Abstract
Salmonella enterica serovar Typhimurium, an intracellular pathogen and leading cause of food-borne illness, encodes a plethora of virulence effectors. Salmonella virulence factors are translocated into host cells and manipulate host cellular activities, providing a more hospitable environment for bacterial proliferation. In this study, we report a new set of virulence factors that is translocated into the host cytoplasm via bacterial outer membrane vesicles (OMV). PagK (or PagK1), PagJ, and STM2585A (or PagK2) are small proteins composed of ∼70 amino acids and have high sequence homology to each other (>85% identity). Salmonella lacking all three homologues was attenuated for virulence in a mouse infection model, suggesting at least partial functional redundancy among the homologues. While each homologue was translocated into the macrophage cytoplasm, their translocation was independent of all three Salmonella gene-encoded type III secretion systems (T3SSs)–Salmonella pathogenicity island 1 (SPI-1) T3SS, SPI-2 T3SS, and the flagellar system. Selected methods, including direct microscopy, demonstrated that the PagK-homologous proteins were secreted through OMV, which were enriched with lipopolysaccharide (LPS) and outer membrane proteins. Vesicles produced by intracellular bacteria also contained lysosome-associated membrane protein 1 (LAMP1), suggesting the possibility of OMV convergence with host cellular components during intracellular trafficking. This study identified novel Salmonella virulence factors secreted via OMV and demonstrated that OMV can function as a vehicle to transfer virulence determinants to the cytoplasm of the infected host cell.
INTRODUCTION
Salmonella enterica serovar Typhimurium utilizes a secretory cascade of virulence effectors to interact with host cells. To date, more than 40 secreted virulence factors have been identified in Salmonella, but the functions and mechanisms of action are largely undefined (35, 52). Effector proteins conduct diverse functional activities, including actin rearrangement, modulation of vesicular trafficking, induction of inflammatory response, and regulation of motility of infected cells (52, 75, 80). Salmonella tightly controls the expression and secretion of virulence determinants to disrupt host cell activities at appropriate times and locations during infection. Failure to properly regulate these effectors in an appropriate manner results in attenuation of Salmonella virulence (14, 55, 57, 61). Thus, finely tuned regulation of virulence effectors allows Salmonella to respond to the host defense system, permitting persistent infection and promoting dissemination of the pathogen to other hosts.
The type III secretion system (T3SS), the best-characterized secretion apparatus, is a specialized needle-like organelle made up of more than 20 components, and it delivers more than 30 Salmonella effectors into host cells (24, 34, 58, 59). Salmonella pathogenicity island 1 (SPI-1) and SPI-2, mapping to separate regions on the S. Typhimurium chromosome, carry genes for distinct T3SSs, in addition to carrying genes encoding regulators, chaperones, and effector proteins. Virulence determinants secreted from SPI-1 T3SS primarily play roles in host cell invasion and enhance intestinal inflammation (24). SPI-2 T3SS-secreted virulence effectors are required for intracellular replication and persistence and inhibit the inflammatory response during a systemic infection (24). The flagellar system is structurally related to T3SS and may secrete virulence factors as well (31, 78, 79). Although Salmonella possesses other secretion mechanisms, including a two-partner secretion system (ZirT/ZirS) (25) and a type VI secretion system (9), the T3SS is considered the major mechanism to deliver virulence factors to the host cell. Therefore, identification of virulence factors secreted independently of these well-studied systems is important to understanding Salmonella pathogenesis.
In this study, we demonstrate that outer membrane vesicles (OMV) can act as a means to deliver previously unknown Salmonella virulence factors. Previous work has demonstrated that OMV are released from the cell envelopes of Gram-negative bacteria and are comprised of a variety of outer membrane and periplasmic constituents, including proteins, phospholipids, lipopolysaccharides (LPSs), and DNA (18, 44, 49, 64). The vesicles can vary in size from 10 to 300 nm in diameter depending on a number of factors and can serve as a vehicle to deliver the vesicular contents to adjacent bacterial and animal cells, as well as to the extracellular milieu (10, 22, 40, 41, 43). Salmonella enterica serovar Typhi encloses ClyA, a pore-forming cytotoxin, in OMV and exports it to the extracellular environment, thereby making contact with eukaryotic cells (74). OMV not only disseminate toxins and enzymes into host cells to protect bacterial survival but also serve as bacterium-like decoys to neutralize the host immune responses and facilitate Gram-negative pathogens' immune evasion (10, 71). OMV delivery of antigens has been a large area of investigation, as OMV could improve vaccines by delivery to the major histocompatibility complex (MHC) class I pathway while simultaneously providing pathogen-associated molecular patterns as adjuvants (2, 4, 62, 73). Based on the accumulative observations of OMV in a variety of microorganisms, multiple biological functions of OMV, including virulence attributes and its biogenesis, including production mechanism and cargo selectivity have been reviewed recently (1, 21, 46). Using a proteomics method to characterize the proteins that Salmonella secretes into culture medium under conditions mimicking the intracellular environment, we identified a group of proteins that are secreted independently of SPI-2 T3SS (58). Here, we demonstrate that a subset of these T3SS-independent secreted proteins is translocated into the host cytoplasm via OMV, including PagK (PagK1), PagJ, and STM2585A (PagK2).
MATERIALS AND METHODS
Bacterial strains and plasmids.
Salmonella enterica serovar Typhimurium 14028s was used as the parent in all strains studied here. All deletion and tagged strains were constructed using the phage λ Red recombination system (17). In brief, the kanamycin resistance (kan) or chloramphenicol acetyltransferase (cat) cassette was amplified by PCR with 40-nucleotide (nt) flanking sequences, homologous to target genes, at both termini. PCR products were introduced into recipient cells harboring pKD46 to replace target genes with a kan or cat cassette. Antibiotic marker genes were subsequently removed by flip recombinase provided from pCP20 to make in-frame deletions that were presumably nonpolar (17).
Tagging of chromosomal genes with cyaA′ was performed in a similar way using λ Red-mediated recombination. Instead of pKD13mod (77) and pKD3 (17) as PCR templates, pMini-Tn5-cycler (30) was used to insert a DNA fragment encoding a carboxy-terminal peptide of CyaA and kanamycin resistance immediately prior to the stop codon sequence of PagJ, PagK1, and PagK2. Construction of chromosomal sipA::cyaA′ and sseJ::cyaA′ was described previously (30).
In order to construct derivatives of PagK homologue-CyaA′ fusions, which lack their N termini or contain their N termini only, an additional flanking sequence providing the ribosome binding site (RBS) and a new start codon was positioned upstream of the truncated cyaA′ fusions on pMJW1753 (30). DNA fragments encoding intact PagK homologue-CyaA′ fusions were also cloned on pMJ1753 by the same strategy and served as the reference for the truncated fusion constructs. All primers used to construct bacterial strains and plasmids are listed in Table S1 in the supplemental material.
For a control for a variety of CyaA′ fusions, the C terminus of β-galactosidase α peptide was fused with CyaA′ on pMJW1753, generating pMJW1791 (30). This fusion construct should not be secreted by either T3SS or OMV as is demonstrated below.
To visualize Salmonella using red fluorescent protein (Tomato [68]), bacteria were transformed with pWKS30-Tomato (29). Bacterial cells were grown in Luria-Bertani (LB) medium or an acidic minimal medium as described previously (AMM1 [77]).
Macrophage infection.
Murine macrophage-like cells (ATCC RAW264.7 [63]) were prepared and infected as described previously (77). Except for bacteria containing a sipA::cyaA′ allele, all Salmonella strains were prepared for infection as described below. Salmonella cells grown overnight in LB were opsonized with 10% mouse serum (Innovative Research) for 20 min prior to infection. Bacterial cells resuspended in Dulbecco's modified Eagle's medium (DMEM) were added to macrophage monolayers at an input multiplicity of infection (MOI) of 100, and infections were initiated by centrifuging at 1,000 × g for 5 min. Infected macrophages were incubated at 37°C with 5% CO2 for 30 min, and extracellular bacteria were subsequently removed by washing the cells with phosphate-buffered saline (PBS) and incubating them in DMEM containing gentamicin (Gibco) at 100 μg/ml for 1 h. After treatment with gentamicin (100 μg/ml), the cells were washed with PBS three times and overlaid with DMEM containing 20 μg/ml gentamicin for the remainder of the experiments. Salmonella strains harboring the sipA::cyaA′ allele were cultivated in LB for 3 h to induce Salmonella pathogenicity island 1 (SPI-1) expression, opsonized with 10% mouse serum, and then added to macrophages at an input MOI of 50.
