibody production with protection in a mucosal vaccination model by using several vaccine candidates that targeted iron acquisition system proteins expressed in vivo by human UPEC strains (2). This suggested that eliciting a humoral immune response to UPEC surface proteins, some encoded by specific accessory genetic sequences, may be beneficial in creating novel UPEC vaccines. Clearly, new candidate vaccine targets will be required to deal with the diverse mechanisms of pathogenesis observed among different UPEC isolates (7).
New insight into the mechanisms of bacterial pathogenesis mediated by proteins synthesized only during the course of an infection can be achieved through the use of in vivo-induced antigen technology (IVIAT) (21). IVIAT utilizes sera from infected patients or experimentally infected animals that have been adsorbed against bacteria cultured in vitro. This process removes antibodies that react to common antigens made during in vitro growth and enriches the sera for those antibodies that may recognize antigens induced only in vivo. These sera are then used to probe libraries that express genes from the pathogen of interest to identify which genes encode antigenic proteins that are synthesized preferentially during the course of an infection (30). This process has been applied successfully to study a diverse collection of important human pathogens, including enterohemorrhagic E. coli (EHEC) (22), Streptococcus pyogenes (31), and Mycobacterium tuberculosis (13, 23). This approach could be particularly effective for addressing urinary tract infections, as the proteins identified by IVIAT elicit a host immune response and are expressed only during the course of an infection, both properties expected to be critical for the formulation of an effective UPEC vaccine (32).
In this study, we generated a genomic expression library by using DNA from the human pyelonephritis isolate E. coli CFT073. This library was screened against antisera that were collected from mice chronically infected with this UPEC strain and that had been adsorbed against bacteria cultured in vitro. In total, more than 40,000 clones were screened, resulting in 93 IVIAT-identified genes. In vivo expression of all of 13 representative genes was confirmed by quantitative PCR (qPCR). Five isogenic mutants of CFT073, deficient in five representative systems identified by IVIAT, were constructed, and one of these, an osmoprotection system, conferred a fitness advantage when competed against wild-type CFT073 in an animal UTI model. Finally, we determined that the tosA gene (originally annotated as upxA [39]), encoding a repeat-in-toxin (RTX) family member (29) and identified here by IVIAT, contributes significantly to the virulence of UPEC. These results not only expand our understanding of the processes that occur during an infection but also expand the list of candidates for the development of novel therapeutics to combat UTI.
MATERIALS AND METHODS
Bacteria and culture conditions.
E. coli CFT073, a prototypic UPEC strain, was isolated from the blood and urine of a patient with acute pyelonephritis (27); its genome has been sequenced and annotated fully (39). E. coli TOP10 (Invitrogen) and BL21(DE3)/pLysS were used for cloning and as the host expression strain, respectively. Isolated colonies were inoculated into LB medium and cultured aerobically overnight. Culture in human urine was conducted as described previously (3). For library screening, dilutions of the expression strain were plated onto LB agar containing 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG), kanamycin (25 μg/ml), and chloramphenicol (34 μg/ml) and incubated at 30°C.
Library construction.
Genomic DNA was purified from uropathogenic E. coli strain CFT073 by use of DNeasy columns (Qiagen) and was partially digested using the enzyme Sau3AI. Digested DNA was size fractionated in a 0.9% agarose gel, and 1.0- to 1.5-kb fragments were isolated and purified using a gel extraction kit (Qiagen). The isolated fragments were ligated into a pool of BamHI-digested pET30a, pET30b, and pET30c plasmids and transformed into competent TOP10 cells. A total of 60,000 colonies from 25 plates were pooled into LB medium and used for plasmid Midi preps (Promega). Purified plasmids were electroporated into BL21(DE3)/pLysS, and all transformants, approximately 100,000 colonies from 35 plates, were pooled into 10 ml of LB medium containing 35% glycerol and stored in 0.5-ml aliquots at −80°C.
Preparation of antisera.
Sera from 20 chronically infected mice (19) were pooled (final volume of 2.75 ml) and adsorbed with CFT073 bacterial cells cultured in LB medium to an optical density at 600 nm (OD600) of 0.5 as follows. Bacterial cells were collected from the LB medium by centrifugation, washed in phosphate-buffered saline (PBS), suspended in PBS, and divided into aliquots containing 1011 CFU of CFT073. Aliquots were used for adsorption directly (whole cells) or lysed in a French pressure cell (whole-cell lysates) as described previously (4). Pooled sera were adsorbed with aliquots of whole cells on a rocking platform at 4°C for 1 h. Unadsorbed antibodies were collected from the supernatant following centrifugation, and adsorption was repeated two additional times with CFT073 cells and three times with BL21 cells. Following adsorption to whole cells, antibodies were incubated with nitrocellulose strips saturated with 1 mg whole-cell lysate from CFT073 on a rocking platform at 4°C for 1 h. Soluble antibodies were recovered and applied to additional whole-cell lysate three times for CFT073, followed by two adsorptions using whole-cell lysate from BL21. Additional whole-cell lysate was electrophoresed in a 12% SDS-polyacrylamide gel and transferred to a polyvinylidene difluoride (PVDF) membrane to prepare denatured whole-cell extracts for absorption. Adsorption was continued using denatured whole-cell extracts as described, for a total of five adsorptions to denatured extracts from CFT073 and three adsorptions to denatured extracts from BL21. Aliquots of the final adsorbed sera were stored at −20°C.
In vivo-induced antigen screening.
IVIAT was performed as described previously (21), with the following modifications. Aliquots of BL21(DE3)/pLysS containing the expression library were thawed, diluted, and plated onto LB medium containing kanamycin to obtain 300 to 400 colonies per plate. Master plates were incubated at 30°C for 16 h before transfer onto nitrocellulose filters. Filters were placed colony side up on LB agar containing 1 mM IPTG and kanamycin (25 μg/ml) and were incubated at 37°C for 3 to 4 h. Following induction, nitrocellulose filters were removed and colonies were lysed in chloroform vapor. Colony immunoblotting was performed using the pooled adsorbed sera as primary antibodies. Reactive colonies were detected using goat anti-mouse–horseradish peroxidase (HRP) secondary antibodies and were visualized using a chemiluminescent substrate (Pierce). Clones identified during primary screening were picked from the master plate and subjected to secondary screening by patching, in an alternating pattern, with the BL21 vector control. Secondary screens were carried out as described for the primary screen. Clones that maintained reactivity on two of three plates were considered positive identifications and subjected to DNA sequencing using T7 primers that flanked the multiple cloning site within the vectors.
