Abstract
Clostridium perfringens type A strains producing enterotoxin (CPE) cause one of the most common bacterial food-borne illnesses, as well as many cases of non-food-borne human gastrointestinal disease. Recent studies have shown that an Agr-like quorum-sensing system controls production of chromosomally encoded alpha-toxin and perfringolysin O by C. perfringens, as well as sporulation by Clostridium botulinum and Clostridium sporogenes. The current study explored whether the Agr-like quorum-sensing system also regulates sporulation and production of two plasmid-encoded toxins (CPE and beta2 toxin) that may contribute to the pathogenesis of non-food-borne human gastrointestinal disease strain F5603. An isogenic agrB null mutant was inhibited for production of beta2 toxin during vegetative growth and in sporulating culture, providing the first evidence that, in C. perfringens, this system can control production of plasmid-encoded toxins as well as chromosomally encoded toxins. This mutant also showed reduced production of alpha-toxin and perfringolysin O during vegetative growth. Importantly, when cultured in sporulation medium, the mutant failed to efficiently form spores and was blocked for CPE production. Complementation partially or fully reversed all phenotypic changes in the mutant, confirming that they were specifically due to inactivation of the agr locus. Western blots suggest that this loss of sporulation and sporulation-specific CPE production for the agrB null mutant involves, at least in part, Agr-mediated regulation of production of Spo0A and alternative sigma factors, which are essential for C. perfringens sporulation.
INTRODUCTION
The Gram-positive, anaerobic, spore-forming bacterium Clostridium perfringens is an important human and medical pathogen, causing both histotoxic infections (e.g., traumatic gas gangrene) and infections originating in the intestines (e.g., human food poisoning). Isolates of this bacterium are commonly assigned to one of five types (A to E) based upon their production of four typing toxins (alpha-, beta-, epsilon-, and iota-toxin). Although not part of the toxinotyping classification system, C. perfringens enterotoxin (CPE) is one of the most important toxins produced by this bacterium. CPE-producing type A strains are responsible for C. perfringens type A food poisoning, which is the second most commonly identified bacterial food-borne disease in the United States, where nearly a million cases occur annually (24). Besides causing food poisoning, CPE-producing type A strains are also responsible for 5 to 15% of all cases of human non-food-borne gastrointestinal (GI) diseases (2). Molecular Koch's postulate studies (22) have shown that CPE is very important for the GI pathogenesis of both C. perfringens type A food poisoning and CPE-associated non-food-borne human GI diseases. However, it has been proposed that beta2 toxin (CPB2) may also contribute to the pathogenesis of some cases of non-food-borne human GI disease (6).
Sporulation offers two critical contributions to the GI diseases caused by type A CPE-positive strains. First, spores are considered important for the transmission of C. perfringens type A food poisoning, as they likely facilitate survival in temperature-abused foods (15); spores may also contribute to transmission of CPE-associated non-food-borne human GI illnesses. Notably, the spores made by many CPE-positive type A food poisoning strains are, relative to spores made by other C. perfringens strains, extremely resistant to environmental stresses, such as temperature or nitrites (11, 15). In part, the exceptional resistance phenotype of those spores is due to their containing a variant small acid-soluble protein (11). Second, sporulation is also crucial for pathogenesis since CPE expression is sporulation specific. During both food poisoning and CPE-associated non-food-borne human GI diseases, CPE is produced in the intestines by sporulating cells of CPE-positive type A strains (15).
