Abstract
The Kdp system is widely distributed among bacteria. In Escherichia coli, the Kdp-ATPase is a high-affinity K+ uptake system and its expression is activated by the KdpDE two-component system in response to K+ limitation or salt stress. However, information about the role of this system in many bacteria still remains obscure. Here we demonstrate that KdpFABC in Staphylococcus aureus is not a major K+ transporter and that the main function of KdpDE is not associated with K+ transport but that instead it regulates transcription for a series of virulence factors through sensing external K+ concentrations, indicating that this bacterium might modulate its infectious status through sensing specific external K+ stimuli in different environments. Our results further reveal that S. aureus KdpDE is upregulated by the Agr/RNAIII system, which suggests that KdpDE may be an important virulence regulator coordinating the external K+ sensing and Agr signaling during pathogenesis in this bacterium.
INTRODUCTION
Staphylococcus aureus is a significant human pathogen that causes a wide range of infections. Its capacity to cause diseases arises from its production of a diverse array of virulence factors during different stages of infection, including secreted proteins, such as serine protease (Ssp), nuclease, hemolysins, enterotoxins, lipase, and coagulase, and proteins exposed on the cell surface, such as protein A (Spa) and fibrinogen-, fibronectin-, and collagen-binding proteins (9, 14). Expression of these factors is regulated by a range of global regulators that mainly comprise two families: two-component regulatory systems, which are sensitive to environmental signals and consist of a sensor histidine kinase and a response regulator protein, and the Sar homologs, a family of DNA-binding proteins homologous to SarA (9, 14, 17, 31). Genomic scans have revealed that there are 16 two-component systems in the genome of S. aureus (28). Among these, several have been revealed to have specific physiological roles, including Agr (accessory gene regulator) and ArlRS, SaeRS, SrrAB, LytRS, VraRS, HssRS, and GraRS, some of which are known as virulence-associated sensor-regulator systems involved in the induced production of toxins and exoproteins and the regulation of biofilm formation (12, 20, 24, 28, 30, 33, 39, 49, 55). The Agr system is also the major quorum sensing system in which agrD encodes the autoinducing peptide pheromone (AIP), which activates the two-component AgrC-AgrA system; the latter two function as sensor and response regulator proteins, respectively (31, 33, 39). As the most important quorum sensing system, Agr controls the expression of many virulence factors and primarily regulates alterations in the gene expression pattern when cells enter the post-exponential phase. In contrast, the physiological functions of several S. aureus two-component systems still remain to be explored. The orthologues of these systems are widely found in Gram-positive bacteria, suggesting that they play important roles in cell physiology (9).
The KdpDE two-component system was first characterized in Escherichia coli, in which proteins KdpD and KdpE regulate the production of the high-affinity K+ transporter Kdp- ATPase (6, 19, 38, 40). In E. coli, Kdp-ATPase is an efficient K+-scavenging system that is expressed when cells are subjected to extreme K+ limitation or osmotic upshock and other low-affinity K+ transporters cannot meet the cellular requirements for K+ (2, 4, 18, 26, 27, 29). The E. coli Kdp system consists of four proteins encoded by a single operon, kdpFABC, and its regulatory element, kdpDE, which is situated downstream of the kdpC gene (2, 19). Under K+ limitation or high osmolarity imposed by a salt, the histidine kinase KdpD autophosphorylates and transfers the phosphoryl group to the response regulator KdpE (51). Phosphorylated KdpE exhibits increased affinity for a 23-bp sequence upstream of the canonical −35 and −10 regions of the kdpFABC promoter and thereby triggers kdpFABC transcription (47). A BLAST search of Kdp protein sequences shows that the Kdp-ATPase system is widely distributed among Gram-negative bacteria (e.g., E. coli, Salmonella enterica serovar Typhimurium LT2, and Clostridium acetobutylicum) and Gram-positive bacteria (e.g., Bacillus cereus E33L, Alicyclobacillus acidocaldarius, and Mycobacterium tuberculosis). In distantly related bacteria, the ordering of the kdpA, kdpB, and kdpC genes is relatively fixed, but the kdpDE genes show different arrangements (6, 44, 52). In S. aureus, the organization of the kdpFABC operon is similar to that of E. coli, but the kdpDE genes are arranged in a reverse orientation upstream of the kdpA gene. Although experimental evidence has shown that the Kdp-ATPase system in several bacteria also functions as a high-affinity K+ transporter (1, 5, 21, 22, 50), the role of the Kdp system in pathogenic bacteria has not been investigated in detail.
Recently, several lines of evidence have shown that the two-component system KdpD-KdpE is involved in virulence in some bacteria. In Mycobacterium tuberculosis, deletion of kdpDE resulted in increased virulence. Mice infected with the M. tuberculosis kdpDE mutant died more rapidly than did those infected with wild-type bacteria (36). In S. aureus, the function of the Kdp system has not yet been clarified, although several reports have shown that the transcript level of kdpDE changes under certain environmental stresses (exposure to neutrophil microbicides or growth under biofilm conditions) (7, 35). Our previous work showed that the transcript level of kdpDE in the luxS mutant increased compared with that in the wild type and the addition of exogenous autoinducer 2 (AI-2) restored the parental phenotype; besides this, the inactivation of kdpDE resulted in a decreased transcript level of cap, indicating that the LuxS/AI-2 signaling system regulates capsular polysaccharide synthetase gene expression via KdpDE (56). All these data suggest that KdpDE might be a functional two-component system in S. aureus; however, detailed information about its physiological role and how it functions needs further exploration.
In the present study, we identified the function of KdpDE in S. aureus. In S. aureus NCTC8325, KdpDE displays a repression effect on the transcription of kdpFABC under all of the different K+ conditions that were tested and KdpFABC is not a major K+ transporter. However, inactivation of kdpDE results in alterations of transcription for a range of virulence genes, including spa, cap, hla, aur, geh, and hlgB. In addition, our electrophoretic mobility shift assay (EMSA) data showed that KdpE can directly bind to the promoter regions of most of these genes so as to regulate their transcription. Besides this, we also revealed that the transcript level of kdpDE was influenced by the external K+ concentration, indicating that this bacterium might modulate its infectious status by sensing specific external K+ stimuli in different environments. Finally, we found that Agr/RNAIII strongly activated the transcript level of kdpDE in the post-exponential phase of the cells, and we confirmed that this regulatory effect was via Rot.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The bacterial strains and plasmids used in this study are described in Table 1. Staphylococcus aureus and E. coli were grown in Luria-Bertani (LB) medium or tryptic soy broth (TSB; soybean-casein digest medium USP; Oxoid) medium with the appropriate antibiotics for plasmid selection and maintenance. The mutants were constructed using a method previously described (11). All primers used in this study are listed in Table S1 in the supplemental material.
Table 1.
Strain and plasmid list
| Strain or plasmid | Relevant genotype | Source or reference |
|---|---|---|
| Strains | ||
| S. aureus | ||
| WTb | NCTC8325, wild type | NARSAa |
| RN4220 | 8325-4, r− | NARSA |
| RN6911 | agr locus in 8325-4 replaced by tetM | NARSA |
| SX8 | 8325 kdpDE::ermB | L. Zhao |
| SX9 | 8325 kdpDE::ermB pLIkdpDE | L. Zhao |
| SX10 | 8325 kdpE::ermB | L. Zhao |
| SX11 | 8325 kdpE::ermB pLIkdpE | L. Zhao |
| SX13 | 8325 kdpFABC::ermB | This study |
| SX14 | 8325 kdpFABC::ermB pLIkdpFABC | This study |
| SX15 | 8325 agr::ermB | This study |
| SX16 | 8325 agr::ermB pLIagr | This study |
| SX17 | 8325 RNAIII::ermB | This study |
| SX18 | 8325 RNAIII::ermB pLIRNAIII | This study |
| SX19 | 8325 rot::ermB | This study |
| SX20 | 8325 rot::ermB pLIrot | This study |
| SX21 | RN6911 rot::ermB, agrrot double mutant | This study |
| E. coli | ||
| DH5α | Clone host strain, supE44 ΔlacU169(φ80 lacZΔM15) hsdR17 recA1 endA1 gyrA96 thi-1 relA1 | Invitrogen |
| BL21 | Expression strain, F−ompT hsdS(rB− mB−) gal dcm (DE3) | Invitrogen |
| Plasmids | ||
| pEASY-Blunt | Clone vector, Kanr Apr | Transgen |
| pET28a(+) | Expression vector | Novagen |
| pGkdpE | pET28a(+) with kdpE gene | This study |
| pGrot | pET28a(+) with rot gene | This study |
| pEC1 | pBluescript derivative; source of ermB gene; Apr | R. Bruckner |
| pBT2 | Shuttle vector, temperature sensitive, Apr Cmr | R. Bruckner |
| pBTkdpFABC | pBT2 derivative, for kdpFABC mutagenesis; Apr Cmr Emr | This study |
| pBTkdpDE | pBT2 derivative, for kdpDE mutagenesis; Apr Cmr Emr | L. Zhao |
| pBTkdpE | pBT2 derivative, for kdpE mutagenesis; Apr Cmr Emr | L. Zhao |
| pBTagr | pBT2 derivative, for agr mutagenesis; Apr Cmr Emr | This study |
| pBTRNAIII | pBT2 derivative, for RNAIII mutagenesis; Apr Cmr Emr | This study |
| pBTrot | pBT2 derivative, for rot mutagenesis; Apr Cmr Emr | This study |
| pLI50 | Shuttle cloning vector, Apr Cmr | Addgene |
| pLIkdpFABC | pLI50 with kdpFABC and its promoter, Apr Cmr | This study |
| pLIkdpDE | pLI50 with kdpDE and its promoter, Apr Cmr | L. Zhao |
| pLIkdpE | pLI50 with kdpE and the promoter of kdp operon, Apr Cmr | L. Zhao |
| pLIagr | pLI50 with agr operon and its promoter, Apr Cmr | This study |
| pLIRNAIII | pLI50 with RNAIII and its promoter, Apr Cmr | This study |
| pLIrot | pLI50 with rot and its promoter, Apr Cmr | This study |
NARSA, Network on Antimicrobial Resistance in Staphylococcus aureus.