Outer membrane vesicle (OMV) isolation.
Salmonella cells were grown on AMM1 medium with a protease inhibitor cocktail (Complete protease inhibitor cocktail tablet; Roche) for 5 h and centrifuged at 5,000 × g for 10 min. The cell-free supernatant was filtered through a 0.45-μm-pore-size polyvinylidene difluoride (PVDF) filter (Millipore) to remove the remaining bacteria, and the filtrate was ultracentrifuged at 153,000 × g for 2 h at 4°C in a SW32 Ti rotor to pellet the vesicles (Beckman Instruments Inc.). The supernatant was carefully removed, leaving 1 ml at the bottom, which was then diluted with 35 ml of PBS and reultracentrifuged as described above. The pellets were resuspended in PBS, and the protein concentration was measured using a bicinchoninic acid (BCA) assay kit (Pierce).
cAMP assay.
To examine translocation of CyaA′-tagged proteins into the cytosol, macrophages infected with Salmonella CyaA′ fusion strains were washed with PBS three times and then lysed with 0.1 M HCl as directed in the manufacturer's instructions for the cyclic AMP (cAMP) enzyme immunoassay (EIA) kit (Assay Designs/Stressgen). Before the assay was performed, the lysate was spun down at 3,000 × g for 5 min to remove macrophages and bacterial debris, and the cytosolic fraction was used to determine the levels of cAMP. All cAMP assays were repeated at least three times using independently infected macrophages, and the averages are shown in this study.
Immunoblot analysis.
For Western blot analysis of isolated OMV, Salmonella cells were grown on AMM1 medium as described previously (77). Vesicles isolated from 80 ml of Salmonella culture were precipitated with trichloroacetic acid (TCA) (10%). The pellet was dissolved in 1× Laemmli sample buffer with heating, and the proteins were separated by SDS-PAGE.
To investigate protein expression inside macrophage cells, the infected RAW264.7 macrophage-like cells were incubated in lysis buffer (50 mM HEPES, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, 100 mM phenylmethylsulfonyl fluoride [PMSF], protease inhibitor cocktail, 50 mM NaF, DNase I [DNase Set; Qiagen], and 2 mM sodium orthovanadate) and centrifuged at 10,000 × g for 5 min to separate macrophage cytosol from the pellet containing host nuclei, cytoskeleton, and bacteria. The pellet was directly resuspended in 1× Laemmli sample buffer, boiled for 5 min, and loaded on an SDS-polyacrylamide gel. Proteins on gels were transferred to PVDF membranes and probed with antibodies. Anti-CyaA (3D1) antibody (Santa Cruz Biotechnology) and anti-DnaK antibody (Assay Designs) were used as the primary antibodies The antibody against OmpA was kindly provided by Roland Lloubès (CNRS, France). Anti-mouse IgG conjugated with peroxidase (Sigma) was used as a secondary antibody in all immunoblot experiments.
Immunofluorescence microscopy.
For CCF4-AM cleavage detection, macrophages were seeded in Lab-Tek II chamber cover glass slides (Nunc) and infected with Salmonella Bla fusion strains as described above. β-Lactamase (Bla) cleaves CCF4-AM to change its emission spectrum from green to blue when it is translocated into the cytoplasm. At 18 h postinfection, macrophages were loaded with membrane-permeable CCF4-AM (Invitrogen) solution for 2 h per the manufacturer's instructions, and the cleavage was examined using an Applied Precision (Issaquah) DeltaVision image restoration system with emission filter sets for green (528-nm) and blue (457-nm) fluorescence by CCF4-AM and in the red spectrum for the fluorescent protein named “Tomato” (617 nm). All Salmonella strains were transformed with pWKS30-Tomato (29). CCF4-AM cleavage assays were performed three times, and the images shown are representatives of three independent assays.
In order to locate CyaA′-tagged PagK homologues inside macrophages, RAW264.7 macrophage-like cells seeded on Lab-Tek II chamber cover glass slides were infected with Salmonella CyaA′ fusion strains for the desired time. Infected cells were fixed in 4% paraformaldehyde (Pierce) for 1 h and permeabilized with 0.1% saponin (Sigma). Rabbit anti-CyaA antibody (Santa Cruz Biotechnology) and Alexa Fluor 647-conjugated goat anti-rabbit IgG (Invitrogen) were used for CyaA′ detection. Mouse anti-LPS antibody (Abcam) and Alexa Fluor 350-conjugated goat anti-mouse IgG (Invitrogen) were used to label OMV and the Salmonella outer membrane. Fluorescein isothiocyanate (FITC)-conjugated cholera toxin B subunit (CTB) (Sigma) was used to identify lipid rafts of the plasma membrane. All antibodies were diluted in 10% goat serum (Abcam) supplemented with 0.1% saponin and 1% bovine serum albumin (BSA) (Sigma) prior to dispensing to the fixed macrophage samples. Bacteria were visualized with pWKS30-Tomato expressing the red fluorescent protein Tomato (29). For statistical analysis, three different fields of RAW264.7 cells were captured, and at least 50 cells were examined in each field. Images shown in this study are representative of more than 90% of cells examined from at least three independent specimen preparations. Images were acquired by an Applied Precision DeltaVision system, and deconvolution was conducted using Softwork Explorer Suite (Applied Precision) image processing software.
Immunogold electron microscopy.
In the analysis of isolated OMV, vesicle pellets were resuspended, fixed in PBS containing 2% paraformaldehyde, and then mounted onto Formvar-coated 300-mesh nickel grids (Electron Microscopy Sciences [EMS]). After 20 min of incubation and attachment of the vesicle samples on the grids, they were washed with PBS twice and blocked with a buffer containing 5% BSA, 0.1% cold water fish skin (CWFS) gelatin (EMS), 15 mM NaN3, 0.05% Tween 20, and 10% goat serum. After the grids were rinsed with a washing buffer containing 0.1% Aurion BSA-c (EMS), 0.1% CWFS gelatin (EMS), 15 mM NaN3, 0.005% Tween 20, and 5% goat serum, they were incubated with rabbit anti-CyaA antibody (Santa Cruz Biotechnology) or rabbit antilipolysaccharide (anti-LPS) antibody (Abcam) at 4°C overnight and sequentially treated with 15-nm-gold-conjugated anti-rabbit IgG (EMS) for 2 h at room temperature. The antibodies were diluted in the washing buffer described above. After the grids were rinsed with the washing buffer and washed with ultrapure water, they were poststained with 8% uranyl acetate and Reynolds' lead citrate (65).
For immunogold microscopy of macrophages, RAW264.7 macrophage-like cells were infected with Salmonella CyaA′ fusion strains and fixed with 4% paraformaldehyde as described above. The cells were dehydrated with a graded ethanol series and embedded in LR-White resin (London Resin Co., Hampshire, United Kingdom) (72), and then the samples were polymerized for 24 h and sectioned. Thin sections were collected on pioloform-coated nickel grids and treated with the blocking buffer described above. The grids were incubated with either rabbit anti-CyaA antibody or rabbit anti-lysosome-associated membrane protein 1 (anti-LAMP1) antibody (Abcam) and mouse anti-LPS antibody (Abcam) at 4°C overnight and sequentially probed with 15-nm-gold-conjugated anti-rabbit IgG and 25-nm-gold-conjugated anti-mouse IgG (EMS) as described above. After serial washing steps with the washing buffer and ultrapure water, the grids were poststained with 8% uranyl acetate and Reynolds' lead citrate. The samples were analyzed using a transmission electron microscope (Morgagni-FEI) at an acceleration voltage of 70.0 kV at calibrated magnifications. At least 50 isolated vesicles or infected macrophages were examined per experimental specimen. The images shown in this study represent the dominant phenotypes detected in more than 80% of the micrographs.