While still incompletely understood, the regulation of C. perfringens sporulation is currently under active study. Previous studies have shown that production of Spo0A, which is the master protein initiator of sporulation in Bacillus, is necessary for both sporulation and CPE production by C. perfringens type A strains (8). More recently, a role for sporulation-associated alternative sigma factors has also been demonstrated in the regulation of both C. perfringens sporulation and CPE production (7, 12). Specifically, isogenic sigma factor null mutants were used to show that production of all four sporulation-associated alternative sigma factors is required for C. perfringens sporulation. However, production of only three of the four alternative sigma factors, i.e., SigE, SigF, and SigK, was found to be necessary for CPE production (12). The involvement of those three alternative sigma factors in CPE production is attributable to (i) SigF controlling production of all other alternative sigma factors, including SigE and SigK, and (ii) SigE and SigK then mediating cpe transcription from SigE-dependent and SigK-dependent promoters upstream from the cpe open reading frame (ORF) (12).
Quorum-sensing (QS) mechanisms allow bacterial populations to sense their environment. The importance of QS systems is well established for regulating the initiation of sporulation by Bacillus spp. (1, 25). Sequencing projects (16, 26) have identified ORFs encoding certain portions of the Staphylococcus aureus Agr QS system in the genomes of several pathogenic Clostridium spp. This includes C. perfringens strains 13 and SM101, which carry ORFs encoding proteins homologous to (i) AgrD, which is the precursor signaling peptide of the Agr system, and (ii) AgrB, a membrane protein that modifies AgrD to the active form. Recent directed and random mutagenesis studies have generated isogenic agrB or agrBD null mutants of strain 13, a cpe-negative and nonsporulating C. perfringens type A strain (19, 27). Studies using those isogenic mutants established that the Agr-like system regulates production of alpha-toxin (CPA) and perfringolysin O (PFO), two chromosomally encoded toxins expressed during vegetative growth. Furthermore, this Agr-like system-mediated regulation of toxin production was shown to involve a diffusible molecule (27), consistent with a QS mechanism.
Another recent study (5) identified the presence of two copies of agrBD loci in the genomes of Clostridium botulinum and Clostridium sporogenes. Using antisense RNA techniques or an isogenic agrB null mutant, respectively, that study also demonstrated agr locus-mediated regulation of sporulation by C. botulinum and C. sporogenes. Additionally, the agr locus was shown to control the ability of C. botulinum to produce botulinum neurotoxin, which peaks during stationary phase.
Given the precedent for the agr locus controlling (i) toxin production by vegetative cells of C. perfringens and C. botulinum and (ii) sporulation in other pathogenic Clostridium spp., the current study has assessed the importance of AgrB when C. perfringens sporulates and produces CPE, both critical events for the development of CPE-mediated human gastrointestinal disease. We also evaluated whether Agr regulates production of CPB2, which has been suggested to contribute to CPE-associated non-food-borne human GI diseases.
MATERIALS AND METHODS
Bacterial strains and media.
F5603, which was isolated in the United Kingdom from a case of sporadic diarrhea during the 1990s, was used throughout this study as the wild-type CPE-positive C. perfringens strain. This type A strain carries the plc and pfo toxin genes on the chromosome and the cpe and cpb2 toxin genes on a large plasmid (6). Escherichia coli Top10 cells (Invitrogen) were used as the cloning host.
Media used in this study for culturing C. perfringens included FTG (fluid thioglycolate medium; Difco Laboratories), TGY (3% tryptic soy broth [Becton-Dickinson], 2% glucose [Fisher scientific], 1% yeast extract [Becton-Dickinson], 0.1% sodium thioglycolate [Sigma Aldrich]), brain heart infusion (BHI) agar (Becton-Dickinson), and, for inducing sporulation, Duncan-Strong medium (13). For culturing E. coli, Luria-Bertani (LB) broth (1% tryptone [Becton-Dickinson], 0.5% yeast extract [Becton-Dickinson], 1% NaCl [Fisher scientific]), and LB agar medium (1.5% agar [Becton-Dickinson]) were used. All antibiotics used in this study were purchased from the Sigma-Aldrich Chemical Company.
Sequencing of the agr locus in F5603.