RNA extraction and quantitative real-time PCR.
For preparation of RNA, bacteria were collected by centrifugation from overnight cultures and washed with sterile PBS, and 106 CFU was used to inoculate prewarmed LB medium or human urine. Cultures were incubated statically at 37°C until exponential phase (OD600 = 1.0 for LB medium and 0.35 for human urine). Following incubation, 1 ml of bacterial culture was added to 0.125 ml of ice-cold phenol-ethanol stop solution (5% phenol in ethanol), and bacteria were collected by centrifugation. Cells were lysed and RNA was extracted using an RNeasy kit (Qiagen) following the manufacturer's protocol. Following elution, nucleic acid concentrations were determined by spectrophotometry (NanoDrop), and residual DNA contamination was removed by incubating the samples with 4 U of Turbo DNase (Ambion). After DNase inactivation, RNA was recovered, quantified, and used as a template for PCR to confirm inactivation of contaminating DNA. The PCR-negative RNA was used for first-strand cDNA synthesis using SuperScript II reverse transcriptase (Invitrogen). For cDNA synthesis, the manufacturer's protocol was followed, starting with 1.35 μg total RNA template and 50 ng random hexamers. Following synthesis, the cDNA was purified on a QIAquick column (Qiagen), quantified using a NanoDrop spectrophotometer, and diluted to 5 ng/μl with nuclease-free water. RNA transcripts were quantified on an MX3000P real-time PCR machine (Stratagene), using Brilliant Sybr green QPCR mix (Stratagene) in 25-μl volumes containing 25 ng cDNA. Optimal primer concentrations were determined empirically. For comparative, quantitative analysis, transcript levels were normalized to the level of gapA (glyceraldehyde 3-phosphate dehydrogenase A gene), and changes were determined using an experiment-specific calibrator (LB) and the MXPro v 3.00 software package (Stratagene). For in vivo quantitative PCR, RNAs were purified as described above from pooled urines from groups of mice experimentally infected with CFT073 to determine transcript levels during UTI.
Construction of mutants in UPEC strain CFT073.
Deletion mutants were generated using the lambda red recombinase system (12). Primers homologous to sequences within the 5′ and 3′ ends of the target genes were designed and used to replace target genes with a nonpolar kanamycin resistance cassette derived from the template plasmid pKD4 (12). Kanamycin (25 μg/ml) was used for selection of all mutant strains. Gene deletions began with the start codon and ended with the stop codon for each gene. To determine whether the kanamycin resistance cassette recombined within the target gene site, primers that flanked the target gene sequence were designed and used for PCR. After amplification, each PCR product was compared to the wild-type PCR product, and in cases where size differences were negligible, PCR products were digested with the restriction enzyme EagI (New England BioLabs). Both the PCR products and restriction digests were visualized in a 0.8% agarose gel stained with ethidium bromide.
Experimental UTI.
A murine model of ascending UTI was used to assess virulence and fitness (20). Six- to 8-week-old female CBA/J mice (20 to 22 g; Jackson Laboratories) were inoculated transurethrally with a 50-μl bacterial suspension per mouse by use of a sterile polyethylene catheter (inner diameter, 0.28 mm; outer diameter, 0.61 mm) connected to an infusion pump (Harvard Apparatus). For independent challenges, overnight LB cultures of CFT073 and an isogenic mutant were collected by centrifugation, resuspended in sterile PBS, and adjusted to deliver 2 × 108 CFU per mouse. To measure the relative fitness of mutants and the wild type, overnight LB cultures of CFT073 and individual mutant strains were collected by centrifugation, resuspended in sterile PBS, mixed 1:1, and adjusted to deliver 4 × 108 CFU per mouse. Dilutions of each inoculum were spiral plated onto LB, with and without kanamycin, by use of an Autoplate 4000 instrument (Spiral Biotech) to determine the input CFU/ml. At 48 h postinoculation, mice from independent challenge or cochallenge groups were sacrificed by overdose with isoflurane, and the bladder and kidneys were aseptically removed, weighed, and homogenized in sterile culture tubes containing 3 ml of PBS by use of an Omni mechanical homogenizer (Omni International). Appropriate dilutions of the homogenized tissue were then spiral plated onto duplicate LB plates, with and without kanamycin, to determine the output CFU/g of tissue, and differences in colonization were determined using the Mann-Whitney significance test. For cochallenge, plate counts obtained with kanamycin plates were subtracted from those for plates lacking antibiotic to determine the number of wild-type bacteria. Statistically significant differences in colonization (P < 0.05) were determined using a two-tailed Wilcoxon matched-pair test. All animal protocols were approved by the University Committee on Use and Care of Animals at the University of Michigan Medical School.
RESULTS
Pooled and adsorbed sera from chronically infected mice do not react with uropathogenic E. coli cultured in vitro.
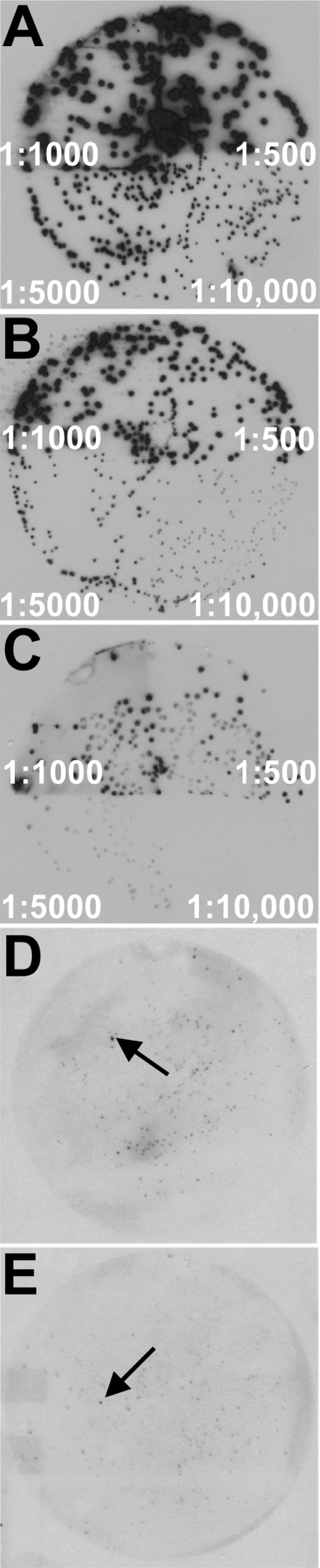
Sera collected from 20 female CBA/J mice inoculated transurethrally with UPEC isolate CFT073 in a chronic UTI infection model were collected as previously described (19). Sera were adsorbed against whole cells of strain CFT073 cultured in vitro and then adsorbed against native and denatured whole-cell lysates of strain CFT073 immobilized on a membrane in order to expose the sera to both surface-exposed and nonexposed proteins synthesized during in vitro growth. This adsorption process was repeated with the host expression strain E. coli BL21(DE3)/pLysS, which carried the genomic expression library used for the IVIAT screening. Multiple rounds of adsorption were conducted until colony immunoblots of both bacterial strains registered negligible background levels of reactivity with the treated sera compared to unabsorbed sera (Fig. 1 A to C).