Competitive infection study.
To examine the fitness of a strain lacking all three PagK homologues during competitive infection in mice, the competitive index (CI) assay was performed. Test strains (ΔpagJ pagK1 pagK2 mutant or S. Typhimurium 14028s) and the reference strain MA6054 (37) were separately grown in LB medium overnight. Salmonella Typhimurium MA6054 carries a gene that encodes an arabinose-inducible β-galactosidase. Prior to mouse infection, the bacterial cells were washed with PBS, and each test strain was mixed with the reference strain at a 1:1 ratio. The bacterial mix was diluted with PBS, and doses of 104 CFU were inoculated intraperitoneally (i.p.) into groups of five female 5- to 6-week-old 129SvJ mice (Jackson Laboratory). Infected animals were euthanized and dissected at 7 days postinfection. The spleen was isolated, homogenized, and plated at different dilutions on LB agar medium containing 40 μg/ml X-Gal (5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) (Sigma) and 1 mM arabinose (Sigma). The competitive index (CI) was calculated as follows: CI = (percentage of test strain recovered/percentage of reference strain recovered)/(percentage of test strain inoculated/percentage of reference strain inoculated).
Ethics statement.
Mouse experiments in this study were performed in accordance with the Guide for the Care and Use of Laboratory Animals (57a). The animal protocol was approved by the Oregon Health & Science University Institutional Animal Care and Use Committee (OHSU IACUC) under permit number A085/2008. All efforts were made to minimize animal suffering during the experiments.
RESULTS
Translocation of PagK homologues is independent of SPI-1/SPI-2 T3SSs and the flagellar system.
PagK and its homologue PagJ were first identified as PhoP/PhoQ-activated genes by Miller et al. (6, 32, 33, 54), but the deletion of these genes individually or in combination did not attenuate Salmonella virulence in mice. We identified another homologue of PagK, STM2585A (STM14_3167 in strain 14028s) and named it PagK2 to distinguish it from PagK (hereafter called PagK1). They are small proteins composed of 66 amino acids (aa) (PagK1 and PagJ) and 75 aa (PagK2) and exhibit high homology to each other (between 83 and 95% amino acid sequence identity; see Fig. S1A in the supplemental material). A Salmonella strain deleted for all three pagK homologues was outcompeted by wild-type Salmonella in mice, which implies that all three proteins are required for full virulence (see Fig. S1B in the supplemental material). Interestingly, PagK2 alone also appeared to be required for Salmonella intracellular growth to some extent, differing from the other PagK homologues despite high sequence homology among them (data not shown). In secretome profiling, these homologue proteins were identified in the culture fluid of acidic nutrient-restricted medium that stimulates the expression of a Salmonella pathogenicity island 2 (SPI-2) type III secretion system (T3SS) (58). However, a strain (ΔssaK mutant) lacking an essential component of this secretion apparatus still secreted the PagK-homologous proteins. Translocation of PagK homologues into the macrophage cytosol was verified by fusion to β-lactamase missing its signal sequence. When macrophages were infected with Salmonella expressing a C-terminally tagged β-lactamase, the tagged proteins were translocated into the cytoplasm, and cleaved fluorescent substrate, with CCF4 changing its fluorescence to blue as shown (see Fig. S2 in the supplemental material).
A number of effector proteins can be secreted by both SPI-1 T3SS and SPI-2 T3SS, which allows for secretion under more-diverse conditions (29, 53). Type III secretion systems are evolutionarily related to the flagellar system, and in fact, SopE can be secreted via the flagellar system, as well as via SPI-1 T3SS (20). In order to test the possibility that these three proteins were secreted via another type III system, we deleted the genes encoding essential components for all three secretion systems (InvA in SPI-1 T3SS, SsaK in SPI-2 T3SS, and FlgB in the flagellar system). A carboxy-terminal translational fusion with CyaA′ was constructed for each PagK-related protein and used to evaluate translocation into the eukaryotic cytoplasm. The fused cyaA′ encodes an active fragment of the calmodulin-dependent adenylyl cyclase from Bordetella pertussis (69). Calmodulin is ubiquitous within animal cells but absent within Salmonella or the Salmonella-containing vacuole (SCV), the main Salmonella reservoir within host cells; therefore, translocation of PagK homologues into the host cytoplasm can be determined by measuring mammalian cyclic AMP (cAMP) levels (19). To verify that all three secretion systems were disrupted by deleting invA, ssaK, and flgB, translocation of SipA (an SPI-1 T3SS-secreted effector) and SseJ (a SPI-2 T3SS-secreted effector) was examined in macrophages infected with ΔinvA and ΔssaK mutants using CyaA′ fusion (Fig. 1 A). SipA-CyaA′ and SseJ-CyaA′ were not translocated in the absence of InvA and SsaK, respectively, and did not increase cytosolic cAMP levels. Similarly, deletion of flgB abolished Salmonella motility on agar plates (data not shown). When intracellular cAMP levels were measured 18 h after macrophage infection, PagK homologue-CyaA′ fusion proteins were translocated even when all three secretion systems were disrupted (SPI-1 T3SS, SPI-2 T3SS, and the flagellar system), indicating that these proteins were translocated by an alternative mechanism (Fig. 1A). In fact, Western hybridization on the macrophage lysate revealed that these proteins all showed measurable increases in protein levels when the three T3SSs were inactivated as discussed below (Fig. 1B). Visual inspection of the sequences of these three effectors did not detect the twin-arginine translocation (TAT) signal for secretion but suggested the possibility of the Sec pathway-dependent secretion as described next.
Fig. 1.
PagK homologues are translocated independent of Salmonella pathogenicity island 1 (SPI-1) or SPI-2 type III secretion system (T3SS) and the flagellar system. Macrophages were infected with Salmonella strains expressing PagK1, PagK2, or PagJ-CyaA′ in the presence (+) or absence (−) of SPI-1/2 T3SS and flagella for 18 h. (A and B) Translocation of CyaA′ fusions into the macrophage cytosol was measured by the cAMP assay (A), and their expression levels were compared in parallel by Western blotting using anti-CyaA antibody (αCyaA) (B). DnaK levels were used to normalize protein levels. In order to confirm that SPI-1 and SPI-2 secretion systems were disrupted by deleting invA and ssaK, SipA (SPI-1 effector) and SseJ (SPI-2 effector) were tagged with CyaA′, and their translocation (A) and expression (B) were examined in the absence of InvA and SsaK, respectively. Note that translocation and expression of SipA-CyaA′ was measured at 1 h postinfection, and the analysis of the other CyaA′-tagged proteins was performed at 18 h postinfection. Salmonella not expressing CyaA′ (strain 14028s) or expressing LacZ-CyaA′ from pMJW1791 (30) did not exhibit an increased cAMP level in panel A as expected. Intriguingly, the level of expression of each PagK homologue increased in the triple mutant background as can be observed.
N-terminal deletion of PagK homologues abolishes their translocation into the host cytoplasm.