DNA was isolated from F5603 using the Master Pure Gram-positive DNA purification kit (Epicentre). The primers agrF1 and agrR1 (27) were used in a PCR to amplify the F5603 agr locus. In detail, 1 μl of each primer (at a 0.5 μM final concentration), 1 μl of purified DNA template, and 25 μl 2× Taq long-range mixture (NEB) were mixed together, and double-distilled water (ddH2O) was added to reach a total reaction volume of 50 μl. The reaction mixtures were then placed in a thermal cycler (Techne) and subjected to the following amplification conditions: 1 cycle of 95°C for 2 min; 35 cycles of 95°C for 30 s, 55°C for 40 s, and 65°C for 3 min; and a single extension of 65°C for 5 min. The resultant 2,893-bp PCR product was cloned into the Invitrogen pCR2.1-TOPO vector according to the manufacturer's instructions, and that insert was then sequenced at the University of Pittsburgh Core Sequencing Facility.
Construction of an F5603 agr null mutant.
The agrB gene in F5603 was insertionally inactivated using a Clostridium-modified TargeTron gene knockout system (3). Using intron insertion sites identified by the Sigma TargeTron algorithm (Sigma Genosys), the 900-bp intron was targeted to insert, in the sense orientation, between agrB ORF nucleotides 566 and 567 in F5603. Primers used for targeting the intron were 566/567s-IBS (5′-AAAAAAGCTTATAATTATCCTTATTCCACACTGAAGTGCGCC-CAGATAGGGTG-3′), 566/567s-EBS1d (5′-CAGATTGTACAAATGTGGTGATAACAGATAAGTCACTGAATATAACTTACC-TTTCTTTGT-3′), and 566/567s-EBS2 (5′-TGAACGCAAGTTTCTAATTTCGGTTTGGAATCGATAGAGGAAAGTGTCT-3′). The 350-bp PCR products produced using those primers were inserted into pJIR750ai (3). This resultant plasmid was named pJIR750agri. The pJIR750agri vector was then electroporated into F5603. The transformation efficiency for F5603 was 10 transformants/μg DNA. Transformants were selected on BHI agar plates containing 15 μg/ml of chloramphenicol and then PCR screened for an intron-disrupted agrB gene by using primers AgrBscF (5′-ATACTTAGAAAATATTCTGGAGG-3′) and AgrBscR (5′-ATTTCCTATGTAAGTTAGAGTAAT-3′). The F5603::agr mutant was shown to carry an agrB intron insertion (see Results) and then grown in FTG medium without antibiotics for 10 days, with daily subculturing, to cure the intron-carrying donor plasmid pJIR750agri. Curing was shown by lack of growth on chloramphenicol-containing BHI plates. The complementing strain of this null mutant (F5603agrBcomp) was prepared by electroporation of the agr-carrying plasmid CPJVp3 (27), and transformants were then selected on BHI agar plates containing 15 μg/ml of chloramphenicol.
Southern hybridization analyses.
DNA was isolated from wild-type F5603 and the F5603 agrB null mutant using the MasterPure Gram-positive DNA purification kit. Each DNA was then digested with EcoRI overnight at 37°C and run on a 1% agarose gel. After the alkali transfer to a nylon membrane (Roche), the blot was hybridized with a digoxigenin-labeled, intron-specific probe as described previously. This probe was prepared using the primer pair KO-IBS and KO-EBS1d (23) and a PCR digoxigenin (DIG) labeling kit purchased from Roche Applied Science. CSPD substrate (Roche Applied Science) was used for detection of DIG-labeled hybridized probes, according to the manufacturer's instructions.
RNA isolation and reverse transcription (RT)-PCR.
Total RNA from wild-type F5603, the F5603::agrB mutant, and the complementing strain F5603agrBcomp was extracted from pelleted cells obtained from 10 ml of a 5-h Duncan-Strong (DS) culture, as described previously (28). Briefly, the pellet was resuspended in 200 μl of acetate buffer before the addition of 200 μl of saturated phenol (Fisher Scientific). That mixture was then incubated at 60°C in a water bath with vigorous shaking for 15 min. After centrifugation, the supernatant was collected and cold ethanol was added to recover RNA. All RNA samples were additionally treated with 2 U of DNase I (Promega) at 37°C for 1 h. DNase I inhibitor (Promega) was used to stop DNase I activity. The purified RNA was quantified by absorbance at 260 nm and stored in a −80°C freezer.