WT, wild type.
Development of the CDM.
To study the effect of external K+ on S. aureus strains under an increasing gradient of K+ conditions, a formulation based on the one by Onoue and Mori was used (34). Initially, all potassium-containing salts of this medium were replaced with their sodium equivalents. The detailed composition of the formulation is described in Table S2 in the supplemental material. Three groups of wild-type bacteria were initially cultivated in chemically defined medium (CDM) with 0.2 mM K+ to an optical density at 600 nm (OD600) of 0.3, and then K+ was added to two groups of these until final concentrations of 4 mM and 100 mM were reached. Each group was divided into three parts on average, and the three parts of the cells were harvested after cultivation for 10 min, 40 min, and a longer time (about 3 h to reach an OD600 of 0.5). The RNA from each group was subsequently extracted for real-time reverse transcription-PCR (RT-PCR) assays.
Measurement of the internal potassium concentration of S. aureus.
The cells were cultivated in CDMs with different K+ concentrations to an OD600 of 0.6, harvested by centrifugation, and then washed with deionized water six times to eliminate any residual K+ in the medium. The cleaned cells were dried and lysed with sulfuric acid (metal-oxide-semiconductor grade), and the internal potassium concentration was assessed using an AAnalyst 800 atomic absorption spectrometer (Perkin-Elmer Corporation).
Total RNA isolation, cDNA generation, real-time RT-PCR, and microarray processing.
For the microarray assays, overnight cultures of S. aureus were diluted 1:100 in LB medium and grown to the late exponential phase (OD600 = 2.0). Cells were collected by centrifugation and resuspended in Tris-EDTA (TE) buffer (pH 8.0) containing 10 g/liter lysozyme and 40 mg/liter lysostaphin. After incubation at 37°C for 5 min, S. aureus cells were prepared for total RNA extraction using the Trizol method (Invitrogen), and any residual DNA was removed with DNase (RNase free; TaKaRa). The cDNAs were synthesized and labeled according to the manufacturer's recommendations for S. aureus antisense genome arrays (Affymetrix Inc., Santa Clara, CA). Further preparation, hybridization, and scanning were conducted by the Biochip Company of Shanghai, China. Microarray data were analyzed with the Affymetrix Microarray Suite software 5.1 (Affymetrix Inc.) and a four-comparison survival method. Real-time RT-PCR was performed with the PrimeScript 1st Strand cDNA synthesis kit and the SYBR Premix Ex Taq (TaKaRa) using the StepOne real-time PCR system (Applied Biosystems). The quantity of cDNA measured by real-time PCR was normalized to the abundance of 16S cDNA. All the real-time PCR assays were repeated at least four times.
Purification of KdpE and Rot.
The same purification methods were used for KdpE and Rot. Plasmid was transformed into E. coli BL21(DE3). The transformant was grown in 1 liter of LB at 37°C to an OD600 of 0.3 and induced with 0.5 mM IPTG (isopropyl-β-d-thiogalactopyranoside) for 3 h. Cells were harvested by centrifugation and washed with cell washing buffer (20 mM Tris-HCl, pH 8.0, and 0.5 M NaCl). The cells were resuspended in 50 ml of lysis buffer (20 mM Tris-HCl, pH 8.0, and 0.5 M NaCl) and were then lysed by sonication and centrifuged at 12,500 rpm for 30 min at 4°C. The supernatant was mixed with 2 ml of Ni-NTA agarose solution (Invitrogen), and the suspension was loaded onto a column at 4°C. After the column was washed with 5 ml of washing buffer I (5 mM imidazole, 20 mM Tris-HCl, pH 8.0, 0.5 M NaCl) and then with 100 ml of washing buffer II (20 mM imidazole, 20 mM Tris-HCl, pH 8.0, 0.5 M NaCl) and 10 ml of washing buffer III (100 mM imidazole, 20 mM Tris-HCl, pH 8.0, 0.5 M NaCl), the Rot protein was eluted with 5 ml of elution buffer (250 mM imidazole, 20 mM Tris-HCl, pH 8.0, 0.5 M NaCl, 10% glycerol). The imidazole in the eluant was removed by using a Centrifuge Biomax-5 column (Millipore), and then the protein solution was stored at −80°C until use. The purity of the protein was analyzed by SDS-PAGE, and the protein concentration was measured using the Bradford assay with bovine serum albumin (BSA) as a standard.
Electrophoretic mobility shift assay.
The DNA fragments containing the promoters were amplified from the S. aureus NCTC8325 chromosome. The PCR products were labeled using the digoxigenin (DIG) gel shift kit (Roche) according to the manufacturer's instructions. The labeled fragment was incubated at 25°C for 15 min with various amounts of purified proteins in 10 μl of incubation buffer (50 mM Tris-HCl, pH 7.5, 100 mM NaCl, 3 mM magnesium acetate, 0.1 mM EDTA, 0.1 mM dithiothreitol). After incubation, the mixtures were electrophoresed in a 4% native polyacrylamide gel in an 0.5× Tris-borate-EDTA (TBE) buffer. The band shifts were detected and analyzed according to the manufacturer's instructions.
DNase I footprinting assay.
The forward primer was synthesized and subsequently 5′ labeled with 6-carboxyfluorescein (6-FAM), resulting in the labeled primer p-kdp-f-FAM. The labeled DNA fragments were prepared by PCR using S. aureus NCTC8325 genomic DNA as the template. The labeled DNA fragments were purified by PAGE. The DNase I footprinting assays were performed with a 3730XL DNA analyzer (Applied Biosystems) using a modified method based on previous studies (54).
S. aureus survival in human blood and in U937 monocytic cells.
Heparinized venous blood samples were collected from healthy donors who provided written informed consent to participate in the study. The bacterial strains were harvested from TSB plates after being cultured at 37°C for 16 h, washed twice in phosphate-buffered saline (PBS), and suspended to an OD600 of 0.8. The heparinized human blood (1 ml) was inoculated with 1 × 106 CFU of S. aureus and incubated at 37°C with shaking (250 rpm). A total of 5 × 106 U937 monocytic cells were mixed with 2 × 106 CFU of S. aureus opsonized with 10% normal human serum and incubated at 37°C under an atmosphere of 5% CO2 with intermittent shaking. The bacteria were diluted to the appropriate concentration for testing at the required intervals, and CFU were calculated by plate counts performed in duplicate on TSB agar. The percentage of S. aureus CFU that survived was determined by comparing the bacterial burden in each sample after the indicated time with the bacterial burden at the start of the assay (0 h).
Microarray data accession number.
The microarray data and detailed protocols have been deposited in the CIBEX database (http://cibex.nig.ac.jp) with accession number CBX136.
RESULTS
KdpE can bind to the promoters of kdpF and kdpD and always represses transcription of the kdpFABC operon.