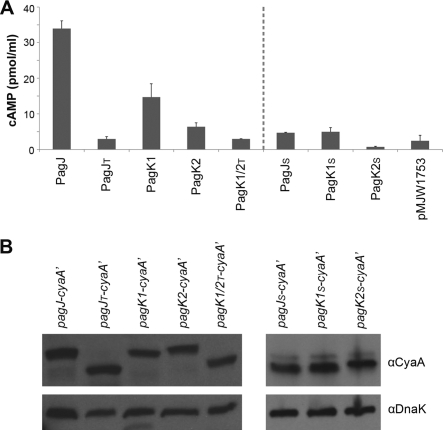
PagJ, PagK1, and PagK2 were translocated into the macrophage cytosol independent of all three type III secretion systems tested. Analysis of amino acid sequences using the SignalP 3.0 server (7) suggested the presence of signal peptides at the amino termini (N termini) of PagJ, PagK1, and PagK2 that would direct these peptides to the periplasm (see Fig. S3 in the supplemental material). To determine whether they were translocated via the signal sequences, we deleted the first 78 nucleotides (PagJ and PagK1) and 105 nucleotides (PagK2) following the initiation codon that encode the putative signal peptides (see Fig. S1A for their truncated derivatives). Due to the high sequence homology between PagK1 and PagK2, deletion of the putative signal sequence in PagK1 produces a sequence identical to that of PagK2 (see Fig. S1 in the supplemental material). Intact and truncated forms of PagJ, PagK1, and PagK2 were fused with CyaA′, and their translocation into the host cytosol was compared at 6 h postinfection (Fig. 2, left side of graph). Translocation of PagK homologues was abolished when their N-terminal signal peptides were removed, yet the expression of each protein, as determined by Western hybridization, was comparable between the intact and truncated constructs. This result suggests that the N-terminal peptides enabled these proteins to be delivered into the host cytoplasm. We further tested whether the signal peptides alone were sufficient to lead to translocation of any bacterial proteins into host cells. The N termini encoding 27 aa of PagK1/PagJ and 36 aa of PagK2 were fused with CyaA′, and the translocation into the macrophage cytoplasm was examined (Fig. 2A, right side of panel). CyaA′ with the signal peptides of PagK homologues significantly decreased intracellular cAMP levels compared with CyaA′ fused with full-length PagK homologues, indicating that the signal sequences are necessary but not sufficient to cause their translocation. The N termini of these proteins is likely to be required to cross the inner membrane through the Sec system, but not enough to lead to secretion across the outer membrane barrier, as described below in Discussion. The differences in expression shown in Fig. 1 and 2 were a consequence of differences in fusion construction: expression of CyaA′ fusions from their native promoters in Fig. 1 versus expression of CyaA′ fusions under the common Plac promoter and Shine-Dalgarno sequence in Fig. 2 (see Materials and Methods).
Fig. 2.
N-terminal deletion of PagK homologues abolishes their translocation into the host cells. The full-length PagK homologues and their truncated forms without their predicted N-terminal signal peptides or with their N-terminal signal peptides only were fused to CyaA′ as described in Materials and Methods. Salmonella strains expressing the full-length PagK homologues (PagJ, PagK1, and PagK2) and the truncated derivatives (PagJT and PagK1/PagK2T lacking N-terminal signal peptides; PagJS, PagK1S, and PagK2S harboring only N-terminal signal peptides) were inoculated into RAW264.7 macrophage-like cells. (A and B) Infected macrophage cells were lysed at 6 h postinfection for cAMP assay (A) and Western blot analysis (B). (A)The translocation levels of PagK homologues and their derivatives were compared by measuring intracellular cAMP levels. (B) In parallel, their expression levels were determined by anti-CyaA antibody using the same macrophage lysates used in the cAMP assay. DnaK levels were measured to normalize protein levels loaded in the different lanes. Note that the sequences of PagK1 and PagK2 are identical if the signal sequences have been deleted; therefore, only a single construct, PagK1/2T, is shown for the N-terminally truncated derivatives of PagK1 and PagK2. Salmonella harboring pMJW1753, which was used as a backbone construct in all CyaA′ fusions in this experiment, was used in the cAMP assay as a negative control.
Immunofluorescence microscopy provided insights into the locations of PagK homologues inside macrophages. Macrophage cells were infected with Salmonella expressing PagK2-CyaA′ for 18 h. Translocated PagK2-CyaA′ was detected by anti-CyaA antibody and Alexa Fluor 647-conjugated secondary antibody as shown in yellow in Fig. 3. Lipopolysaccharide (LPS), a major component of the Salmonella outer membrane, was visualized in blue with anti-Salmonella LPS antibody and secondary Alexa Fluor 350-labeled antibody. Salmonella cells expressed the red fluorescent protein named “Tomato” from plasmid pWKS30-Tomato (30). The macrophage plasma membrane was labeled in green with fluorescein isothiocyanate (FITC)-conjugated cholera toxin B subunit (CTB), which binds to the glucosphingolipid GM1 ganglioside of the plasma membrane (15). The merged image demonstrated that PagK2 was colocalized with LPS, forming irregular punctate spots (see spots harboring yellow and blue signals, which are indicated with white arrows in Fig. 3F). Moreover, these spots were distinct from intracellular bacteria and were smaller than bacteria that were fluorescent due to the red fluorescent protein Tomato. The observation of PagK2 in punctate compartments that are distinguishable from bacteria raised the possibility that PagK2 was translocated into the cytosol enclosed within vesicles. Macrophages infected with wild-type bacteria not expressing CyaA′ showed LPS-containing punctate structures as well, but they were not labeled with anti-CyaA′ antibody, while uninfected macrophages were labeled only with FITC-conjugated CTB (see Fig. S4 in the supplemental material). PagK1 and PagJ showed similar punctate fluorescence patterns to that observed for PagK2 (Fig. S4).
Fig. 3.
PagK2 is colocalized with LPS but not with Salmonella inside macrophages. Macrophages were infected with Salmonella expressing PagK2-CyaA′ for 18 h. (A to D) The fixed cells were incubated with rabbit anti-CyaA antibody (B) and mouse anti-LPS antibody (A) overnight and treated with Alexa Fluor 647-conjugated anti-rabbit IgG (B), Alexa Fluor 350-conjugated anti-mouse IgG (A), and FITC-conjugated cholera toxin B subunit (CTB) (D). Salmonella expresses the red fluorescent protein “Tomato” from pWKS30-Tomato plasmid (C). (A) LPS staining in the blue channel; (B) CyaA′ staining in the yellow channel; (C) Salmonella with fluorescence from the the red fluorescent protein Tomato; (D) plasma membrane staining in the green channel using CTB; (E) merged image of LPS, CyaA′, and Salmonella; (F) merged image of LPS, CyaA′, CTB, and Salmonella. The sites of colocalization of PagK2 with LPS are indicated by white arrows in panel F. Note that a bleb-like protuberance from the host cell (indicated by the white arrow in panel D) encloses PagK2 and LPS (as shown in panel F), while the bacteria are present in the spacious vacuole seen as a large red vacuole at the top of the cell as depicted.
PagK homologues are enclosed within OMV secreted into the culture medium.
Pathogenic Gram-negative bacteria produce outer membrane vesicles (OMV) composed of outer membrane proteins, specific periplasmic proteins, lipopolysaccharide, and phospholipids and can deliver virulence determinants to adjacent bacterial or eukaryotic cells (10, 18, 41, 43, 44, 49, 74). Spano et al. observed that CdtB, an S. Typhi exotoxin, was exported extracellularly within a compartment described as small “puncta” inside the host cell cytoplasm that they speculated could be OMV (70). We also observed that PagK-homologous proteins were translocated in a punctate pattern, colocalized with LPS (Fig. 3; see Fig. S4 in the supplemental material). Because of the possibility that these punctate particles were vesicles released from the Salmonella outer membrane, we performed a direct test to determine whether PagK homologues were carried within outer membrane vesicles.
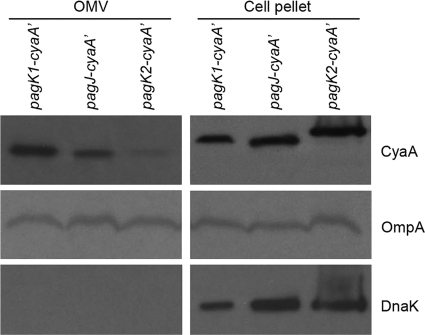
Salmonella strains expressing CyaA′-tagged PagK homologues were cultivated in an acidic minimal medium (AMM1 [77]) for 5 h, and the culture medium was filter sterilized following centrifugation to remove bacterial cells. Sterile supernatant was ultracentrifuged and concentrated in a small volume. The constituents within the isolated vesicles included PagK1, PagJ, and PagK2, as analyzed by protein immunoblotting (Fig. 4). Colocalization of OmpA, a major component of vesicles as well as the outer membrane, and the lack of cytosolic protein DnaK in the vesicle fractions support the hypothesis of OMV-mediated translocation of these proteins. Intriguingly, the molecular weight of PagK2 appeared to be identical to that of PagK1 and PagJ in the vesicle fractions, while it was larger than the other two when cell pellets were analyzed. One explanation for this result could be that the signal sequence had been removed in the vesicle fractions but not inside the bacteria, resulting in peptides of comparable molecular weights when secreted into vesicles.