Fig. 1.
Identification of antigens induced by UPEC in vivo. Colony Western blots were performed on E. coli CFT073 colonies, using pooled sera from chronically infected mice (A), adsorbed sera (B), or BL21/pET30a probed with adsorbed sera (C). Representative primary screening results showing reactive BL21 clones expressing CFT073 in vivo-induced antigens are indicated with arrows in panels D and E. Dilutions of primary sera are indicated in panels A to C.
Screening of a CFT073 genomic expression library with adsorbed sera identified candidate in vivo-expressed genes.
Fragments of DNA (1.0 to 1.5 kb) were size fractionated from genomic DNA isolated from the UPEC isolate CFT073, ligated into an IPTG-inducible vector that permits cloning in all three possible open reading frames, and transformed into E. coli BL21(DE3)/pLysS. The CFT073 genome contains 5,079 predicted protein coding sequences; to achieve adequate representation, it was desirable to obtain 50,000 clones with an insert size of 1.0 kb. After multiple transformations and selection, we obtained 60,000 clones, with 24 of 24 representative clones having inserts of >1.0 kb. The resulting genomic expression library is predicted to cover most of the repertoire of protein antigens contained in the CFT073 genome.
Primary screening of the adsorbed sera was conducted using colony immunoblots of the genomic expression library. A total of 461 clones that exhibited reactivity significantly above background (Fig. 1D and E) were isolated and subjected to a second round of screening. Three replicate colony immunoblots were conducted during secondary screening to minimize false-positive results. A total of 83 clones remained reactive to the adsorbed sera in at least two of the three replicate screens. Plasmids from these clones were isolated and sequenced to identify the gene fragments identified by IVIAT screening (Table 1). Ten inserts contained DNA that spanned two genes, bringing the total number of candidate in vivo-expressed genes to 93.
Table 1.
Genes identified by IVIAT as induced in uropathogenic E. coli during experimental urinary tract infection
| Genea | Open reading frame | Function of gene product |
|---|---|---|
| Transport genes | ||
| yddQ* | c5079 | Hypothetical dipeptide ABC transporter, permease subunit |
| proW*-proX* | c3231-c3232 | Glycine betaine transporter |
| yjcQ | c5086 | Multidrug efflux system protein MdtO |
| kpsS* | c3691 | Capsule polysaccharide export |
| cirA | c2690 | Ferric iron-catecholate outer membrane receptor |
| fliY | c2335 | Cystine transporter subunit |
| lolD* | c1392 | Lipoprotein transporter ATP-binding subunit |
| lolA* | c1028 | Outer membrane lipoprotein carrier protein |
| ybeX | c0743 | Magnesium and cobalt efflux protein CorC |
| sapB | c1770 | Peptide transport system permease protein SapB |
| btuD-btuE | c2105-c2106 | Vitamin B12 transporter |
| malM-lamB | c5007-c5006 | Maltose regulon periplasmic protein; maltoporin |
| iucC | c3625 | Aerobactin siderophore synthesis |
| fhuC | c0186 | Iron-hydroxamate transporter ATP-binding subunit |
| c4487 | Putative fructose-specific phosphotransferase system | |
| ydjK | c2179 | Hypothetical metabolite transport protein |
| yphD | c3068 | Hypothetical ABC transporter permease |
| ydiM | c2085 | Hypothetical transport protein |
| yieO | c4682 | Hypothetical transport protein |
| ydhP | c2051 | Hypothetical transport protein |
| ybhR | c0875 | Hypothetical ABC transporter |
| yijE | c4901 | Hypothetical transport protein |
| yajQ-yajR | c0537 | Hypothetical transporter |
| yehZ | c2661 | Hypothetical transporter |
| Regulatory genes | ||
| fhlA | c3291 | Formate hydrogen lyase transcriptional activator |
| cytR | c4887 | DNA-binding transcriptional regulator |
| hupB | c0556 | Transcriptional regulator HU subunit beta |
| yaeG | c0199 | Carbohydrate diacid transcriptional activator CdaR |
| nlp | c3946 | DNA-binding transcriptional activator of maltose metabolism |
| yijO | c4913 | Hypothetical transcriptional regulator |
| ybiH | c0879 | Putative DNA-binding transcriptional regulator |
| Secreted gene | ||
| tosA*b | c0363 | Putative RTX family exoprotein A gene |
| Genes involved in metabolism | ||
| accA-ldcC | c0223-c0224 | Acetyl-CoA carboxylase; constitutive lysine decarboxylase |
| adhP* | c1911 | Alcohol dehydrogenase |
| nanA-nanT | c3979-c3978 | N-Acetylneuraminate lyase; sialic acid transporter |
| phnJ | c5104 | Phosphate metabolism |
| c2432* | Putative thioesterase | |
| aroG | c0830 | Phospho-2-dehydro-3-deoxyheptonate aldolase |
| hemA | c1668 | Glutamyl-tRNA reductase |
| citC | c0709 | [Citrate [pro-3S]-lyase] ligase |
| cdd | c2675 | Cytidine deaminase |
| celA | c1955 | 6-Phospho-beta-glucosidase |
| astD | c2146 | Arginine succinyltransferase |
| c2147 | Succinylglutamic semialdehyde dehydrogenase | |
| fdhF | c5625 | Formate dehydrogenase H |
| adhE | c1705 | Acetaldehyde dehydrogenase |
| metH | c4976 | Vitamin B12-dependent methionine synthase |
| asnA | c4672 | Asparagine synthetase AsnA |
| yphC | c3067 | Hypothetical zinc-type alcohol dehydrogenase |
| ygjK | c3838 | Predicted glycosyl hydrolase |
| rhaB | c4853 | Rhamnulokinase |
| phnM | c5101 | Carbon-phosphate lyase |
| accC | c4012 | Acetyl-CoA carboxylase biotin carboxylase subunit |
| prsA | c1665 | Ribose-phosphate pyrophosphokinase |
| nudF | c3780 | ADP-ribose pyrophosphatase NudF |