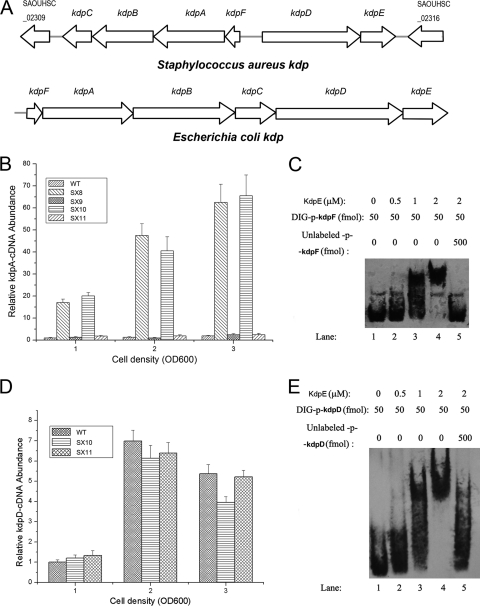
The available genomic information shows that the organization of the S. aureus Kdp system is different from that in E. coli. In E. coli, the kdpDE operon is located downstream of kdpC, while in S. aureus, kdpDE is arranged in a reverse orientation upstream of kdpA (Fig. 1A). Since the E. coli kdpDE and other investigated kdpDE operons all activated the transcription of kdpFABC, it was reasonable for us to first investigate whether S. aureus kdpDE also has the same regulatory effect on the transcription of kdpFABC. The transcript levels of kdpFABC in the wild type, the kdpDE mutant, and the kdpE mutant were measured by real-time reverse transcription-PCR (RT-PCR) analysis. Unexpectedly, the transcript levels of kdpFABC in both the kdpDE mutant and the kdpE mutant displayed an increase compared with that in the wild type, whether the cells were grown in LB medium to an optical density at 600 nm (OD600) of 1, 2, or 3 (Fig. 1B), suggesting that, in contrast to all of the other investigated kdpDE operons, the S. aureus kdpDE represses transcription of kdpFABC throughout the growth phase. Since KdpE is the response regulator containing a helix-turn-helix DNA-binding domain, we supposed that its regulatory effect on the transcription of kdpFABC might be through direct binding to the promoter regions of the kdpFABC operon. In order to determine this, we carried out EMSAs. The intergenic region between kdpF and kdpD was divided into two parts, which were used as p-kdpF and p-kdpD, respectively. As expected, KdpE appeared to have a strong ability to bind to the promoter regions of kdpFABC in vitro (Fig. 1C).
Fig. 1.
Regulatory effect of KdpE on the kdp operon and kdpDE transcription. (A) Organization of the kdp operons in S. aureus and E. coli. The arrows indicate the directions of translation as determined from the nucleotide sequence. (B) The regulatory effect of KdpDE on the transcription of kdpFABC in cells grown in LB medium. The transcript levels of kdpFABC were compared using real-time RT-PCR in wild type (WT; S. aureus NCTC8325), SX8 (kdpDE mutant), SX9 (kdpDE mutant with a plasmid encoding KdpDE), SX10 (kdpE mutant), and SX11 (kdpE mutant with a plasmid encoding KdpE). The strains were grown in LB medium to OD600s of 1, 2, and 3. (C) The ability of KdpE to bind to the kdpFABC promoter as determined by EMSAs. (D) The regulatory effect of KdpE on the transcription of kdpD. The transcript levels of kdpD were compared between WT, SX10 (kdpE mutant), and SX11 (kdpE mutant with a plasmid encoding KdpE). (E) The ability of KdpE to bind to the kdpDE promoter as determined by EMSAs. All the real-time PCR assays were repeated five times with similar results. Error bars indicate standard deviations.
Most of the response regulators of the two-component system can bind to the promoter region of their own operons and regulate transcription. Therefore, it was appropriate to investigate whether S. aureus KdpE also has this common feature. We performed real-time RT-PCR experiments and EMSAs for this purpose. Interestingly, our results showed that although KdpE can directly bind to the promoter region of kdpDE (Fig. 1E), it displayed no apparent influence on the transcription of this operon when the cells were grown in LB medium to different growth phases (Fig. 1D).
KdpFABC is not a major K+ transporter in S. aureus.
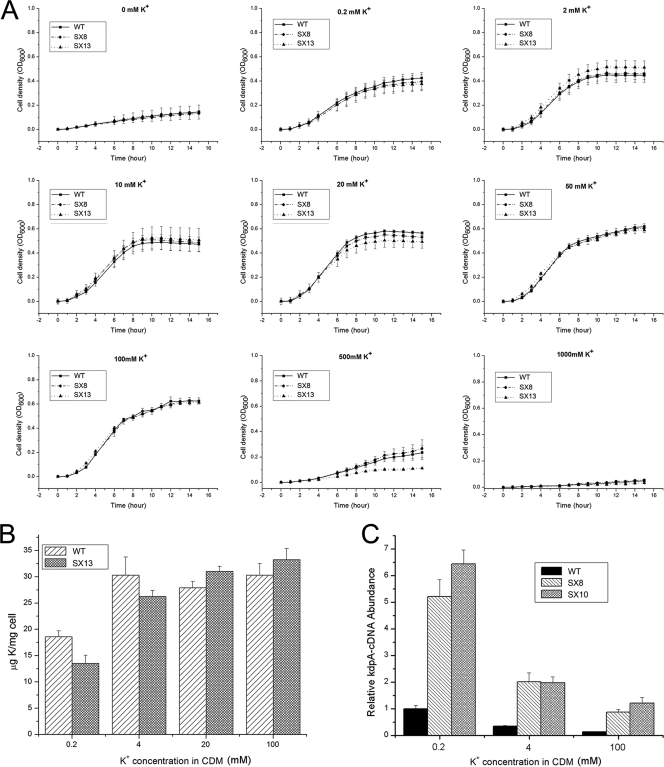
In E. coli, KdpFABC is a highly efficient K+ transporter and inactivation of kdpFABC will result in notable growth inhibition of the cells when the external K+ concentration is low. However, according to the above data, the regulatory effect of S. aureus KdpDE on the transcription of kdpFABC appears to be contrary to that of its homolog in E. coli, which led us to suspect that the KdpFABC in S. aureus may not function like that in E. coli. We designed a series of experiments in order to explore whether the S. aureus KdpFABC is associated with K+ transport. We first compared the growth rates of the wild type, the kdpDE mutant, and the kdpFABC mutant when they were grown in chemically defined medium (CDM) with different K+ concentrations. The results showed that no remarkable difference was observed between the growth rates of the three strains, whether the external K+ concentration was low or high (Fig. 2 A). After this, we assessed the internal K contents of the wild type and the kdpFABC mutant strain when they were grown under different external K+ conditions. As shown in Fig. 2B, the internal K contents of the kdpFABC mutants were quite high no matter which external K+ condition they were grown under. When the external K+ concentration was 0.2 mM or 4 mM, the internal K content of the kdpFABC mutant was a little lower than that of the wild type. Under the other external K+ conditions, the two strains showed no remarkable difference in the internal K content. We further investigated the regulatory effect of KdpDE on kdpFABC transcription when the cells were grown under different external K+ conditions. Our results still confirmed that transcription of kdpFABC is repressed by KdpDE (Fig. 2C). Compared with the activation of kdpFABC transcription by KdpDE in E. coli, the sustained repression effect of KdpDE on kdpFABC in S. aureus strongly suggests that KdpFABC is not a major K+ transporter.
Fig. 2.
KdpFABC is not a major K+ transporter in S. aureus. (A) Comparison of growth rates of WT, SX8 (kdpDE mutant), and SX13 (kdpFABC mutant) in CDM with different K+ concentrations. (B) Measurement of the internal K contents of the WT and SX13 (kdpFABC mutant) at different K+ concentrations of 0.2 mM, 4 mM, 20 mM, and 100 mM. (C) The regulatory effect of KdpDE on the transcription of kdpFABC in cells grown under different external K+ conditions. The transcript levels of kdpFABC were compared between WT, SX8 (kdpDE mutant), and SX10 (kdpE mutant) cells at different external K+ concentrations of 0.2 mM, 4 mM, and 100 mM. The real-time PCR assay was repeated four times with similar results. Error bars indicate standard deviations.
Transcription of kdpDE is influenced by external K+ concentration.