Fig. 4.
PagK homologues are present in isolated outer membrane vesicles (OMV). Salmonella strains expressing CyaA′-tagged PagK1, PagJ, and PagK2 (C-terminal translational fusions) were cultivated on AMM1 medium for 5 h, and vesicles were isolated as described in Materials and Methods. Vesicles produced from 80 ml of bacterial culture (∼24 μg/80 ml) were precipitated with TCA to concentrate proteins. (Left) The vesicle-containing precipitate produced from the initial 80-ml culture supernatant was resuspended in 1× Laemmli sample buffer and loaded in a lane of the SDS-polyacrylamide gel (OMV fraction). (Right) Bacterial cells (5 × 107) were pelleted and loaded onto the SDS-polyacrylamide gel (cell pellet fraction). PagK1, PagJ, and PagK2 were immunoblotted using anti-CyaA antibody. OmpA, a highly expressed outer membrane protein, was used as a control for the presence of outer membrane vesicles. DnaK, a cytosolic protein, was used to verify that no bacterial lysis occurred during the OMV isolation procedure. Note that the molecular weight of PagK2 is approximately the same as PagK1 and PagJ in OMV fraction but not within whole bacterial cells (compare the molecular weight of PagK2 with those of PagK1 and PagJ in the OMV and cell pellet fractions). We speculate that signal sequences present in these effectors may contribute to the difference in molecular weights between the left- and right-hand panels.
In order to further investigate the location of these proteins within OMV, the vesicles were isolated from the culture supernatant as described above and examined by transmission electron microscopy (TEM). The vesicle fractions were incubated, allowed to attach to Formvar-coated grids, and processed for immunogold labeling without sectioning. CyaA′-tagged PagK homologues and LPS were identified using gold-conjugated antibodies. By this technique, LPS was observed on all vesicles, while PagK homologues were not observed on the surfaces of unsectioned vesicles (less than 2% of examined vesicles contained CyaA′-tagged PagK homologues on their surface; Fig. 5). This observation suggested that OMV contained LPS on their outer surfaces but enclosed PagK homologues inside the vesicles, as tested further below.
Fig. 5.
Localization of PagK homologues in intact unsectioned OMV. Vesicles were isolated from the culture of Salmonella pagK2-cyaA′ strain grown on AMM1 medium as described in Materials and Methods. Isolated vesicles were fixed and loaded onto Formvar/carbon-coated grids without sectioning vesicles. (A and B) The grids were incubated with a solution containing rabbit anti-CyaA antibody (A) or rabbit anti-LPS antibody (B) overnight and then treated with 15-nm-gold-conjugated anti-rabbit IgG. The micrographs are representative of the vesicles examined. (A) PagK2-CyaA′; (B) LPS. Vesicles isolated from cultures of pagJ-cyaA′ and pagK1-cyaA′ strains also showed similar distribution patterns of anti-CyaA and anti-LPS antibodies.
PagK homologues are translocated into the host cytoplasm via OMV.
The failure to find PagK homologues on the surfaces of vesicles, which were isolated in acidic minimal medium, prompted us to test whether the proteins were located inside vesicles and whether the vesicle properties varied for intracellular and extracellular bacteria. In fact, increased bacterial vesiculation has been previously observed near infected host cells and tissues (21, 39, 51, 71, 76). To investigate the events that take place within host cells, macrophages were infected with Salmonella CyaA′ fusion strains for 18 h and then embedded, sectioned, and employed as specimens in TEM analysis using gold-labeled antibodies. Antibodies directed against the CyaA′ peptide and LPS bound to their targets specifically, showing that LPS molecules were mainly distributed along the Salmonella outer membrane and near vesicles, whereas CyaA′ peptides were not detectable in uninfected and wild-type Salmonella-infected macrophages. TEM revealed that PagJ, PagK1, and PagK2 (all shown as smaller beads) were colocalized with LPS (shown as larger beads) on vesicular compartments (Fig. 6). Comparing the CyaA′ signal distribution between cellular organelles, 25.5% ± 5.4% of anti-CyaA antibody was found on OMV-like structures, 37.6% ± 11.9% was on the Salmonella moiety, including the envelope and the interior compartments, and 36.9% ± 14.5% was distributed in the vacuolar interior or the macrophage cytoplasm. In these micrographs, OMV harboring PagK homologues appeared to be either fused to the bacterial outer membrane (Fig. 6C, D, and E) or floating in the vicinity of the Salmonella-containing vacuole (SCV) membrane (Fig. 6A, D, and F). Moreover, OMV that were transported to the macrophage cytoplasm across the SCV membrane were occasionally clustered and were colabeled with an antibody detecting LAMP1, a lysosomal glycoprotein enriched on the SCV membrane (27) (Fig. 7). This observation raises the possibility that OMV are modified by cellular components during exocytosis. Overall, these results suggest that OMV serve as an important secretion mechanism for virulence determinants by Salmonella when it is intracellular.
Fig. 6.
Immunogold electron microscopy to locate PagK homologues inside macrophages. Macrophages were infected with Salmonella CyaA′ fusion strains for 10 h and fixed with 4% paraformaldehyde for transmission electron microscopy. The methods are described in detail in Materials and Methods. CyaA′-tagged PagK homologues were detected using rabbit anti-CyaA antibody and 15-nm-gold-conjugated anti-rabbit IgG (small black beads indicated by small black arrows). LPS was labeled with 25-nm-gold-conjugated anti-LPS antibody (large black beads). Salmonella and OMV are indicated by S and V, respectively. (A to F) The micrographs show macrophages infected with Salmonella strain pagJ-cyaA′ (A and B), pagK1-cyaA′ (C and D), and pagK2-cyaA′ (E and F).
Fig. 7.
Modification of OMV during translocation into the cytoplasm. (A and B) Uninfected macrophages (A) and Salmonella-infected macrophages (B) (10 h after infection) were examined after ultrathin sectioning. The grids were treated with 15-nm-gold-conjugated anti-LAMP1 antibody and 25-nm-gold-conjugated anti-LPS antibody as described in Materials and Methods. Based on the examination of 100 cells from the thin sections prepared in parallel, LAMP1 molecules (small black beads indicated by black arrows) were more plentiful in Salmonella-infected macrophages and appeared to be enriched in OMV released from the Salmonella-containing vacuole. Salmonella and OMV are labeled with S and V, respectively. (A) RAW264.7 macrophage-like cells; (B) RAW264.7 cells infected with S. Typhimurium 14028s.
DISCUSSION
The type III secretion system (T3SS) is one of the major mechanisms for transporting virulence factors in Gram-negative pathogenic bacteria. However, a subset of proteins including PagJ, PagK1, and PagK2 was translocated into the host cytosol independent of the three type III secretion systems tested. Interestingly, the expression and translocation of all three effectors (PagJ, PagK1, and PagK2) were increased in strains in which Salmonella pathogenicity island 1 (SPI-1) T3SS, SPI-2 T3SS, and the flagellar system were inactivated (Fig. 1). When the T3SSs were individually mutated, only a mutation in the SPI-2 gene-encoded T3SS increased their protein levels inside macrophages, even though bacterial growth was restrained due to the lack of SPI-2 T3SS (unpublished data). Expression of a wild-type copy of ssaK in the SPI-2 T3SS-defective strains (ΔssaK mutant) restored their expression to those of the parent strain, which showed that the effects were not due to second site mutations or to polar effects (data not shown). The mRNA levels of the pagJ, pagK1, and pagK2 genes were not affected by a mutation in SPI-2 T3SS inside macrophages, indicating that the increase of PagK-homologous proteins in SPI-2 T3SS-defective strains was caused by posttranscriptional regulation (data not shown).