| katG* | c4900 | Peroxidase/catalase |
| ispB | c3945 | Octaprenyl diphosphate synthase |
| ivbL-ivbB | c5498-c4596 | IlvB operon leader peptide; acetolactate synthase catalytic subunit |
| hyaF | c1118 | Hydrogenase-1 operon protein |
| hypE | c3290 | Hydrogenase isoenzyme formation protein HypE |
| narJ*-narI* | c1687-c1688 | Respiratory nitrate reductase 1 |
| yhjA | c4329 | Probable cytochrome c peroxidase |
| pnp | c3920 | Polynucleotide phosphorylase/polyadenylase |
| wecC-wecB | c4707-c4706 | UDP-N-acetyl-d-mannosamine dehydrogenase; UDP-N-acetylglucosamine 2-epimerase |
| prfC | c5456 | Peptide chain release factor 3 |
| rpsS | c4083 | 30S ribosomal protein S19 |
| ycfH | c1372 | Predicted metallodependent hydrolase |
| holB | c1371 | DNA polymerase III subunit delta′ |
| yebU | c2244 | rRNA (cytosine-C(5)-)-methyltransferase RsmF |
| fusA | c4112 | Elongation factor G |
| fhiA | c0377 | FhiA protein (FhlA homolog) |
| yegD | c2596 | Putative chaperone |
| ygbJ | c3297 | Hypothetical oxidoreductase |
| yajO | c0530 | Hypothetical oxidoreductase |
| ydhX* | c2065 | Putative ferredoxin-like protein |
| yjgB | c5370 | Hypothetical zinc-type alcohol dehydrogenase-like protein |
| Hypothetical protein genes | ||
| yahJ | c0445 | Hypothetical deaminase |
| c1590/3154 | Phage-related protein | |
| yedE | c2344 | Predicted inner membrane protein |
| c5347-c5348 | Hypothetical protein | |
| c3664 | Hypothetical protein | |
| yfeK | c2954 | Hypothetical protein |
| c2770 | Hypothetical protein | |
| c4743 | Hypothetical protein | |
| yqiB | c3779 | Hypothetical protein |
| c3293-c3291 | Hypothetical protein | |
| c0446 | Hypothetical protein | |
| yjjJ | c5470 | Hypothetical protein |
| aroM | c0498 | Hypothetical protein |
| yjiN | c5419 | Hypothetical protein |
| yedJ | c2381 | Hypothetical protein |
| yicH | c4480 | Hypothetical protein YicH |
| ycfC | c1511 | Hypothetical protein |
*, genes selected for qPCR validation of expression levels in vivo.
Also known as upxA (39).
A subset of the IVIAT-identified genes have known roles during UPEC colonization of the urinary tract and were not investigated further in this study. iucC and fhuC, both involved in hydroxymate sideophore transport, were identified in our screen, consistent with iron acquisition being a key virulence trait for UPEC during UTI. NanA, involved in the transport and catabolism of sialic acid, was previously shown to be upregulated in human urine; however, a mutant had no apparent fitness defect during UTI (4). Interestingly, the same mutant strain demonstrated a significant fitness defect in vivo in a bacteremia model (34). Thus, finding these antigens by use of IVIAT suggests that other genes identified in the screen may also have important roles in UPEC pathogenesis.
Validation of in vivo expression pattern by qPCR of bacterial RNAs from pooled urines from the murine model.
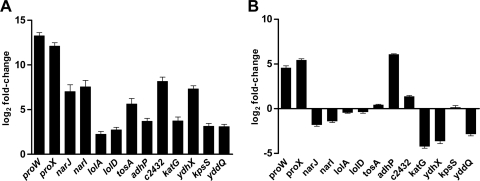
To verify that the adsorbed sera identified genes that were expressed in vivo but not in vitro, 13 of the genes identified in this study were selected for validation of in vivo expression by qPCR (Table 1). These genes, yddQ, proWX, kpsS, lolA, lolD, tosA (upxA), adhP, c2432, katG, narJI, and ydhX, were selected as representative genes from six categories, including those involved in transport, regulation, secreted proteins, metabolism, cellular processes, and hypothetical function. The predicted dipeptide transport component gene yddQ was selected because the known peptide transport genes dppA and oppA are required for UPEC fitness in vivo (4). Similarly, the roles of osmoprotection within the urinary tract (proWX) and of the capsule (kpsS) have previously been shown to be important for UPEC pathogenesis in vivo (5, 11). The remaining genes selected for qPCR validation represent putative or known metabolism or transport genes that have unknown contributions to UPEC virulence or fitness during UTI.
Pooled urines from three replicate groups, each containing five female CBA/J mice that had been inoculated transurethrally with strain CFT073, were collected three times a day for 3 days postinoculation. RNAs were isolated from bacteria from within each pooled urine sample for qPCR analysis, and data were compared to results for RNA extracted from strain CFT073 cultured in vitro under the same conditions used initially to adsorb the sera. The results demonstrated that each of the 13 genes was expressed more highly in vivo than in bacteria cultured under standard laboratory conditions (Fig. 2 A). The greatest increase in expression in vivo over in vitro was an increase of >2,000-fold for the proW and proX genes. With the exception of lolA and lolD, the increase in expression was always ≥4-fold for all 11 other genes examined by qPCR. This demonstrates the ability of the IVIAT screening approach to identify genes that are expressed manifold higher in vivo than under standard in vitro culture conditions.
Fig. 2.
Quantitative real-time qPCR for transcripts of CFT073 in vivo-induced antigen genes during experimental ascending UTI (A) or during growth in human urine (B). Data were normalized to gapA (glyceraldehyde 3-phosphate dehydrogenase) expression levels, and changes were determined using expression levels during growth in vitro in LB medium as the calibrator to generate fold change values.