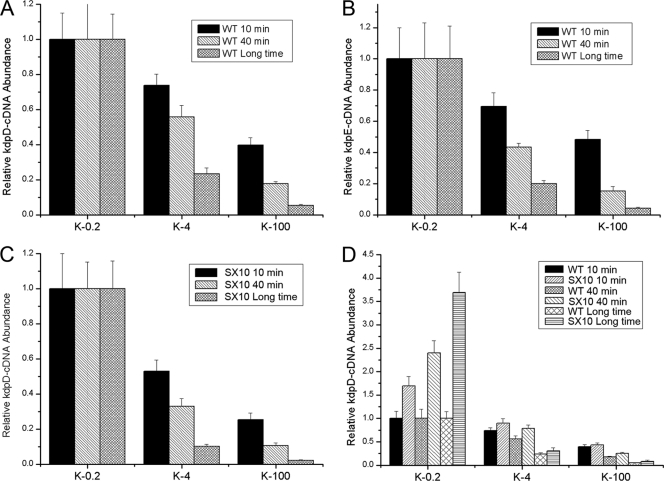
In E. coli, the transcript level of kdpDE changes with fluctuations in external K+ concentrations. Therefore, we performed experiments to determine whether or not the transcription of S. aureus kdpDE can also be influenced by external K+ concentrations. We tested the effects of three different K+ concentrations: 0.2 mM, which is close to the usual K+ concentration in the natural environment; 4 mM, which is almost equal to the K+ concentration in human blood and serum; and 100 mM, which is similar to the normal K+ concentration in host cells. Three groups of wild-type bacteria were initially cultivated in CDM with 0.2 mM K+ to an OD600 of 0.3, and then K+ was added to two groups of these to reach final K+ concentrations of 4 mM and 100 mM, respectively. In order to integrate the time factor, we cultivated the bacteria under specific K+ conditions for different times and then measured the transcript levels of kdpD and kdpE by using real-time RT-PCR analysis. As shown in Fig. 3 A and B, when the external K+ concentration was 4 mM, the transcript levels of kdpD and kdpE remarkably decreased compared with those of the cells cultivated under 0.2 mM K+ conditions; when the external K+ concentration was 100 mM, this tendency to decrease became much more obvious. These data suggest that the transcription of both kdpD and kdpE can be influenced by alterations in the external K+ concentrations. Besides this, it was notable that these influences always existed whether the cultivation time was short (10 min and 40 min) or long (about 3 h), indicating that the external K+ concentration has an instant effect on the transcription of kdpD and kdpE.
Fig. 3.
Influence of external K+ on kdpDE transcription. (A and B) Influence of K+ stimuli on the transcription of kdpDE. The transcript levels of kdpD and kdpDE in the WT were tested when the cells were grown under different K conditions for different times. Three groups of wild-type bacteria were initially cultivated in CDM with 0.2 mM K+ to an OD600 of 0.3, and then K+ was added to two groups of these until final concentrations of 4 mM and 100 mM were reached. Each group was divided into three parts on average, and the three parts of the cells were harvested after cultivation for 10 min, 40 min, and a longer time (about 3 h to reach an OD600 of 0.5). (C) Experiments to explore whether or not the K+ stimuli in the environment influenced the transcription of kdpDE through KdpE. The transcript levels of kdpD in SX10 (kdpE mutant) were tested with cells grown under different K+ conditions for different times. (D) Comparison of the transcript levels of kdpD between WT and SX10 (kdpE mutant) under different K+ conditions. All the real-time PCR assays were repeated four times with similar results. Error bars indicate standard deviations.
Furthermore, we measured kdpD transcript levels in the kdpE mutants growing in CDM at different external K+ concentrations, and the results showed that inactivation of kdpE did not affect the influence of external K+ concentration on the transcription of kdpD (Fig. 3C), suggesting that alterations in the transcript level of kdpD in response to changes in the external K concentration are not dependent on KdpE.
As mentioned above, our data showed that KdpE exhibited no regulatory effects on the transcription of kdpD in the LB medium. To exclude the specific influence of the high K+ concentration of the LB medium, we compared the transcript levels of kdpD between the wild type and the kdpE mutant under different K+ conditions. As shown in Fig. 3D, when the external K+ concentration was 0.2 mM, the transcript level of kdpD in the kdpE mutant was much higher than that of the wild type; however, when the external K+ concentration was above 4 mM, this difference was no longer apparent. These results suggest that KdpE can repress the transcription of kdpD, but only when the cells are under the lowest external K+ conditions.
KdpDE is a global regulator of virulence genes.
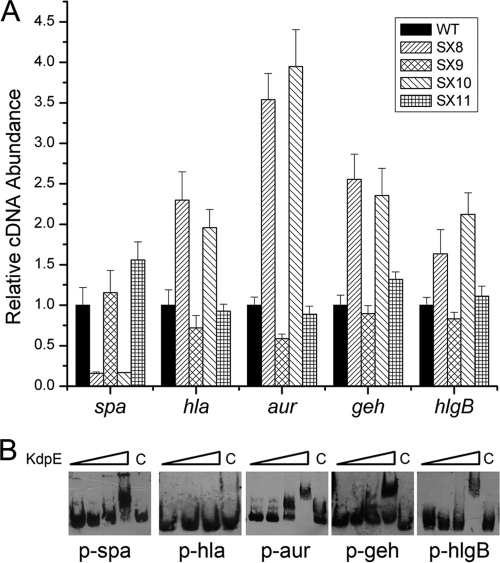
To characterize the gene transcriptional profiling influence of KdpDE, DNA microarray assays were performed using the parental strain NCTC8325 and the kdpDE deletion mutant strain. The cells were grown in LB medium to an OD600 of 2.0. A 2-fold induction ratio was used as the cutoff limit for comparing the transcriptional profiling of the wild type and the kdpDE mutant strain. Microarray data indicated that 48 genes were induced and 58 genes were repressed in the kdpDE mutant strain (Table 2 ). Of importance, the transcript levels of a range of virulence factor genes, such as spa, cap, hla, aur, geh, and hlgB, were altered in the kdpDE mutant strain (3, 8, 10, 13, 15, 25, 37, 42, 45, 46). Real-time RT-PCR experiments were conducted to further analyze the regulatory effect of KdpDE on the transcription of these genes. Our previous work showed that the inactivation of kdpDE resulted in a decreased transcript level of cap operon (36). Here, our results showed that the transcript levels of spa, hla, aur, geh, and hlgB displayed apparent alterations in the kdpDE mutant compared with those in the parental strain (Fig. 4 A). Among these genes, spa and cap belong to the group of genes encoding cell wall-associated proteins and polysaccharides that play roles in bacterial colonization, while hla, aur, geh, and hlgB belong to the group of genes encoding the toxin proteins that facilitate local invasion. Interestingly, our results showed that the transcript levels of spa and cap decreased, whereas the transcript levels of hla, aur, geh, and hlgB all increased in the kdpDE mutant compared to those in the parental strain.
Table 2.
Main genes affected by KdpDE
| Gene function and identifier | Gene product | Fold change, mutant vs WTc |
|---|---|---|
| Metabolism genes | ||
| SAOUHSC_01389 | Thioredoxin reductase | 2 |
| SAOUHSC_00113 | AdhE product = alcohol-acetaldehyde dehydrogenase | 2 |
| SAOUHSC_01619 | Probable exodeoxyribonuclease VII small subunit | 2.14 |
| SAOUHSC_01825 | Aminotransferase, class V | 2 |
| SAOUHSC_02281 | IlvD product = dihydroxy-acid dehydratase | 2.63 |
| SAOUHSC_02282 | IlvB product = acetolactate synthase large subunit | 2.14 |
| SAOUHSC_02283 | Similar to acetolactate synthase small subunit | 2.63 |
| SAOUHSC_02671 | NarK product = nitrite extrusion protein | 2 |
| SAOUHSC_02679 | Similar to nitrate reductase delta chain | 3.73 |
| SAOUHSC_02680 | NarH product = nitrate reductase beta chain | 3.03 |
| SAOUHSC_02682 | NasF product = uroporphyrin III C-methyltransferase | 2.29 |
| SAOUHSC_02684 | NasD product = nitrite reductase | 2 |
| SAOUHSC_02849 | Pyruvate oxidase | 2.14 |
| SAOUHSC_02945 | Precorrin-2 dehydrogenase | 2 |
| SAOUHSC_02969 | ArcA product = arginine deiminase | 2.46 |
| SAOUHSC_03011 | HisB product = imidazole glycerolphosphate dehydratase | 2.14 |
| SAOUHSC_03012 | Hypothetical protein | 2.29 |
| SAOUHSC_03013 | Histidinol dehydrogenase | 2 |
| SAOUHSC_01030 | Putative NrdH-redoxin | 2 |