In the past decade, OMV have been discovered as a new vehicle to deliver virulence molecules, although the process by which Gram-negative organisms produce vesicles is an area of active investigation. Deatherage et al. recently demonstrated that the cell envelope controls OMV release properties, including production rate, size distribution, and protein content via conserved protein associations such as outer membrane protein-peptidoglycan-inner membrane protein interactions (18). We suggest that OMV-mediated secretion of PagK homologues is a two-step process in which proteins first cross the inner membrane through the Sec system and then are enclosed within a spherically extruded outer membrane vesicle, although the requirement for the Sec system has not been directly tested. Putative signal peptides were observed at the N termini in PagJ, PagK1, and PagK2 (see Fig. S3 in the supplemental material), and deletion of 26 (PagJ and PagK1) and 35 (PagK2) N-terminal amino acids abolished their translocation into the macrophage cytoplasm, as predicted by the two-step model (Fig. 2). Furthermore, the peptides observed within vesicles were smaller than those observed in whole bacteria, which is suggestive of signal peptide cleavage during the Sec-dependent secretion pathway, although the fact may also suggest that cleavage occurs concomitantly with extrusion into vesicles. In the same context, translational fusions of green fluorescent protein (GFP) at the C termini of PagK homologues did not fluoresce (unpublished observation), as would be expected for the Sec pathway: GFP fusions secreted through the Sec pathway do not fold correctly, while GFP fusions secreted through the twin-arginine translocation (TAT) pathway are exported as properly folded proteins and remain fluorescent (23, 67).
All three PagK homologues studied in this work possess signal sequences and appeared to be transported to the periplasm via the Sec pathway. However, not all periplasmic proteins are secreted in OMV. Others have demonstrated the selectivity in OMV contents (18, 38, 42, 45–48). Lee et al. reported that some outer membrane and periplasmic proteins with low abundance were specifically enriched in OMV fractionation, whereas dominant periplasmic proteins were excluded (47). Deatherage et al. observed that the protein contents within OMV varied depending on the location in which the vesicle was released; OMV released from a septal region between dividing cells were enriched with proteins localized in the division septa, although this study was carried out using a filamentous strain to induce differential OMV populations (18). The fact that OMV production is increased under harsh conditions such as the host environment during infection (21, 39, 71) suggests that OMV production is related to a pathogenic bacterial virulence attribute responding to the host immune system (21, 46). The possibility that the composition of the released outer membrane material is influenced by nutrients present in the medium has been suggested for Escherichia coli (50). Proteins close to the inner leaflet of the outer membrane may be more likely to be enclosed within OMV than proteins with an affinity for the inner membrane. In support of the selectivity in OMV cargo, we verified that several periplasmic proteins, including β-lactamase, periplasmic dipeptidase (PdgL), and two putative periplasmic proteins (YiiQ and STM1263) were not translocated to the infected-cell cytoplasm despite the fact that they carry periplasmic signal sequences and, in the case of β-lactamase and PdgL, their location within the periplasm is well documented (36, 56; unpublished data). Furthermore, CyaA′ tagged with N-terminal signal sequences of PagK homologues were not translocated, indicating that their signal peptides are necessary but not sufficient to enable their secretion into OMV (Fig. 2).
Beatty et al. observed that mycobacterial cell wall lipids were liberated from phagosomes containing bacteria and accumulated in late endosomal/lysosomal compartments and speculated that the endocytic pathway was responsible for the trafficking of bacterial cell wall constituents (5). The Salmonella-containing vacuole (SCV) is a modified late endosomal compartment that has been shown to be blocked in fusion with specific terminal lysosome components, thereby providing a replicative niche for Salmonella within host cells (16, 26, 28). However, the SCV can fuse with specific late vesicular bodies, and it maintains interaction with the endocytic pathway (19, 60). Electron micrographs revealed that OMV traversing SCV contained LAMP1, a protein abundant on the late endosomal/lysosomal membrane as well as the SCV membrane (Fig. 7). Moreover, uninfected macrophages harboring LPS signals were frequently observed in microscopic analyses (see the panel of S. Typhimurium 14028s in Fig. S4 in the supplemental material), raising the possibility of OMV trafficking between macrophages. Dynamic interactions of SCVs with lysosomal contents suggest the possibility that the OMV cross the SCV membrane and hijack lysosomal exocytosis to be transported outside the infected host cell. Exocytosis of lysosomes and lysosomal contents has been observed in response to the increase in cytosolic calcium caused by an invading pathogen, such as Salmonella (66), Trypanosoma cruzi (12, 13) and Neisseria (3). The question of how LAMP1 associates with bacterial outer membrane vesicles is still open. To examine the possibility of cell-to-cell transfer of OMV, the vesicles were isolated from conditioned medium containing Salmonella-infected macrophages by filter sterilizing the medium and centrifuging it, and then the vesicles were added directly to uninfected macrophages. Following the addition of sterile vesicle fractions, we observed that CyaA′-tagged PagK homologues were either free in the cytoplasm or colocalized with LPS, forming a punctate pattern in juxtaposition with the macrophage plasma membrane (unpublished observation). This observation suggests that vesicles may be exocytosed from infected macrophages and taken up by “bystander” macrophages. Bhatnagar et al. demonstrated that exosomes liberated from Salmonella-infected macrophages could transfer Salmonella-derived LPS to uninfected macrophages and stimulate proinflammatory responses, which we have also observed (8).
PagJ, PagK1, and PagK2 were translocated into the host cytoplasm enclosed within OMV, and they were required for Salmonella fitness within mice. Ellis et al. demonstrated that the combined action of vesicular proteins and LPS triggered upregulation of proinflammatory cytokines in macrophages infected by Pseudomonas aeruginosa (22). To gain insights into the virulence mechanism of PagK homologues in Salmonella infection, we examined whether the vesicles isolated from a strain lacking PagK homologues altered host inflammatory responses. However, the levels of cytokine induction were comparable for wild-type OMV- and PagK homologue-deficient OMV-treated macrophages, indicating that PagK-related proteins play a yet unidentified role during Salmonella proliferation inside the host (unpublished data). Considering the high sequence homology among these proteins, these three peptides are likely to function in a redundant manner, as observed for other virulence effectors (11).
It remains unclear how specific proteins become enclosed in OMV and how these vesicles deliver their contents across the SCV. The apparent feedback loop between SPI-2 type III secretion and expression of OMV proteins discussed above (Fig. 1) also merits additional investigation. What is clear is that Salmonella secretes multiple virulence factors within outer membrane vesicles. The three highly related peptides described in this report are only 39 amino acids long when the putative signal peptides are removed, and hence, they are unlikely to have any enzymatic activity. Although the functions of these redundant proteins have yet to be established, this study discovered a new subset of Salmonella virulence effectors that is translocated via outer membrane vesicles.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Rebecca Tempel, George Niemann, Roslyn Brown, Liang Shi, and Penny Colton for helpful discussions. We thank Aurelie Snyder for technical assistance with immunofluorescence microscopy and Michael Webb for technical assistance with TEM.
This work was supported by the National Institute of Allergy and Infectious Diseases NIH/DHHS through interagency agreement Y1-AI-8401 (project website www.SysBEP.org).
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 4 April 2011.