Bacteria incubated in pooled filter-sterilized human urine, a condition that may mimic in vivo conditions, showed increased expression of 4 of the 13 genes examined by qPCR, >2-fold over that for growth in LB (proW, proX, adhP, and c2432) (Fig. 2B). The remaining 9 genes were expressed at comparable or lower levels than those in bacteria cultured aerobically to exponential phase (OD600 = 1.0) in LB. katG, ydhX, and yddQ were downregulated significantly in human urine compared to their levels in LB. While the expression in this specialized growth medium more closely resembled expression patterns in vivo, the IVIAT approach was clearly more successful at identifying genes specific for growth in vivo during the course of an infection and identified many genes that would have been overlooked by methods relying strictly on in vitro growth to mimic in vivo conditions.
IVIAT identified the putative virulence gene tosA.
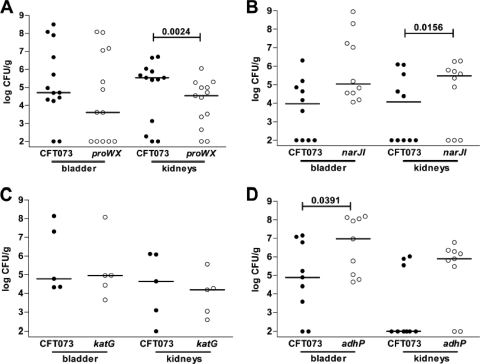
Five isogenic mutant strains of CFT073 were constructed using lambda red recombinase methodology (tosA, proWX, katG, adhP, and narJI) (12) and were tested for colonization in a murine model of ascending UTI. Of the five genes under study, narJI and tosA had high in vivo expression levels by qPCR, despite little or no upregulation in human urine, proWX demonstrated the highest in vivo expression levels, katG was upregulated in vivo and downregulated in human urine, and adhP displayed substantial upregulation both in vivo and in human urine (Fig. 2). CFT073 mutants in each of these genes were competed against wild-type CFT073 in cochallenge infections or tested by independent challenge in the murine model of ascending UTI. CFU levels in the bladder and kidney were quantified for each strain at 48 h postinoculation (Fig. 3 and 4).
Fig. 3.
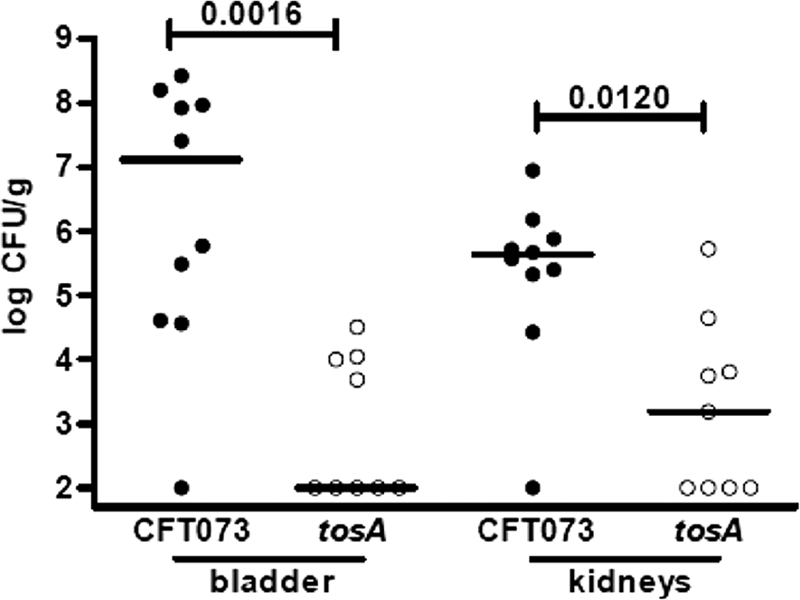
A putative RTX family exoprotein gene, tosA, is required for virulence of UPEC. Bladder and kidney colonization levels were determined at 48 h post-transurethral inoculation following independent challenge with wild-type uropathogenic E. coli CFT073 and its ΔtosA mutant. Circles represent the log10 CFU/g from individual mice, and horizontal bars represent the median CFU/g. P values were determined using the nonparametric Mann-Whitney significance test.
Fig. 4.
Assessment of fitness for selected in vivo-induced antigen mutants in cochallenge infections. Individual female mice were inoculated transurethrally with 2 × 108 CFU of a 1:1 mixture of wild-type and mutant bacteria. In vivo fitness was determined at 48 h postinoculation for UPEC mutants lacking proWX (A), narJI (B), katG (C), and adhP (D). At 48 h postinoculation, bladders and kidneys were removed aseptically, homogenized, and plated on LB and LB containing kanamycin to determine viable counts of wild-type and mutant strains, respectively. In all cochallenges depicted, 37 of 37 mice had detectable CFU of the wild type and/or mutant in the bladder; all but 5 (2 for the adhP mutant, 1 for the proWX mutant, and 2 for the narJI mutant) of 37 mice had detectable CFU of the wild type and/or mutant in the kidneys. Each filled circle represents the log10 CFU/g from an individual animal. Bars represent the median CFU/g, and the limit of detection was 100 CFU. Significant differences in colonization levels (P < 0.05) are indicated and were determined using a two-tailed Wilcoxon matched-pair test.
In a previous study, our group reported on the contribution of the 100-kb pathogenicity-associated island PAICFT073-aspV, containing the RTX family member tosA, to fitness in the murine urinary tract (26). In the current study, IVIAT independently identified TosA as an antigen that is expressed preferentially in vivo (Table 1 and Fig. 2A). tosA expression was determined to be specific to the in vivo environment, and expression above background levels was not observed when bacteria were cultured in vitro in pooled filter-sterilized human urine (Fig. 2B). A ΔtosA mutant of CFT073 was tested in the murine model of ascending UTI, and colonization levels in the bladder and kidneys were compared to colonization levels observed when the parental wild-type strain was tested independently (Fig. 3). The ΔtosA mutant was severely attenuated in the animal model of infection; significantly smaller numbers of bacteria were recovered from both bladder (P = 0.0016) and kidney (P = 0.0120) tissue than those for the wild-type strain. Thus, the IVIAT approach employed in this study identified a new virulence factor of UPEC that is specifically induced in vivo and contributes significantly to the pathogenesis of this strain of E. coli in an animal model of urinary tract infection.
proWX, identified by IVIAT, contributes to fitness in a murine model of UTI.