| SAOUHSC_00465 | Veg product = VEG protein homolog | 2.29 |
| SAOUHSC_00898 | ArgH product = argininosuccinate lyase | 2.63 |
| SAOUHSC_00899 | ArgG product = argininosuccinate synthase | 2.46 |
| SAOUHSC_02118 | Glutamyl-tRNAGln amidotransferase subunit C | 2.29 |
| SAOUHSC_01191 | RpmB product = 50S ribosomal protein L28 | 0.47 |
| SAOUHSC_01216 | SucC product = succinyl-CoAa synthetase subunit beta | 0.5 |
| SAOUHSC_01218 | SucD product = succinyl-CoA synthetase alpha subunit | 0.5 |
| SAOUHSC_00195 | Acetyl-CoA acetyltransferase homolog | 0.5 |
| SAOUHSC_00196 | Putative 3-hydroxyacyl-CoA dehydrogenase FadB | 0.47 |
| SAOUHSC_00197 | Putative acyl-CoA dehydrogenase FadD | 0.5 |
| SAOUHSC_00198 | Putative acyl-CoA synthetase FadE | 0.44 |
| SAOUHSC_00199 | Putative acetyl-CoA/acetoacetyl-CoA transferase | 0.47 |
| SAOUHSC_00206 | LctE product = l-lactate dehydrogenase | 0.44 |
| SAOUHSC_00365 | AhpC product = alkyl hydroperoxide reductase subunit C | 0.5 |
| SAOUHSC_01002 | Quinol oxidase polypeptide II QoxA | 0.5 |
| SAOUHSC_02366 | FbaA product = fructose-bisphosphate aldolase | 0.5 |
| Cell surface protein genes | ||
| SAOUHSC_01385 | PstB product = phosphate ABC transporter | 3.24 |
| SAOUHSC_01386 | Similar to phosphate ABC transporter | 2.63 |
| SAOUHSC_02311 | KdpB product = probable potassium-transporting ATPase B | 2 |
| SAOUHSC_02310 | KdpC product = probable potassium-transporting ATPase C | 2.14 |
| SAOUHSC_01990 | Glutamate ABC transporter ATP-binding protein | 4 |
| SAOUHSC_00636 | Similar to ABC transporter, permease protein | 2.14 |
| SAOUHSC_00637 | Similar to ABC transporter ATP-binding protein | 2.14 |
| SAOUHSC_02661 | ScrA product = PTSb system, sucrose-specific IIBC component | 0.43 |
| Regulator genes | ||
| SAOUHSC_01384 | Similar to negative regulator PhoU | 2.46 |
| SAOUHSC_01490 | Hu product = DNA-binding protein II | 0.44 |
| SAOUHSC_02314 | KdpD product = sensor protein | 0.0059 |
| SAOUHSC_02261 | Accessory gene regulator protein B | 0.5 |
| SAOUHSC_02264 | Accessory gene regulator protein C | 0.5 |
| SAOUHSC_02262 | Accessory gene regulator protein D | 0.41 |
| SAOUHSC_02810 | Transcriptional regulator, MerR family | 0.44 |
| SAOUHSC_00794 | GapR product = glycolytic operon regulator | 0.35 |
| Virulence genes | ||
| SAOUHSC_02629 | Similar to multidrug resistance protein | 2 |
| SAOUHSC_02851 | LrgA family protein | 2 |
| SAOUHSC_02855 | Similar to secretory antigen precursor SsaA | 2.63 |
| SAOUHSC_02971 | Aur product = zinc metalloproteinase aureolysin | 2.29 |
| SAOUHSC_00069 | Spa product = immunoglobulin G binding protein A precursor | 0.31 |
| SAOUHSC_00114 | CapA | 0.38 |
| SAOUHSC_00115 | CapB | 0.35 |
| SAOUHSC_00116 | CapC | 0.35 |
| SAOUHSC_00117 | CapD | 0.38 |
| SAOUHSC_00118 | CapE | 0.44 |
| SAOUHSC_00119 | CapF | 0.41 |
| SAOUHSC_00120 | CapG | 0.44 |
| SAOUHSC_00121 | CapH | 0.35 |
| SAOUHSC_00122 | CapI | 0.43 |
| SAOUHSC_00123 | CapJ | 0.43 |
| SAOUHSC_00124 | CapK | 0.46 |
| SAOUHSC_00125 | CapL | 0.5 |
| SAOUHSC_00126 | CapM | 0.5 |
| SAOUHSC_00127 | CapN | 0.5 |
| SAOUHSC_00300 | Geh product = glycerol ester hydrolase | 0.41 |
| SAOUHSC_02260 | Hld product = delta-hemolysin | 0.38 |
| SAOUHSC_02709 | HlgC product = gamma-hemolysin component C | 0.47 |
| SAOUHSC_02710 | HlgB product = gamma-hemolysin component B | 0.41 |
| Hypothetical protein genes | ||
| SAOUHSC_02384 | Hypothetical protein | 2 |
| SAOUHSC_02523 | Hypothetical protein | 2 |
| SAOUHSC_02858 | Hypothetical protein | 2 |
| SAOUHSC_01296 | Hypothetical protein | 2.46 |
| SAOUHSC_01729 | Hypothetical protein | 2.14 |
| SAOUHSC_01991 | Hypothetical protein | 2.82 |
| SAOUHSC_00202 | Hypothetical protein | 2 |
| SAOUHSC_02850 | Hypothetical protein | 2 |
| SAOUHSC_03047 | Hypothetical protein | 2 |
| SAOUHSC_01032 | Hypothetical protein | 2.14 |
| SAOUHSC_01072 | Hypothetical protein | 2.14 |
| SAOUHSC_00704 | Hypothetical protein | 2.29 |
| SAOUHSC_02886 | Hypothetical protein | 2.29 |
| SAOUHSC_01557 | Hypothetical protein | 0.38 |
| SAOUHSC_02521 | Hypothetical protein | 0.5 |
| SAOUHSC_02838 | Hypothetical protein | 0.38 |
| SAOUHSC_00091 | Hypothetical protein | 0.5 |
| SAOUHSC_00094 | Hypothetical protein | 0.44 |
| SAOUHSC_01675 | Hypothetical protein | 0.5 |
| SAOUHSC_01918 | Hypothetical protein | 0.5 |
| SAOUHSC_02781 | Hypothetical protein | 0.47 |
| SAOUHSC_02788 | Hypothetical protein | 0.33 |
| SAOUHSC_02805 | Hypothetical protein | 0.36 |
| SAOUHSC_00401 | Hypothetical protein | 0.5 |
| SAOUHSC_00413 | Hypothetical protein | 0.47 |
| SAOUHSC_01956 | Hypothetical protein | 0.47 |
| SAOUHSC_00414 | Hypothetical protein | 0.47 |
| SAOUHSC_00257 | Hypothetical protein | 0.5 |
| SAOUHSC_01109 | Hypothetical protein | 0.47 |
| SAOUHSC_01956 | Hypothetical protein | 0.47 |
| SAOUHSC_02796 | Hypothetical protein | 0.088 |
CoA, coenzyme A.
PTS, phosphotransferase system.
WT, wild type.
Fig. 4.
KdpDE is a global virulence regulator. (A) Comparative measurements of a range of virulence gene transcripts by real-time RT-PCR in WT, SX8 (kdpDE mutant), SX9 (kdpDE mutant with a plasmid encoding KdpDE), SX10 (kdpE mutant), and SX11 (kdpE mutant with a plasmid encoding KdpE). All of the strains were grown in LB medium to an OD600 of 1.7. The relative transcription of each gene compared to that of the constitutively expressed 16S rRNA gene in SX8, SX9, SX10, and SX11 was compared with that in the wild type, to which we assigned a value of 1. All the real-time PCR assays were repeated four times with similar results. Error bars indicate standard deviations. (B) The ability of KdpE to bind to these gene promoters as determined by EMSAs. The concentrations of KdpE used for p-hla EMSA were 0 μM, 0.5 μM, 1 μM, and 2 μM (from left to right, respectively); the concentrations of KdpE used for other EMSAs with other promoters were 0 μM, 0.5 μM, 1 μM, 2 μM, and 2 μM (from left to right, respectively). Tenfold unlabeled probes were used for the negative-control assays.
In addition, the transcript levels of spa, hla, aur, geh, and hlgB in the wild type and the kdpE mutant were also compared using real-time RT-PCR analysis. The results showed that KdpE displayed the same regulatory tendency as did KdpDE on the transcription of these virulence genes (Fig. 4A). Since KdpE is a DNA-binding protein, we further performed EMSAs to investigate whether or not KdpE can regulate the transcription of these genes by directly binding to their promoter regions. As shown in Fig. 4B, KdpE can specifically bind to the promoter regions of all of these genes, except hla, in vitro. Meanwhile, we also measured the transcript levels of these virulence factors in the kdpFABC mutant, and the results showed that the transcription of these genes (spa, cap, hla, aur, geh, and hlgB) almost did not change in the mutant compared with the wild type (data not shown). These results suggest that, in S. aureus, KdpE is a global regulator which can bind to many virulence targets and regulate their transcription.
Modulatory effect of KdpDE on expression of Spa.