RT-PCRs were performed on the purified RNA samples using the AccessQuick RT-PCR system (Promega). Briefly, 100 ng of each RNA sample was reverse transcribed to cDNA at 45°C for 1 h. That cDNA was then used as template for PCRs with primers (AgrBscF and AgrBscR) targeting agrB sequences (as above), primers (plcKOF [5′-GATTTGTAAGGCGCTTATTTGTG-3′] and plcKOR [5′-CCATTCATATCTAGCTAATGCTG-3′]) targeting the cpa gene, primers (pfoAKOF [5′-TTTATGAACTTAACAAATGAGGGG-3′] and pfoAKOR [5′-CTACTCCAAGTGAGTTTTCAAGG-3′]) targeting the pfoA gene, primers (cpeF [5′-GGAGATGGTTGGATATTAGG-3′] and cpeR [5′-GGACCAGCAGTTGTAGATA-3′]) targeting the cpe gene, or primers (cpb2univF and cpb2univR [6]) targeting the cpb2 gene.
Growth rate measurement for wild-type, mutant, and complementing strains.
Measurement of vegetative growth for wild-type, agrB null mutant, and complementing strains in TGY medium was determined as described previously (13).
Measurement of sporulation.
A 0.2-ml aliquot of overnight FTG cultures of the wild-type, agrB null mutant, or complementing strains was added to 10 ml of DS medium. After an overnight incubation at 37°C, each DS culture was heated to 75°C for 20 min to kill vegetative cells and enhance spore germination. These heat-shocked suspensions were then serially diluted from 10−2 to 10−7 with sterile water and plated onto BHI agar plates, which were incubated anaerobically overnight at 37°C prior to colony counting.
Western blot analyses.
A 0.2-ml aliquot of an 8-h FTG culture of the wild-type F5603 parent, isogenic agrB null mutant, or complementing strain was inoculated into 10 ml of TGY broth or DS medium. To perform PFO, CPA, CPE, and CPB2 Western blots, supernatants of 16-h TGY or DS cultures were used. Samples were collected, and each supernatant was mixed with SDS loading buffer and boiled for 10 min. Those mixtures were electrophoresed on a 10% polyacrylamide gel containing SDS and then subjected to Western blotting using the appropriate antibodies, as described below. To perform Spo0A, SigF, and SigG Western blotting, the pellets of 5-h DS cultures were used. Samples were collected and pellets were mixed with SDS loading buffer and boiled for 10 min. The boiled samples were then microcentrifuged, and 25 μl of the supernatant was subjected to Western blotting using appropriate antibodies, as previously described (12).
Antibodies.
The CPE, CPA, CPB2, SigF, SigG, and Spo0A antibodies used in this study have been described previously (6, 8, 12). To prepare PFO antibodies, the pfoA ORF was amplified from C. perfringens type C strain CN3685 by PCR using primers rPFO-L (TTGGATCCTTCAAAGGATATAACAGATAAAAATC) and rPFO-R (TTGGTACCTTAATTGTAAGTAATACTAGATCCAGGGT). The pfo PCR product was then cloned in the prokaryotic expression plasmid pTrcHisA (Invitrogen). After that vector was transformed into DH5α, recombinant PFO (rPFO) tagged on its N terminus with a peptide containing an His6 sequence was expressed in the transformants and then purified by affinity chromatography using Ni-nitrilotriacetic acid (NTA) agarose (Qiagen), according to the manufacturer's protocol. Briefly, the bacterial transformants were grown to an optical density at 600 nm (OD600) of 0.5 to 0.6 and induced with isopropyl-d-thiogalactoside (IPTG) at a final concentration of 1 mM. Cells were harvested by centrifugation and resuspended in 0.01 M phosphate-buffered saline (PBS), pH 7.4. Lysozyme was added to a final concentration of 2 mg/ml, and cells were lysed by sonication. To obtain purified rPFO, the soluble cell lysate was incubated with Ni-NTA agarose at 4°C overnight. Bound fusion protein was then eluted from the resin according to the manufacturer's protocol. The purity of the purified rPFO was verified by SDS-PAGE and staining with Coomassie blue.