The proWX genes are involved in osmoregulation by encoding the proline betaine transport system. An isogenic mutant that contained a single deletion covering the proW and proX genes, corresponding to the region of the genome contained in a single expression clone identified by IVIAT, was outcompeted by the parental strain in the kidney (P = 0.0024) (Fig. 4A). The data from this cochallenge illustrate that the IVIAT screen may identify genes and systems that confer an advantage during UPEC colonization of the urinary tract.
The mutant lacking catalase, encoded by katG, was recovered in numbers equal to those for the wild type (Fig. 4B) and thus did not have a fitness advantage, so katG could not be considered a virulence factor. Surprisingly, the ΔadhP mutant, lacking an alcohol peroxidase (Fig. 4C) which was very highly expressed in human urine (Fig. 2B), and the ΔnarJI mutant, lacking an anaerobic nitrate reductase I (Fig. 4D), were both recovered in significantly larger numbers than the wild type in the bladder (P = 0.0391) and kidneys (P = 0.0156), respectively. While each of these genes was properly identified by the IVIAT approach as being expressed at higher levels in vivo (Fig. 2A) and eliciting a host immune response, these results indicate that not all of the identified genes confer a fitness advantage in the host environment. Taken together with the other animal model results, the remaining IVIAT-identified genes would need to be studied independently in greater detail to understand the importance of each gene to the pathogenesis of UTI.
DISCUSSION
Two of the genes identified by the IVIAT screening approach employed in this study, encoding a putative secreted RTX family exoprotein (tosA) and an osmoprotection system that transports glycine betaine (proWX), contribute to fitness of UPEC in the host urinary tract (Fig. 3 and 4). In addition, more than 90 additional genes were identified by IVIAT (Table 1), and many of these may also contribute to the pathogenesis of UTI. Of particular note is the observation that of the 93 identified genes, 23 genes have not been studied previously and were assigned a function based only on sequence homology to known proteins. Seventeen additional genes did not demonstrate sufficient homology to be assigned a function. The putative RTX family member tosA (originally annotated as upxA [39]), which was determined in this study to contribute significantly to the ability of the human pyelonephritis isolate CFT073 to colonize a murine UTI model (Fig. 3), is just one example from this group of genes of unknown function. Because the IVIAT method relies on antibodies made by the host during an infection (30), this list also represents UPEC antigens that the host immune system recognized and against which an immune response was mounted. While additional studies will be needed to clarify the roles of those genes not pursued further in this study, this list may contain useful targets for the development of a UPEC vaccine.
Several limitations of the project must be addressed to clarify the results of this study. Due to the limited quantity of murine antiserum available, only 40,000 clones of the genomic expression library were screened. Because our group calculated that ∼50,000 clones would be needed to obtain 95% genome coverage, Table 1 represents only a partial but nevertheless substantial list of genes preferentially expressed in vivo and their encoded proteins that are recognized as antigenic by the host. In addition, for a gene product to be recognized by IVIAT, the antibody titer must be sufficiently high to obtain a consistently strong signal on immunoblots in both primary and secondary screening. Next, any proteins produced during in vitro growth, even those with known contributions to fitness in the urinary tract, would be available to remove specific antibodies during the serum adsorption process and are not predicted to be recognized by IVIAT. Finally, there was difficulty in constructing a stable plasmid containing the 4.8-kb tosA gene, and thus complementation was not confirmed in the independent challenge (Fig. 3). These limitations may explain why many well-characterized UPEC virulence factors, such as P fimbriae, which are known to be expressed during in vitro growth on LB agar (10), were not identified by this screen.
Despite these caveats, the results of the IVIAT screen clarify and reinforce previous observations about general metabolic processes that UPEC employs to colonize the host urinary tract. Our findings also revealed some overlap with a similar screen of enterohemorrhagic E. coli (EHEC) carried out using convalescent-phase sera from patients following recovery from hemolytic-uremic syndrome (22). Both the current results and the EHEC study identified adhP, osmolarity homeostasis genes, components of the maltose regulon, antigens representing enzymes involved in acetyl-coenzyme A (acetyl-CoA) metabolism (accC), and hydrogenase and additional respiratory components involved in the reduction of nitrate (22). It is not surprising that these core or backbone components of the E. coli chromosome were identified in both screens. For UPEC, finding respiratory components as in vivo-induced antigens is consistent with the fitness requirement for the tricarboxylic acid (TCA) cycle during UTI (4) and the utilization of alternate electron acceptors during “overflow” metabolism that occurs from rapid growth in vivo (18).
While many of the processes identified by IVIAT may be active during the course of a UTI, not all of the systems studied in the murine model demonstrated a clear fitness advantage. For example, isogenic mutants of CFT073 deficient in the adhP, katG, narJ, and narI genes were all successful in competing against wild-type CFT073 during cochallenge experiments (Fig. 4). Each of these genes was determined to be upregulated in vivo compared to growth in LB medium, and expression of the narJ, narI, and katG genes was specific to the in vivo environment and not induced during growth in pooled filtered human urine. While the results for the ΔkatG strain were unexpected, precedence exists in descriptions of full virulence of catalase-defective strains of Neisseria gonorrhoeae and Listeria monocytogenes in animal models of infection (24, 36). It is not clear why these mutants were able to compete equally with wild-type bacteria, but this does illustrate the importance of following up an IVIAT screen with specific experiments to determine the relative importance of the genes identified in a large genome-wide screen such as IVIAT to the fitness of an organism during an infection.
The proW and proX genes, part of the proU osmoprotection system, increased the fitness of the human pyelonephritis isolate CFT073 in the kidneys of infected mice (Fig. 4). A trend toward lower CFU of the ΔproWX mutant was also observed in the bladder on cochallenge with wild-type CFT073. Previous research identified the major substrates of the proU system in E. coli as glycine betaine and proline betaine (17). Both compounds are present in human urine, and the presence of either compound increases the tolerance of E. coli to higher concentrations of osmotic agents than those tolerated without the compounds present (9), suggesting an important role for the transport of these compounds as an adaptation by UPEC to survive the environment of the host urinary tract. Additional studies on the role of the proU system in the human pyelonephritis strains CFT073 and HU734 found that a ΔproU mutant of each strain colonized the murine model of ascending UTI at the same levels as the wild type in both the bladder and kidneys of infected mice (11). Additional osmoprotection systems have been identified in E. coli (11, 41), and this functional redundancy may help to explain why a strain of UPEC that lacks the proU system can still efficiently colonize the urinary tract. In contrast to the current study, which mixed wild-type CFT073 at an equal ratio with the mutant and coinoculated the two strains into the same animal, previous experiments were conducted as independent challenges of each strain's ability to colonize infected mice in separate infections. Indeed, we consider cochallenge a more sensitive index of fitness than independent challenge. Considered together, the results of this study help to clarify the role of the proU osmoprotection system during an experimental UTI. While this system is not necessary for colonization, the expression of the system is strongly induced in both an in vivo infection model and growth in vitro in human urine, and the presence of the system contributes to enhanced fitness of UPEC in the kidneys of infected animals.