According to the above data, KdpDE can modulate the transcription of spa. Since Spa is a representative cell wall-associated exoprotein and a major determinant of virulence in S. aureus (14, 16, 45), it was of importance to further investigate this modulatory effect. First, we examined this effect during the different growth phases of this bacterium. Three groups of strains (the wild type, the kdpDE mutant, the kdpDE mutant with the complementing plasmid, the kdpE mutant, and the kdpE mutant with the complementing plasmid) were cultivated in LB medium to OD600s of 1, 2, and 3, respectively. The results of real-time RT-PCR revealed that the transcript levels of spa in the kdpDE mutant and the kdpE mutant were decreased compared to those in the wild type and the strains with the complementing plasmids, no matter which OD the cells were grown to (Fig. 5A), indicating that kdpDE can strongly activate the transcription of spa throughout the whole growth phase. We also performed Western blot assays to compare the protein levels of Spa between the kdpDE mutant and the wild type when they were grown to different growth phases, and the results confirmed that the expression of Spa in the kdpDE mutant was always lower than that in the wild type (see Fig. S1 in the supplemental material). In addition, previous studies showed that the expression of Spa enhanced bacterial virulence in a mouse bacteremia model and in macrophages due to its antiphagocytic nature. Therefore, we examined the influences of the inactivation of kdpDE or kdpE on the survival of S. aureus in human whole blood and human U937 monocytic cells. As shown in Fig. 5B and C, the kdpDE and kdpE mutants exhibited significantly lower survival rates than did the wild type and the strain with the complementing plasmid in both human whole blood and human U937 monocytic cells, further demonstrating that inactivation of kdpDE repressed the expression of Spa.
Fig. 5.
Regulatory effect of KdpDE on spa expression. (A) Analysis of the transcriptional regulation of spa by KdpDE. The transcript levels of spa were compared using real-time RT-PCR in WT, SX8 (kdpDE mutant), SX9 (kdpDE mutant with a plasmid encoding KdpDE), SX10 (kdpE mutant), and SX11 (kdpE mutant with a plasmid encoding KdpE). Three groups of strains were grown in LB medium to OD600s of 1, 2, and 3, respectively. (B) Comparative measurements of survival rates of WT, SX8, SX9, SX10, and SX11 in heparinized human blood. Results are from five separate blood donors. (C) Comparative measurements of survival rates of WT, SX8, SX9, SX10, and SX11 when cultured with U937 monocytic cells. The percentage of S. aureus CFU that survived was determined as described in Materials and Methods. (D) Influence of K+ stimuli on the transcription of spa and kdpD. The transcript levels of kdpD and spa in WT were tested in cells grown under different K+ conditions for different times. Three groups of wild-type bacteria were cultivated in CDM with 0.2 mM, 4 mM, and 100 mM K+. All the real-time PCR assays were repeated four times with similar results. Error bars indicate standard deviations.
As noted above, the transcription of kdpDE decreased with an increase in the external K+ concentration. We also assessed the transcript levels of spa under different external K+ conditions. Interestingly, the real-time RT-PCR data showed that the changing trend in the transcription of spa was the same as that of kdpD in transition from low to high external K+ conditions (Fig. 5D). This observation, together with the regulatory effect of KdpDE on the transcription of spa as investigated previously, further demonstrated that KdpDE in S. aureus can regulate the transcription of spa in response to K+ stimuli in the environment and that KdpDE will not always stimulate the transcription of spa when the external K+ concentration increases.
Identification of the KdpE binding sequence.
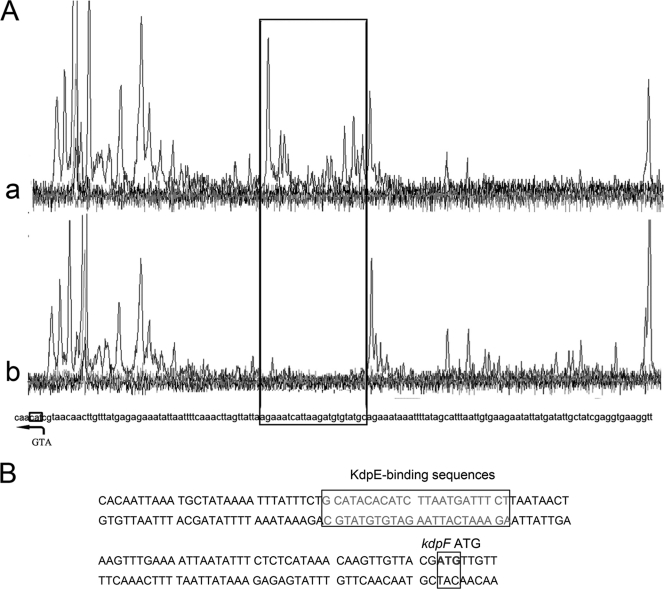
To further identify the precise KdpE binding sequence, DNase I footprinting was performed. As shown in Fig. 6 A, one region from −51 to −73 bp, relative to the translation start site of the kdpF gene, is protected, which is indicated by the disappearing nucleotide peaks in Fig. 6A, panel a, compared to Fig. 6A, panel b. These footprinting results demonstrated that KdpE binds to a 23-bp region (GCATACACATCTTAATGATTTCT) of the kdpF promoter, which is probably near the −35 to −10 transcriptional box of kdpF (Fig. 6B). This evidence led us to conclude that the sustained repression effect of KdpE on kdpFABC transcription is most likely due to the fact that KdpE binds to the −35 to −10 transcriptional box of kdpFABC, thereby competitively inhibiting the binding of its transcriptional factors, such as σ factor and RNA polymerase.
Fig. 6.
Identification of the KdpE-binding sequences by DNase I footprinting. (A) Identification of the KdpE-binding site on the promoter of kdpFABC by DNase I footprinting assays. The black frame indicates the DNA region protected from DNase I by KdpE. (B) KdpE-binding sequences on kdpF promoter regions.
Agr/RNAIII activates the transcription of kdpDE by Rot.
We observed that the transcript levels of both kdpD and kdpE significantly increased when the cells were grown to the post-exponential phase in LB medium (Fig. 7A), suggesting that the transcription of kdpDE might be associated with the Agr quorum sensing system. In order to determine whether or not Agr/RNAIII is involved in the regulation of kdpDE transcription, we first made the agr mutant, the RNAIII mutant, and the strains with the complementing plasmids and subsequently compared the transcript levels of kdpDE of these strains with that of the NCTC8325 parental strain. The results showed that the transcript levels of kdpDE in the agr mutant and the RNAIII mutant were much lower than that of the parental strain, while the strains with the complementing plasmids were restored to the parental phenotype (Fig. 7B), indicating that the Agr system can activate kdpDE transcription through RNAIII. We also cultivated the wild type and the agr mutant in CDMs with different K+ concentrations and tested the transcript levels of kdpD of the two strains, and the results showed that the transcript level of kdpD of the agr mutant was also always much lower than that of the wild type under different K+ conditions (see Fig. S2A in the supplemental material). Since RNAIII cannot directly activate target gene transcription, there certainly exists an intermediate component in this regulatory pathway. As described previously, the mask of rot translation by RNAIII is a key feature of the Agr function (23, 42). Therefore, we proposed that Agr upregulates kdpDE transcription probably through repressing rot translation. Furthermore, we tested the transcript levels of kdpD in the wild-type strain NCTC8325, the rot mutant, and the rot mutant with complementing plasmid when they were in different growth phases. The transcript level of kdpD in the rot mutant was much higher than that in the wild type when the cells were grown to an OD600 of 1 (early exponential phase), indicating that Rot strongly represses kdpDE transcription in this growth phase. The difference in transcript levels between the wild type and the rot mutant was not that apparent (Fig. 7C) when the cells were grown to the exponential phase (OD600 of 2 and 3), which was in accordance with the phenomenon that huge amounts of the RNAIII transcript accumulate in the wild type during the transition from the early exponential to the exponential phase. The accumulation of the RNAIII transcript inhibited the translation of rot, which, in return, diminished the Rot effect on kdpDE transcription. As shown in Fig. 7D, we also tested the transcript levels of kdpD in the wild type, RN6911 (agr mutant), and RN6911 with rot deletion (agr rot double mutant) when they were in different growth phases. The transcript level of kdpD in the agr rot double mutant was similar to that in the rot mutant but different from that in the agr mutant. These data demonstrated that Agr regulates kdpDE transcription through Rot. Furthermore, our EMSAs confirmed that Rot can specifically bind to the promoter of kdpD in vitro (Fig. 7E).
Fig. 7.
Agr/RNAIII activates the transcription of kdpDE by Rot. (A) Transcript levels of kdpD and kdpE in different growth phases. (B) Characterization of the regulatory effect of Agr/RNAIII on kdpDE transcription. The transcript levels of kdpD were measured using real-time RT-PCR in WT, SX15 (agr mutant), SX16 (agr mutant with a plasmid encoding the Agr system), SX17 (RNAIII mutant), and SX18 (RNAIII mutant with a plasmid encoding RNAIII). Three groups of strains were grown in LB medium to OD600s of 1, 2, and 3, respectively. (C) Effect of Rot on kdpDE transcription. The transcript levels of kdpD were measured using real-time RT-PCR in WT, SX19 (rot mutant), and SX20 (rot mutant with a plasmid encoding Rot). Three groups of strains were grown in LB medium to OD600s of 1, 2, and 3. (D) Effect of Rot on kdpDE transcription. The transcript levels of kdpD were measured using real-time RT-PCR in WT, RN6911 (agr mutant), and SX21 (agr rot double mutant). Three groups of strains were grown in LB medium to OD600s of 1, 2, and 3. All the real-time RT-PCR assays were repeated four times with similar results. Error bars indicate standard deviations. (E) The ability of Rot to bind to the kdpDE promoter as determined by EMSAs.