REFERENCES
- 1. Amano A., Takeuchi H., Furuta N. 2010. Outer membrane vesicles function as offensive weapons in host-parasite interactions. Microbes Infect. 12:791–798 [DOI] [PubMed] [Google Scholar]
- 2. Asensio C. J., et al. 2011. Outer membrane vesicles obtained from Bordetella pertussis Tohama expressing the lipid A deacylase PagL as a novel acellular vaccine candidate. Vaccine 29:1649–1656 [DOI] [PubMed] [Google Scholar]
- 3. Ayala B. P., et al. 2001. The pilus-induced Ca2+ flux triggers lysosome exocytosis and increases the amount of Lamp1 accessible to Neisseria IgA1 protease. Cell. Microbiol. 3:265–275 [DOI] [PubMed] [Google Scholar]
- 4. Bakke H., et al. 2001. Meningococcal outer membrane vesicle vaccine given intranasally can induce immunological memory and booster responses without evidence of tolerance. Infect. Immun. 69:5010–5015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Beatty W. L., et al. 2000. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic 1:235–247 [DOI] [PubMed] [Google Scholar]
- 6. Belden W. J., Miller S. I. 1994. Further characterization of the PhoP regulon: identification of new PhoP-activated virulence loci. Infect. Immun. 62:5095–5101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bendtsen J. D., Nielsen H., von Heijne G., Brunak S. 2004. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 340:783–795 [DOI] [PubMed] [Google Scholar]
- 8. Bhatnagar S., Shinagawa K., Castellino F. J., Schorey J. S. 2007. Exosomes released from macrophages infected with intracellular pathogens stimulate a proinflammatory response in vitro and in vivo. Blood 110:3234–3244 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bingle L. E., Bailey C. M., Pallen M. J. 2008. Type VI secretion: a beginner's guide. Curr. Opin. Microbiol. 11:3–8 [DOI] [PubMed] [Google Scholar]
- 10. Bomberger J. M., et al. 2009. Long-distance delivery of bacterial virulence factors by Pseudomonas aeruginosa outer membrane vesicles. PLoS Pathog. 5:e1000382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bruno V. M., et al. 2009. Salmonella Typhimurium type III secretion effectors stimulate innate immune responses in cultured epithelial cells. PLoS Pathog. 5:e1000538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Caler E. V., Morty R. E., Burleigh B. A., Andrews N. W. 2000. Dual role of signaling pathways leading to Ca2+ and cyclic AMP elevation in host cell invasion by Trypanosoma cruzi. Infect. Immun. 68:6602–6610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Caler E. V., Vaena de Avalos S., Haynes P. A., Andrews N. W., Burleigh B. A. 1998. Oligopeptidase B-dependent signaling mediates host cell invasion by Trypanosoma cruzi. EMBO J. 17:4975–4986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coombes B. K., Wickham M. E., Lowden M. J., Brown N. F., Finlay B. B. 2005. Negative regulation of Salmonella pathogenicity island 2 is required for contextual control of virulence during typhoid. Proc. Natl. Acad. Sci. U. S. A. 102:17460–17465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Cuatrecasas P. 1973. Interaction of Vibrio cholerae enterotoxin with cell membranes. Biochemistry 12:3547–3558 [DOI] [PubMed] [Google Scholar]
- 16. Cuellar-Mata P., et al. 2002. Nramp1 modifies the fusion of Salmonella typhimurium-containing vacuoles with cellular endomembranes in macrophages. J. Biol. Chem. 277:2258–2265 [DOI] [PubMed] [Google Scholar]
- 17. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deatherage B. L., et al. 2009. Biogenesis of bacterial membrane vesicles. Mol. Microbiol. 72:1395–1407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Drecktrah D., Knodler L. A., Howe D., Steele-Mortimer O. 2007. Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic 8:212–225 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Ehrbar K., Winnen B., Hardt W. D. 2006. The chaperone binding domain of SopE inhibits transport via flagellar and SPI-1 TTSS in the absence of InvB. Mol. Microbiol. 59:248–264 [DOI] [PubMed] [Google Scholar]
- 21. Ellis T. N., Kuehn M. J. 2010. Virulence and immunomodulatory roles of bacterial outer membrane vesicles. Microbiol. Mol. Biol. Rev. 74:81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ellis T. N., Leiman S. A., Kuehn M. J. 2010. Naturally produced outer membrane vesicles from Pseudomonas aeruginosa elicit a potent innate immune response via combined sensing of both lipopolysaccharide and protein components. Infect. Immun. 78:3822–3831 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Feilmeier B. J., Iseminger G., Schroeder D., Webber H., Phillips G. J. 2000. Green fluorescent protein functions as a reporter for protein localization in Escherichia coli. J. Bacteriol. 182:4068–4076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galan J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53–86 [DOI] [PubMed] [Google Scholar]
- 25. Gal-Mor O., Gibson D. L., Baluta D., Vallance B. A., Finlay B. B. 2008. A novel secretion pathway of Salmonella enterica acts as an antivirulence modulator during salmonellosis. PLoS Pathog. 4:e1000036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Garcia-del Portillo F., Finlay B. B. 1995. Targeting of Salmonella typhimurium to vesicles containing lysosomal membrane glycoproteins bypasses compartments with mannose 6-phosphate receptors. J. Cell Biol. 129:81–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Garcia-del Portillo F., Zwick M. B., Leung K. Y., Finlay B. B. 1993. Salmonella induces the formation of filamentous structures containing lysosomal membrane glycoproteins in epithelial cells. Proc. Natl. Acad. Sci. U. S. A. 90:10544–10548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Garvis S. G., Beuzon C. R., Holden D. W. 2001. A role for the PhoP/Q. regulon in inhibition of fusion between lysosomes and Salmonella-containing vacuoles in macrophages. Cell. Microbiol. 3:731–744 [DOI] [PubMed] [Google Scholar]
- 29. Geddes K., Cruz F., Heffron F. 2007. Analysis of cells targeted by Salmonella type III secretion in vivo. PLoS Pathog. 3:e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Geddes K., Worley M., Niemann G., Heffron F. 2005. Identification of new secreted effectors in Salmonella enterica serovar Typhimurium. Infect. Immun. 73:6260–6271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghelardi E., et al. 2002. Requirement of flhA for swarming differentiation, flagellin export, and secretion of virulence-associated proteins in Bacillus thuringiensis. J. Bacteriol. 184:6424–6433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gunn J. S., Alpuche-Aranda C. M., Loomis W. P., Belden W. J., Miller S. I. 1995. Characterization of the Salmonella typhimurium pagC/pagD chromosomal region. J. Bacteriol. 177:5040–5047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Gunn J. S., Belden W. J., Miller S. I. 1998. Identification of PhoP-PhoQ activated genes within a duplicated region of the Salmonella typhimurium chromosome. Microb. Pathog. 25:77–90 [DOI] [PubMed] [Google Scholar]
- 34. Hansen-Wester I., Hensel M. 2001. Salmonella pathogenicity islands encoding type III secretion systems. Microbes Infect. 3:549–559 [DOI] [PubMed] [Google Scholar]
- 35. Haraga A., Ohlson M. B., Miller S. I. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 36. Hilbert F., Garcia-del Portillo F., Groisman E. A. 1999. A periplasmic D-alanyl-D-alanine dipeptidase in the gram-negative bacterium Salmonella enterica. J. Bacteriol. 181:2158–2165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ho T. D., et al. 2002. Identification of GtgE, a novel virulence factor encoded on the Gifsy-2 bacteriophage of Salmonella enterica serovar Typhimurium. J. Bacteriol. 184:5234–5239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Horstman A. L., Kuehn M. J. 2000. Enterotoxigenic Escherichia coli secretes active heat-labile enterotoxin via outer membrane vesicles. J. Biol. Chem. 275:12489–12496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Irazoqui J. E., et al. 2010. Distinct pathogenesis and host responses during infection of C. elegans by P. aeruginosa and S. aureus. PLoS Pathog. 6:e1000982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kadurugamuwa J. L., Beveridge T. J. 1997. Natural release of virulence factors in membrane vesicles by Pseudomonas aeruginosa and the effect of aminoglycoside antibiotics on their release. J. Antimicrob. Chemother. 40:615–621 [DOI] [PubMed] [Google Scholar]
- 41. Kadurugamuwa J. L., Beveridge T. J. 1999. Membrane vesicles derived from Pseudomonas aeruginosa and Shigella flexneri can be integrated into the surfaces of other gram-negative bacteria. Microbiology 145:2051–2060 [DOI] [PubMed] [Google Scholar]