Antibody against PFO was raised in rabbits by the Pocono Rabbit Farm and Laboratory using the purified His-rPFO fusion protein as the antigen. This immunization followed a standard protocol recommended by Pocono Rabbit Farm and Laboratory, under their approved IACUC permit. Briefly, the rabbits were intradermally injected with 100 to 200 μg rPFO/rabbit in complete Freund's adjuvant (CFA). Four weeks later, 50 to 100 μg rPFO mixed with incomplete Freund's adjuvant (IFA) was given intradermally. After two boosts with purified rPFO administered subcutaneously, rabbits were terminally bled from the carotid artery.
Nucleotide sequence accession number.
The obtained F5603 agr locus sequence was deposited in GenBank under accession number JF343435.
RESULTS
Analysis of the agr locus in F5603.
The current study first confirmed that strain F5603 possesses a similar agr locus as that identified previously in other C. perfringens strains. Sequencing analyses demonstrated that the AgrB and AgrD proteins encoded by F5603 share 99 to 100% homology with the AgrB and AgrD proteins encoded by three previously sequenced C. perfringens strains, including strain 13 (GenBank accession number 989871 CPE1561), JGS1721 (GenBank accession number ED770177), and JGS1495 (GenBank accession number EDS81182.1), and 93% homology with the AgrB and AgrD proteins encoded by C. perfringens strain SM101 (GenBank accession number ABG86149). In addition, the F5603 AgrB and AgrD proteins share 43% homology with those produced by Clostridium sporogenes and 34 to 43% homology with the AgrB and AgrD proteins produced by various Clostridium botulinum strains (5).
Construction and genotypic characterization of an isogenic F5603 agrB null mutant.
It has been shown (19, 27) that, in C. perfringens type A strain 13, PFO and CPA production are regulated by the agr locus. However, since (i) sporulation and CPE production are important for CPE-associated food-borne and non-food-borne human GI diseases (22) and (ii) CPB2 toxin may contribute to the pathogenesis of CPE-positive, CPB2-positive non-food-borne human GI disease isolates (6), the current study constructed an isogenic agrB null mutant of the type A sporadic diarrhea isolate F5603 to directly evaluate whether the agr locus regulates C. perfringens sporulation or CPE and CPB2 synthesis.
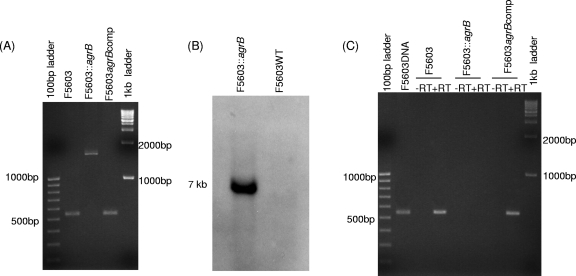
The identity of an F5603 agrB null mutant (F5603::agrB), which was constructed using a Clostridium-modified TargeTron-mediated insertional mutagenesis method (3), was first demonstrated by PCR using primers specific for internal agrB ORF sequences (Fig. 1 A). Using DNA from wild-type F5603, these internal PCR primers specifically amplified a PCR product of 540 bp. However, consistent with the insertion of a 900-bp intron into the agrB ORF, the same primers amplified a PCR product of ∼1.5 kb using DNA from the putative agrB mutant.