DISCUSSION
All of the bacterial kdp operons investigated previously were found to be repressed during growth in media with a high external K+ concentration and activated when the external K+ concentration becomes lower than a threshold value (6, 26, 41). These events are controlled by the KdpDE two-component system. For instance, in E. coli, when the external K+ concentration falls below 2 mM, KdpD is autophosphorylated and activates KdpE, forming a product which binds to the promoter region of kdpFABC and activates its transcription (38, 48). In contrast, our results showed that, in S. aureus, KdpDE always repressed the transcription of kdpFABC, whether under high or low external K+ conditions. In E. coli, KdpFABC is the major K+ transporter when the cells are subjected to K+ limitation. Therefore, inactivation of kdpFABC in E. coli would result in notable growth inhibition of the cells when the external K+ concentration is low (43). However, we found that the growth rates of the S. aureus kdpFABC mutant and the parental strain showed no apparent differences when the cells were grown in CDM with different K+ concentrations. In addition, by carrying out atomic absorption spectrometry assays, we observed that the internal potassium concentration of the S. aureus kdpFABC mutant was almost equal to that of the parental strain whether the K+ concentration of the medium was low or high. Thus, we conclude that KdpFABC is not a major K+ transporter in S. aureus and that another highly efficient K+ transporter which is functional in K+ transport must exist in this bacterium. This would also help to explain why the transcriptions of kdpFABC were always repressed by KdpE to quite low levels in S. aureus and indicates that the regulatory effect of KdpDE on KdpFABC ATPase in S. aureus is extremely different from that in E. coli. However, our real-time RT-PCR data showed that the transcription of kdpDE exhibits notable changes in response to fluctuations of the external K+ concentrations. Taking all of these data into account, we suggest that the S. aureus KdpDE functions in other aspects through sensing the external K+ concentration.
The pathogenic mechanisms of S. aureus infections are highly complex (9, 14, 31). It is very likely that distinct networks of multiple virulence genes are expressed in response to distinct host signals, including those found in blood and specific target tissues and those related to innate host defense factors that emerge during the infectious process. A high K+ concentration is also a host-specific signal which differs from potassium signals in the natural environment. In this study, we found that the transcript levels of kdpDE decreased as the external K+ concentration increased. When the external K+ concentration was 0.2 mM, which is close to the usual K+ concentration found in the natural environment, the transcript levels of kdpD were relatively high. Comparatively, when the external K+ concentration increased to 4 mM, which is almost equal to the K+ concentration in host blood and tissue fluid, or to 100 mM, which is similar to the K+ concentration in the host cells, the transcription of kdpD was largely repressed. As noted above, we showed that KdpE can activate the transcription of genes encoding cell wall-associated proteins and polysaccharides and that it can repress the transcription of toxin genes. Collectively, these interesting data led us to propose that the two-component system KdpDE might be an important virulence gene regulator in response to changes in the environment. In the natural environment, a high level of KdpDE transcription helps to activate the expression of cell wall proteins and polysaccharides, which are beneficial to colonization. However, in transition from the natural environment to the host, which has a higher K+ concentration, transcript levels of kdpDE decrease, causing a low expression of cell wall proteins but a high production of extracellular toxins and enzymes which facilitate local invasion. The capability of KdpDE in regulating alterations in the gene expression pattern indicates that KdpDE plays an important role in the pathogenesis of S. aureus.
It is interesting that the Agr system activates the transcription of kdpDE. Agr is the best-characterized quorum sensing system in S. aureus and regulates specific physiological functions when the population density of the community reaches a threshold (32, 39, 53). Thus, we can conclude that the high transcript level of kdpDE is also dependent on high cell density. This phenomenon is of special significance for the pathogenesis of S. aureus because the pathogenic processes are unproductive when undertaken by an individual bacterium acting alone but become beneficial when carried out simultaneously by a large number of cells. Only when the cell number in the community reaches a high level does it become meaningful for KdpDE to regulate virulence gene expression via sensing of the external K+ concentration in the environment. We cultivated the wild type and the agr mutant in CDM at different K+ concentrations and measured the transcript levels of kdpD and spa of the two strains, and the results showed that the transcript level of kdpD in the agr mutant was also always much lower than that of the wild type under different K+ conditions (see Fig. S2A in the supplemental material). However, the transcript level of spa in the agr mutant showed a fold increase of tens to hundreds compared with that in the wild type, suggesting that Agr acts as a main regulator of the spa transcription. We suggest that because the transcription of kdpD in the agr mutant could not be activated by the Agr system, the influence of Kdp on spa transcription was not obvious under this condition.
Collectively, we think that the pathogenesis of S. aureus is determined by the coordinated gene regulation in response to the self-secreting signal and other specific stimuli in the environment and that KdpDE, as a two-component system that can respond to both, is the partial embodiment of this coordination (Fig. 8).
Fig. 8.
Proposed regulation scheme of KdpDE. The pathogenesis of S. aureus is determined by the coordinated gene regulation in response to the self-secreting signal and other specific stimuli in the environment, and KdpDE, as a two-component system which can respond to both, is the concrete embodiment of this coordination.
Supplementary Material
ACKNOWLEDGMENTS
We thank J. Sun for providing the U937 cells and the Network on Antimicrobial Resistance in Staphylococcus aureus (NARSA) for providing the bacterial strains.
This work was supported by the National Natural Science Foundation of China (grants 31070116 and 31021061).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 21 March 2011.
The authors have paid a fee to allow immediate free access to this article.
REFERENCES
- 1. Alahari A., Ballal A., Apte S. K. 2001. Regulation of potassium-dependent Kdp-ATPase expression in the nitrogen-fixing cyanobacterium Anabaena torulosa. J. Bacteriol. 183:5778–5781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Altendorf K., et al. 1998. Structure and function of the Kdp-ATPase of Escherichia coli. Acta Physiol. Scand. Suppl. 643:137–146 [PubMed] [Google Scholar]
- 3. Arvidson S., Tegmark K. 2001. Regulation of virulence determinants in Staphylococcus aureus. Int. J. Med. Microbiol. 291:159–170 [DOI] [PubMed] [Google Scholar]
- 4. Asha H., Gowrishankar J. 1993. Regulation of kdp operon expression in Escherichia coli: evidence against turgor as signal for transcriptional control. J. Bacteriol. 175:4528–4537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bakker E. P., Borchard A., Michels M., Altendorf K., Siebers A. 1987. High-affinity potassium uptake system in Bacillus acidocaldarius showing immunological cross-reactivity with the Kdp system from Escherichia coli. J. Bacteriol. 169:4342–4348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ballal A., Basu B., Apte S. K. 2007. The Kdp-ATPase system and its regulation. J. Biosci. 32:559–568 [DOI] [PubMed] [Google Scholar]
- 7. Beenken K. E., et al. 2004. Global gene expression in Staphylococcus aureus biofilms. J. Bacteriol. 186:4665–4684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bhakdi S., Tranum-Jensen J. 1991. Alpha-toxin of Staphylococcus aureus. Microbiol. Rev. 55:733–751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bronner S., Monteil H., Prevost G. 2004. Regulation of virulence determinants in Staphylococcus aureus: complexity and applications. FEMS Microbiol. Rev. 28:183–200 [DOI] [PubMed] [Google Scholar]
- 10. Bronner S., Stoessel P., Gravet A., Monteil H., Prevost G. 2000. Variable expressions of Staphylococcus aureus bicomponent leucotoxins semiquantified by competitive reverse transcription-PCR. Appl. Environ. Microbiol. 66:3931–3938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruckner R. 1997. Gene replacement in Staphylococcus carnosus and Staphylococcus xylosus. FEMS Microbiol. Lett. 151:1–8 [DOI] [PubMed] [Google Scholar]