- 42. Kato S., Kowashi Y., Demuth D. R. 2002. Outer membrane-like vesicles secreted by Actinobacillus actinomycetemcomitans are enriched in leukotoxin. Microb. Pathog. 32:1–13 [DOI] [PubMed] [Google Scholar]
- 43. Kesty N. C., Mason K. M., Reedy M., Miller S. E., Kuehn M. J. 2004. Enterotoxigenic Escherichia coli vesicles target toxin delivery into mammalian cells. EMBO J. 23:4538–4549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kolling G. L., Matthews K. R. 1999. Export of virulence genes and Shiga toxin by membrane vesicles of Escherichia coli O157:H7. Appl. Environ. Microbiol. 65:1843–1848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kuehn M. J., Kesty N. C. 2005. Bacterial outer membrane vesicles and the host-pathogen interaction. Genes Dev. 19:2645–2655 [DOI] [PubMed] [Google Scholar]
- 46. Kulp A., Kuehn M. J. 2010. Biological functions and biogenesis of secreted bacterial outer membrane vesicles. Annu. Rev. Microbiol. 64:163–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Lee E. Y., et al. 2007. Global proteomic profiling of native outer membrane vesicles derived from Escherichia coli. Proteomics 7:3143–3153 [DOI] [PubMed] [Google Scholar]
- 48. Lee E. Y., Choi D. S., Kim K. P., Gho Y. S. 2008. Proteomics in gram-negative bacterial outer membrane vesicles. Mass Spectrom. Rev. 27:535–555 [DOI] [PubMed] [Google Scholar]
- 49. Lindmark B., et al. 2009. Outer membrane vesicle-mediated release of cytolethal distending toxin (CDT) from Campylobacter jejuni. BMC Microbiol. 9:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Loeb M. R., Kilner J. 1979. Effect of growth medium on the relative polypeptide composition of cellular outer membrane and released outer membrane material in Escherichia coli. J. Bacteriol. 137:1031–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. McBroom A. J., Kuehn M. J. 2007. Release of outer membrane vesicles by Gram-negative bacteria is a novel envelope stress response. Mol. Microbiol. 63:545–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. McGhie E. J., Brawn L. C., Hume P. J., Humphreys D., Koronakis V. 2009. Salmonella takes control: effector-driven manipulation of the host. Curr. Opin. Microbiol. 12:117–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miao E. A., et al. 1999. Salmonella typhimurium leucine-rich repeat proteins are targeted to the SPI1 and SPI2 type III secretion systems. Mol. Microbiol. 34:850–864 [DOI] [PubMed] [Google Scholar]
- 54. Miller S. I., Kukral A. M., Mekalanos J. J. 1989. A two-component regulatory system (phoP phoQ) controls Salmonella typhimurium virulence. Proc. Natl. Acad. Sci. U. S. A. 86:5054–5058 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Miller S. I., Mekalanos J. J. 1990. Constitutive expression of the phoP regulon attenuates Salmonella virulence and survival within macrophages. J. Bacteriol. 172:2485–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Minsky A., Summers R. G., Knowles J. R. 1986. Secretion of beta-lactamase into the periplasm of Escherichia coli: evidence for a distinct release step associated with a conformational change. Proc. Natl. Acad. Sci. U. S. A. 83:4180–4184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Mouslim C., Delgado M., Groisman E. A. 2004. Activation of the RcsC/YojN/RcsB phosphorelay system attenuates Salmonella virulence. Mol. Microbiol. 54:386–395 [DOI] [PubMed] [Google Scholar]
- 57a. National Research Council 1996. Guide for the care and use of laboratory animals. National Academy Press, Washington, DC [Google Scholar]
- 58. Niemann G. S., et al. 2011. Discovery of novel secreted virulence factors from Salmonella enterica serovar Typhimurium by proteomic analysis of culture supernatants. Infect. Immun. 79:33–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ochman H., Soncini F. C., Solomon F., Groisman E. A. 1996. Identification of a pathogenicity island required for Salmonella survival in host cells. Proc. Natl. Acad. Sci. U. S. A. 93:7800–7804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Oh Y. K., et al. 1996. Rapid and complete fusion of macrophage lysosomes with phagosomes containing Salmonella typhimurium. Infect. Immun. 64:3877–3883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Patel J. C., Hueffer K., Lam T. T., Galan J. E. 2009. Diversification of a Salmonella virulence protein function by ubiquitin-dependent differential localization. Cell 137:283–294 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Pierson T., et al. 2011. Proteomic characterization and functional analysis of outer membrane vesicles of Francisella novicida suggests possible role in virulence and use as a vaccine. J. Proteome Res. 10:954–967 [DOI] [PubMed] [Google Scholar]
- 63. Raschke W. C., Baird S., Ralph P., Nakoinz I. 1978. Functional macrophage cell lines transformed by Abelson leukemia virus. Cell 15:261–267 [DOI] [PubMed] [Google Scholar]
- 64. Renelli M., Matias V., Lo R. Y., Beveridge T. J. 2004. DNA-containing membrane vesicles of Pseudomonas aeruginosa PAO1 and their genetic transformation potential. Microbiology 150:2161–2169 [DOI] [PubMed] [Google Scholar]
- 65. Reynolds E. S. 1963. The use of lead citrate at high pH as an electron-opaque stain in electron microscopy. J. Cell Biol. 17:208–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Roy D., et al. 2004. A process for controlling intracellular bacterial infections induced by membrane injury. Science 304:1515–1518 [DOI] [PubMed] [Google Scholar]
- 67. Santini C. L., et al. 2001. Translocation of jellyfish green fluorescent protein via the Tat system of Escherichia coli and change of its periplasmic localization in response to osmotic up-shock. J. Biol. Chem. 276:8159–8164 [DOI] [PubMed] [Google Scholar]
- 68. Shaner N. C., et al. 2004. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat. Biotechnol. 22:1567–1572 [DOI] [PubMed] [Google Scholar]
- 69. Sory M. P., Boland A., Lambermont I., Cornelis G. R. 1995. Identification of the YopE and YopH domains required for secretion and internalization into the cytosol of macrophages, using the cyaA gene fusion approach. Proc. Natl. Acad. Sci. U. S. A. 92:11998–12002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Spano S., Ugalde J. E., Galan J. E. 2008. Delivery of a Salmonella Typhi exotoxin from a host intracellular compartment. Cell Host Microbe 3:30–38 [DOI] [PubMed] [Google Scholar]
- 71. Tan T. T., Morgelin M., Forsgren A., Riesbeck K. 2007. Haemophilus influenzae survival during complement-mediated attacks is promoted by Moraxella catarrhalis outer membrane vesicles. J. Infect. Dis. 195:1661–1670 [DOI] [PubMed] [Google Scholar]
- 72. Timms B. G. 1986. Postembedding immunogold labeling for electron microscopy using “LR White” resin. Am. J. Anat. 175:267–275 [DOI] [PubMed] [Google Scholar]
- 73. van de Waterbeemd B., et al. 2010. Improved OMV vaccine against Neisseria meningitidis using genetically engineered strains and a detergent-free purification process. Vaccine 28:4810–4816 [DOI] [PubMed] [Google Scholar]
- 74. Wai S. N., et al. 2003. Vesicle-mediated export and assembly of pore-forming oligomers of the enterobacterial ClyA cytotoxin. Cell 115:25–35 [DOI] [PubMed] [Google Scholar]
- 75. Worley M. J., Nieman G. S., Geddes K., Heffron F. 2006. Salmonella typhimurium disseminates within its host by manipulating the motility of infected cells. Proc. Natl. Acad. Sci. U. S. A. 103:17915–17920 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yashroy R. C. 2007. Mechanism of infection of a human isolate Salmonella (3,10:r:-) in chicken ileum: ultrastructural study. Indian J. Med. Res. 126:558–566 [PubMed] [Google Scholar]
- 77. Yoon H., McDermott J. E., Porwollik S., McClelland M., Heffron F. 2009. Coordinated regulation of virulence during systemic infection of Salmonella enterica serovar Typhimurium. PLoS Pathog. 5:e1000306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Young B. M., Young G. M. 2002. YplA is exported by the Ysc, Ysa, and flagellar type III secretion systems of Yersinia enterocolitica. J. Bacteriol. 184:1324–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Young G. M., Schmiel D. H., Miller V. L. 1999. A new pathway for the secretion of virulence factors by bacteria: the flagellar export apparatus functions as a protein-secretion system. Proc. Natl. Acad. Sci. U. S. A. 96:6456–6461 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Zhou D., Mooseker M. S., Galan J. E. 1999. Role of the S. typhimurium actin-binding protein SipA in bacterial internalization. Science 283:2092–2095 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.