Fig. 1.
Intron-based mutagenesis to create an isogenic F5603 agrB mutant and construction of a complementing strain. (A) PCR confirmation of the isogenic agrB null mutant and complementing strain. Using internal agrB primers, this PCR assay amplifies an ∼500-bp product from the wild-type agrB gene, but due to the presence of a 900-bp intron insertion, it amplifies an ∼1.5-kb PCR product from an intron-disrupted agrB gene. (B) Southern blot hybridization of an intron-specific probe to DNA from wild-type F5603 or the agrB null mutant. DNA from each strain was digested with EcoRI prior to electrophoresis on a 1% agarose gel prior to blotting and hybridization with an intron-specific probe. Size of DNA fragments, in kilobases (kb), is shown at the left. (C) RT-PCR analyses for agrB expression by F5603, the F5603::agrB mutant, and the complementing F5603agrBcomp strain grown for 5 h in DS medium. First and last lanes show size markers. Lanes labeled with a plus sign (+) were from samples receiving reverse transcriptase, while lanes labeled with a minus sign (−) lacked reverse transcriptase to show the absence of DNA contamination.
After the intron delivery plasmid was cured, DNA was isolated from the putative null mutant and subjected to Southern blot analysis using an intron-specific probe (Fig. 1B). This Southern blot analysis detected no hybridization of the intron-specific probe to wild-type DNA, as expected. In contrast, the presence of a single intron insertion was visible on these Southern blots using DNA from the F5603::agrB mutant.
An RT-PCR assay was then used (Fig. 1C) to assess agrB expression by 5-h DS sporulation medium cultures of wild-type F5603 and the F5603::agrB mutant. This analysis confirmed that wild-type F5603 expressed agrB transcripts, but no agrB transcription was detectable for the mutant. The RT-PCR product amplified from wild-type F5603 was dependent upon reverse transcription of RNA, since no product was observed in the absence of reverse transcriptase.
A complementing strain, F5603agrBcomp, was prepared using a plasmid where the agr locus had been cloned into the C. perfringens/E. coli shuttle plasmid pJIR750 (27). That agr-carrying plasmid was then transformed into the F5603::agrB mutant by electroporation. PCR detected the presence of the wild-type agrB ORF in the complementing strain (Fig. 1A). RT-PCR confirmed transcription of the agrB locus in 5-h DS cultures of F5603agrBcomp (Fig. 1C). This agrB RT-PCR product obtained for the complementary strain was transcribed RNA since its presence was dependent upon transcription of RNA and no product was observed in the absence of reverse transcriptase (Fig. 1C).
Phenotypic characterization of vegetative cultures of F5603 agr null mutants.
To initiate phenotypic comparisons of the wild-type parent, the agrB null mutant, and the complementing strain, the vegetative growth rates of these three strains were first compared in TGY medium. This analysis determined (data not shown) that the vegetative growth rates of these three strains are nearly identical.