- 12. Brunskill E. W., Bayles K. W. 1996. Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J. Bacteriol. 178:611–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chan P. F., Foster S. J. 1998. Role of SarA in virulence determinant production and environmental signal transduction in Staphylococcus aureus. J. Bacteriol. 180:6232–6241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Cheung A. L., Bayer A. S., Zhang G., Gresham H., Xiong Y. Q. 2004. Regulation of virulence determinants in vitro and in vivo in Staphylococcus aureus. FEMS Immunol. Med. Microbiol. 40:1–9 [DOI] [PubMed] [Google Scholar]
- 15. Cheung A. L., Chien Y. T., Bayer A. S. 1999. Hyperproduction of alpha-hemolysin in a sigB mutant is associated with elevated SarA expression in Staphylococcus aureus. Infect. Immun. 67:1331–1337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cheung A. L., Eberhardt K., Heinrichs J. H. 1997. Regulation of protein A synthesis by the sar and agr loci of Staphylococcus aureus. Infect. Immun. 65:2243–2249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Cheung A. L., Zhang G. 2002. Global regulation of virulence determinants in Staphylococcus aureus by the SarA protein family. Front. Biosci. 7:d1825–d1842 [DOI] [PubMed] [Google Scholar]
- 18. Epstein W. 1992. Kdp, a bacterial P-type ATPase whose expression and activity are regulated by turgor pressure. Acta Physiol. Scand. Suppl. 607:193–199 [PubMed] [Google Scholar]
- 19. Epstein W., et al. 1990. The bacterial Kdp K(+)-ATPase and its relation to other transport ATPases, such as the Na+/K(+)- and Ca2(+)-ATPases in higher organisms. Philos. Trans. R. Soc. Lond. B Biol. Sci. 326:479–486 [DOI] [PubMed] [Google Scholar]
- 20. Fournier B., Klier A., Rapoport G. 2001. The two-component system ArlS-ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol. Microbiol. 41:247–261 [DOI] [PubMed] [Google Scholar]
- 21. Frymier J. S., Reed T. D., Fletcher S. A., Csonka L. N. 1997. Characterization of transcriptional regulation of the kdp operon of Salmonella typhimurium. J. Bacteriol. 179:3061–3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Garcia-Cuellar C., et al. 1995. Kdp-like system in Salmonella typhimurium LT-2. Rev. Latinoam. Microbiol. 37:227–236 [PubMed] [Google Scholar]
- 23. Geisinger E., Adhikari R. P., Jin R., Ross H. F., Novick R. P. 2006. Inhibition of rot translation by RNAIII, a key feature of agr function. Mol. Microbiol. 61:1038–1048 [DOI] [PubMed] [Google Scholar]
- 24. Giraudo A. T., Calzolari A., Cataldi A. A., Bogni C., Nagel R. 1999. The sae locus of Staphylococcus aureus encodes a two-component regulatory system. FEMS Microbiol. Lett. 177:15–22 [DOI] [PubMed] [Google Scholar]
- 25. Giraudo A. T., Cheung A. L., Nagel R. 1997. The sae locus of Staphylococcus aureus controls exoprotein synthesis at the transcriptional level. Arch. Microbiol. 168:53–58 [DOI] [PubMed] [Google Scholar]
- 26. Heermann R., Jung K. 2010. The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. FEMS Microbiol. Lett. 304:97–106 [DOI] [PubMed] [Google Scholar]
- 27. Jung K., Krabusch M., Altendorf K. 2001. Cs(+) induces the kdp operon of Escherichia coli by lowering the intracellular K(+) concentration. J. Bacteriol. 183:3800–3803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Kuroda M., et al. 2003. Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol. Microbiol. 49:807–821 [DOI] [PubMed] [Google Scholar]
- 29. Laimins L. A., Rhoads D. B., Epstein W. 1981. Osmotic control of kdp operon expression in Escherichia coli. Proc. Natl. Acad. Sci. U. S. A. 78:464–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Meehl M., Herbert S., Gotz F., Cheung A. 2007. Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 51:2679–2689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Novick R. P. 2003. Autoinduction and signal transduction in the regulation of staphylococcal virulence. Mol. Microbiol. 48:1429–1449 [DOI] [PubMed] [Google Scholar]
- 32. Novick R. P., et al. 1995. The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol. Gen. Genet. 248:446–458 [DOI] [PubMed] [Google Scholar]
- 33. Novick R. P., et al. 1993. Synthesis of staphylococcal virulence factors is controlled by a regulatory RNA molecule. EMBO J. 12:3967–3975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Onoue Y., Mori M. 1997. Amino acid requirements for the growth and enterotoxin production by Staphylococcus aureus in chemically defined media. Int. J. Food Microbiol. 36:77–82 [DOI] [PubMed] [Google Scholar]
- 35. Palazzolo-Ballance A. M., et al. 2008. Neutrophil microbicides induce a pathogen survival response in community-associated methicillin-resistant Staphylococcus aureus. J. Immunol. 180:500–509 [DOI] [PubMed] [Google Scholar]
- 36. Parish T., et al. 2003. Deletion of two-component regulatory systems increases the virulence of Mycobacterium tuberculosis. Infect. Immun. 71:1134–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pohlmann-Dietze P., et al. 2000. Adherence of Staphylococcus aureus to endothelial cells: influence of capsular polysaccharide, global regulator agr, and bacterial growth phase. Infect. Immun. 68:4865–4871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Polarek J. W., Williams G., Epstein W. 1992. The products of the kdpDE operon are required for expression of the Kdp ATPase of Escherichia coli. J. Bacteriol. 174:2145–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Recsei P., et al. 1986. Regulation of exoprotein gene expression in Staphylococcus aureus by agar. Mol. Gen. Genet. 202:58–61 [DOI] [PubMed] [Google Scholar]
- 40. Rhoads D. B., Laimins L., Epstein W. 1978. Functional organization of the kdp genes of Escherichia coli K-12. J. Bacteriol. 135:445–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Roe A. J., McLaggan D., O'Byrne C. P., Booth I. R. 2000. Rapid inactivation of the Escherichia coli Kdp K+ uptake system by high potassium concentrations. Mol. Microbiol. 35:1235–1243 [DOI] [PubMed] [Google Scholar]
- 42. Said-Salim B., et al. 2003. Global regulation of Staphylococcus aureus genes by Rot. J. Bacteriol. 185:610–619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sardesai A. A., Gowrishankar J. 2001. Improvement in K+-limited growth rate associated with expression of the N-terminal fragment of one subunit (KdpA) of the multisubunit Kdp transporter in Escherichia coli. J. Bacteriol. 183:3515–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Schleussinger E., Schmid R., Bakker E. P. 2006. New type of kdp region with a split sensor-kinase kdpD gene located within two divergent kdp operons from the thermoacidophilic bacterium Alicyclobacillus acidocaldarius. Biochim. Biophys. Acta 1759:437–441 [DOI] [PubMed] [Google Scholar]
- 45. Shang F., et al. 2009. The Staphylococcus aureus GGDEF domain-containing protein, GdpS, influences protein A gene expression in a cyclic diguanylic acid-independent manner. Infect. Immun. 77:2849–2856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Smeltzer M. S., Hart M. E., Iandolo J. J. 1993. Phenotypic characterization of xpr, a global regulator of extracellular virulence factors in Staphylococcus aureus. Infect. Immun. 61:919–925 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Sugiura A., Nakashima K., Mizuno T. 1993. Sequence-directed DNA curvature in activator-binding sequence in the Escherichia coli kdp ABC promoter. Biosci. Biotechnol. Biochem. 57:356–357 [DOI] [PubMed] [Google Scholar]
- 48. Sugiura A., Nakashima K., Tanaka K., Mizuno T. 1992. Clarification of the structural and functional features of the osmoregulated kdp operon of Escherichia coli. Mol. Microbiol. 6:1769–1776 [DOI] [PubMed] [Google Scholar]
- 49. Torres V. J., et al. 2007. A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1:109–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Treuner-Lange A., Kuhn A., Durre P. 1997. The kdp system of Clostridium acetobutylicum: cloning, sequencing, and transcriptional regulation in response to potassium concentration. J. Bacteriol. 179:4501–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Voelkner P., Puppe W., Altendorf K. 1993. Characterization of the KdpD protein, the sensor kinase of the K(+)-translocating Kdp system of Escherichia coli. Eur. J. Biochem. 217:1019–1026 [DOI] [PubMed] [Google Scholar]
- 52. Walderhaug M. O., Litwack E. D., Epstein W. 1989. Wide distribution of homologs of Escherichia coli Kdp K+-ATPase among gram-negative bacteria. J. Bacteriol. 171:1192–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Wright J. S., III, et al. 2005. The agr radiation: an early event in the evolution of staphylococci. J. Bacteriol. 187:5585–5594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Xue T., Zhao L., Sun H., Zhou X., Sun B. 2009. LsrR-binding site recognition and regulatory characteristics in Escherichia coli AI-2 quorum sensing. Cell Res. 19:1258–1268 [DOI] [PubMed] [Google Scholar]
- 55. Yarwood J. M., McCormick J. K., Schlievert P. M. 2001. Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J. Bacteriol. 183:1113–1123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Zhao L., Xue T., Shang F., Sun H., Sun B. 2010. Staphylococcus aureus AI-2 quorum sensing associates with the KdpDE two-component system to regulate capsular polysaccharide synthesis and virulence. Infect. Immun. 78:3506–3515 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.