The IVIAT-identified gene tosA (upxA) was originally described in the genome annotation of CFT073 as a UPEC-specific gene not present in the genomes of commensal E. coli or other E. coli pathotypes (39). The 4.8-kb gene appears as part of a 9.5-kb locus on PAICFT073-aspV that appears to be linked to a type I secretion system (29). While the components of the type I secretion system are well conserved with other RTX family members, the predicted secreted target of the system, TosA, is poorly conserved with other known RTX family members (29). Further differences from the hlyA locus are noted in the organization of the locus in both gene order and gene content in CFT073 (39), suggesting that tosA may function differently from hlyA. In a previous study, our group identified PAICFT073-aspV containing the tosA gene as contributing enhanced fitness to CFT073 in an animal model of infection (26). In the current study, the IVIAT screening approach independently identified TosA as an antigenic protein synthesized in the host urinary tract.
Expression of the RTX family member tosA was determined to be specific to the in vivo environment of the host urinary tract. Unlike the case for the genes of the proU system, tosA expression was measurable only in bacteria that had been growing in vivo (i.e., in the urine of infected mice). Expression of tosA was not induced under any in vitro condition studied, including growth in human urine (Fig. 2B). This limited expression pattern stands in contrast to the pattern for hlyA, the gene that encodes the prototype RTX toxin alpha-hemolysin, which is expressed during in vitro growth in LB medium (25), the same conditions utilized for the reverse transcription-qPCR (RT-qPCR) study presented in Fig. 2B. While the UPEC strain CFT073 contains the two RTX family members tosA and hlyA, the regulatory mechanisms governing expression of each system appear to be different. This raises the possibility that a specific host factor not present in human urine may have triggered the expression of tosA observed in this study.
A ΔtosA strain of CFT073 was attenuated in both the bladder and kidneys of infected mice following independent challenge (Fig. 3). While our previous study identified a role for tosA in enhancing fitness in the murine model of infection during cochallenge with wild-type bacteria (26), the results of the current study indicate that the gene serves a more critical function during infection. In an independent challenge, approximately half of the mice infected with the ΔtosA strain had undetectable levels of bacteria in bladder and kidney tissue (≤102 CFU/g tissue), as opposed to only 1 of 10 mice infected with the wild-type strain (P = 0.043). In addition to the gene regulation differences, this result also indicates a role for tosA that is different from that for hlyA. While ΔhlyA mutants of UPEC are attenuated in animal models of infection outside the urinary tract, notably in several bacteremia models (7, 40), no defect in colonization of bladder or kidney tissue was observed in a similar murine UTI model (35). Both in the urinary tract and during in vitro assays, alpha-hemolysin mediates damage to host cells (35, 37) but is not necessary for successful colonization of the host urinary tract. Additional studies will be necessary to elucidate the function of tosA, but the results of this study implicate a different and important role for this novel RTX family member during a UTI mediated by E. coli.
The results of this study demonstrate the utility of the IVIAT approach to clarify the roles of genes with known functions (e.g., the proU system) as well as to identify genes of unknown function (e.g., tosA) that play an important role in the success of bacterial pathogens in the host environment. The results of the IVIAT screen hold particular importance for the development of new experimental vaccines, as the genes identified are known to be expressed preferentially in vivo and elicit a host immune response in the formation of the antibodies utilized by the IVIAT screen. While many mechanisms of UPEC pathogenesis have been well characterized, the results of the IVIAT screen presented here indicate that much remains to be learned about the specific mechanisms utilized by E. coli during a UTI.
ACKNOWLEDGMENTS
This work was supported in part by Public Health Service grant AI43363 from the National Institutes of Health.
We thank Sara Smith for assistance in conducting the animal studies.
Footnotes
Published ahead of print on 21 March 2011.
REFERENCES
- 1. Aboderin O. A., Abdu A. R., Odetoyin B. W., Lamikanra A. 2009. Antimicrobial resistance in Escherichia coli strains from urinary tract infections. J. Natl. Med. Assoc. 101:1268–1273 [DOI] [PubMed] [Google Scholar]
- 2. Alteri C. J., Hagan E. C., Sivick K. E., Smith S. N., Mobley H. L. 2009. Mucosal immunization with iron receptor antigens protects against urinary tract infection. PLoS Pathog. 5:e1000586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alteri C. J., Mobley H. L. 2007. Quantitative profile of the uropathogenic Escherichia coli outer membrane proteome during growth in human urine. Infect. Immun. 75:2679–2688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Alteri C. J., Smith S. N., Mobley H. L. 2009. Fitness of Escherichia coli during urinary tract infection requires gluconeogenesis and the TCA cycle. PLoS Pathog. 5:e1000448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bahrani-Mougeot F. K., et al. 2002. Type 1 fimbriae and extracellular polysaccharides are preeminent uropathogenic Escherichia coli virulence determinants in the murine urinary tract. Mol. Microbiol. 45:1079–1093 [DOI] [PubMed] [Google Scholar]
- 6. Bours P. H., et al. 2010. Increasing resistance in community-acquired urinary tract infections in Latin America, five years after the implementation of national therapeutic guidelines. Int. J. Infect. Dis. 14:e770–e774 [DOI] [PubMed] [Google Scholar]
- 7. Brzuszkiewicz E., et al. 2006. How to become a uropathogen: comparative genomic analysis of extraintestinal pathogenic Escherichia coli strains. Proc. Natl. Acad. Sci. U. S. A. 103:12879–12884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chakupurakal R., Ahmed M., Sobithadevi D. N., Chinnappan S., Reynolds T. 2010. Urinary tract pathogens and resistance pattern. J. Clin. Pathol. 63:652–654 [DOI] [PubMed] [Google Scholar]
- 9. Chambers S. T., Kunin C. M. 1987. Isolation of glycine betaine and proline betaine from human urine. Assessment of their role as osmoprotective agents for bacteria and the kidney. J. Clin. Invest. 79:731–737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Culham D. E., et al. 1998. Osmoregulatory transporter ProP influences colonization of the urinary tract by Escherichia coli. Microbiology 144:91–102 [DOI] [PubMed] [Google Scholar]
- 11. Culham D. E., et al. 2001. The osmotic stress response and virulence in pyelonephritis isolates of Escherichia coli: contributions of RpoS, ProP, ProU and other systems. Microbiology 147:1657–1670 [DOI] [PubMed] [Google Scholar]
- 12. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deb D. K., Dahiya P., Srivastava K. K., Srivastava R., Srivastava B. S. 2002. Selective identification of new therapeutic targets of Mycobacterium tuberculosis by IVIAT approach. Tuberculosis (Edinburgh) 82:175–182 [DOI] [PubMed] [Google Scholar]
- 14. Dielubanza E. J., Schaeffer A. J. 2011. Urinary tract infections in women. Med. Clin. North Am. 95:27–41 [DOI] [PubMed] [Google Scholar]
- 15. Foxman B. 2002. Epidemiology of urinary tract infections: incidence, morbidity, and economic costs. Am. J. Med. 113(Suppl. 1A):5S–13S [DOI] [PubMed] [Google Scholar]
- 16. Griebling T. L. 2005. Urologic diseases in America project: trends in resource use for urinary tract infections in women. J. Urol. 173:1281–1287 [DOI] [PubMed] [Google Scholar]
- 17. Haardt M., Kempf B., Faatz E., Bremer E. 1995. The osmoprotectant proline betaine is a major substrate for the binding-protein-dependent transport system ProU of Escherichia coli K-12. Mol. Gen. Genet. 246:783–786 [DOI] [PubMed] [Google Scholar]
- 18. Hagan E. C., Lloyd A. L., Rasko D. A., Faerber G. J., Mobley H. L. 2010. Escherichia coli global gene expression in urine from women with urinary tract infection. PLoS Pathog. 6:e1001187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hagan E. C., Mobley H. L. 2007. Uropathogenic Escherichia coli outer membrane antigens expressed during urinary tract infection. Infect. Immun. 75:3941–3949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagberg L., et al. 1983. Ascending, unobstructed urinary tract infection in mice caused by pyelonephritogenic Escherichia coli of human origin. Infect. Immun. 40:273–283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hang L., et al. 2003. Use of in vivo-induced antigen technology (IVIAT) to identify genes uniquely expressed during human infection with Vibrio cholerae. Proc. Natl. Acad. Sci. U. S. A. 100:8508–8513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. John M., et al. 2005. Use of in vivo-induced antigen technology for identification of Escherichia coli O157:H7 proteins expressed during human infection. Infect. Immun. 73:2665–2679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kumar M., et al. 2011. Identification of Mycobacterium tuberculosis genes preferentially expressed during human infection. Microb. Pathog. 50:31–38 [DOI] [PubMed] [Google Scholar]
- 24. Leblond-Francillard M., Gaillard J. L., Berche P. 1989. Loss of catalase activity in Tn1545-induced mutants does not reduce growth of Listeria monocytogenes in vivo. Infect. Immun. 57:2569–2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Leeds J. A., Welch R. A. 1996. RfaH enhances elongation of Escherichia coli hlyCABD mRNA. J. Bacteriol. 178:1850–1857 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lloyd A. L., Henderson T. A., Vigil P. D., Mobley H. L. 2009. Genomic islands of uropathogenic Escherichia coli contribute to virulence. J. Bacteriol. 191:3469–3481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mobley H. L., et al. 1990. Pyelonephritogenic Escherichia coli and killing of cultured human renal proximal tubular epithelial cells: role of hemolysin in some strains. Infect. Immun. 58:1281–1289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nielubowicz G. R., Mobley H. L. 2010. Host-pathogen interactions in urinary tract infection. Nat. Rev. Urol. 7:430–441 [DOI] [PubMed] [Google Scholar]
- 29. Parham N. J., et al. 2005. Prevalence of pathogenicity island II CFT073 genes among extraintestinal clinical isolates of Escherichia coli. J. Clin. Microbiol. 43:2425–2434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rollins S. M., et al. 2005. In vivo induced antigen technology (IVIAT). Cell. Microbiol. 7:1–9 [DOI] [PubMed] [Google Scholar]
- 31. Salim K. Y., et al. 2005. Identification of group A streptococcus antigenic determinants upregulated in vivo. Infect. Immun. 73:6026–6038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sivick K. E., Mobley H. L. 2009. An “omics” approach to uropathogenic Escherichia coli vaccinology. Trends Microbiol. 17:431–432 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sivick K. E., Mobley H. L. 2010. Waging war against uropathogenic Escherichia coli: winning back the urinary tract. Infect. Immun. 78:568–585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Smith S. N., Hagan E. C., Lane M. C., Mobley H. L. 2010. Dissemination and systemic colonization of uropathogenic Escherichia coli in a murine model of bacteremia. MBio 1:e00262–10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Smith Y. C., Rasmussen S. B., Grande K. K., Conran R. M., O'Brien A. D. 2008. Hemolysin of uropathogenic Escherichia coli evokes extensive shedding of the uroepithelium and hemorrhage in bladder tissue within the first 24 hours after intraurethral inoculation of mice. Infect. Immun. 76:2978–2990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Soler-Garcia A. A., Jerse A. E. 2007. Neisseria gonorrhoeae catalase is not required for experimental genital tract infection despite the induction of a localized neutrophil response. Infect. Immun. 75:2225–2233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Trifillis A. L., et al. 1994. Binding to and killing of human renal epithelial cells by hemolytic P-fimbriated E. coli. Kidney Int. 46:1083–1091 [DOI] [PubMed] [Google Scholar]
- 38. Warren J. (ed.). 1996. Urinary tract infections: molecular pathogenesis and clinical management. ASM Press, Washington, DC [Google Scholar]
- 39. Welch R. A., et al. 2002. Extensive mosaic structure revealed by the complete genome sequence of uropathogenic Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 99:17020–17024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Welch R. A., Dellinger E. P., Minshew B., Falkow S. 1981. Haemolysin contributes to virulence of extra-intestinal E. coli infections. Nature 294:665–667 [DOI] [PubMed] [Google Scholar]
- 41. Wood J. M. 1999. Osmosensing by bacteria: signals and membrane-based sensors. Microbiol. Mol. Biol. Rev. 63:230–262 [DOI] [PMC free article] [PubMed] [Google Scholar]