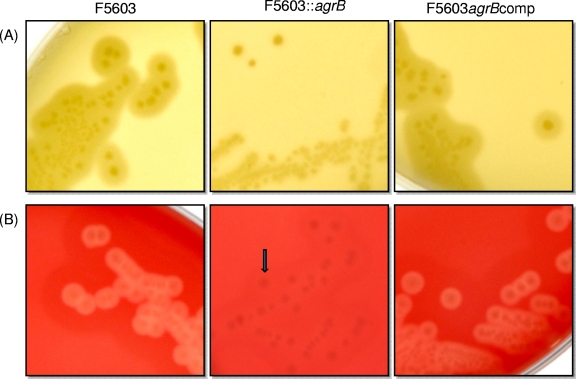
Previous studies (19, 27) had shown that the agr locus regulates CPA and PFO production by vegetative cells of C. perfringens type A strain 13 growing on egg yolk agar and blood agar, respectively. Similar conclusions were obtained when the F5603 wild-type parent, the isogenic agrB null mutant, and the complementing strain were grown on egg yolk agar plates (for detection of CPA) or sheep blood agar plates (for detection of PFO) (23). Specifically, colonies of the wild-type F5603 parent grown on egg yolk agar plates (Fig. 2 A) were surrounded by the characteristic halo zone, indicative of lecithin breakdown due to the phospholipase C activity of CPA. In contrast, the F5603::agrB null mutant strain lost this zone around its colonies, indicating a lack of CPA production. This loss of phospholipase C production was specifically due to Agr regulation, since colonies of the complementing strain exhibited a restored halo production (Fig. 2A). As also shown in Fig. 2B, when grown on sheep blood agar plates, colonies of the wild-type parent, but not the agrB null mutant, were surrounded by a beta-hemolytic zone due to PFO production. The loss of PFO production by the mutant was specifically due to agr locus disruption, since colonies of the complementing strain were β-hemolytic on these blood agar plates.
Fig. 2.
Production of CPA and PFO by vegetative cells of wild-type F5603, the agrB null mutant, and the complementing strain growing on agar plates. The phospholipase C activity of CPA was detected in colonies growing on egg yolk agar plates (A), and the beta-hemolytic activity of PFO was detected on blood agar plates (B). Colonies of wild-type F5603 and the complementing strain showed a characteristic CPA-induced halo zone on egg yolk agar plates and PFO-induced β-hemolysis surrounding colonies grown on blood agar plates. However, the mutant failed to produce these zones.
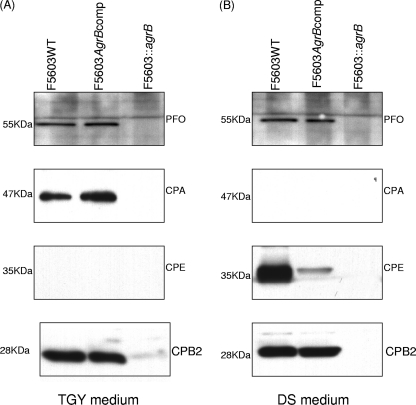
Western blot analyses were then performed to specifically assess the ability of Agr to regulate PFO and CPA production by TGY vegetative cultures of F5603. As shown in Fig. 3 A, Western blot analyses of 16-h TGY cultures demonstrated that wild-type F5603 produced both toxins. However, neither PFO nor CPA was detectable after the agrB mutant was grown similarly in TGY broth. This loss of PFO and CPA production for TGY cultures of the agrB mutant was completely reversible by complementation (Fig. 3A).
Fig. 3.
Western blot analysis of toxin production by wild-type F5603, the isogenic agrB null mutant, and the complementing strain growing in TGY vegetative culture broth (A) or DS sporulation medium (B). Each strain was grown for 16 h in TGY broth or DS medium, as indicated. Panel A shows production of PFO, CPA (alpha-toxin), CPE, and CPB2 toxin expression in TGY broth. Panel B shows production of PFO, CPA, CPE, and CPB2 in DS sporulation medium. The molecular weight of each band is indicated on the left.
Western blot analysis was also performed to examine, for the first time, whether production of CPB2 during vegetative growth is Agr regulated (Fig. 3A). Results from those analyses demonstrated strong CPB2 production by wild-type F5603 when grown for 16 h in TGY broth. However, CPB2 production was nearly absent in similar cultures of the agrB null mutant. This effect was specifically attributable to inactivation of the agr locus in the mutant, since complementation fully restored CPB2 production to wild-type levels under these vegetative growth conditions.
As expected, since expression of CPE is sporulation specific, Western blot analyses detected no CPE expression in 16-h TGY vegetative cultures, even for wild-type F5603 (Fig. 3A).
The agr locus is involved in regulation of C. perfringens sporulation.
To address whether a functional agr locus is required for C. perfringens sporulation, wild-type F5603, the isogenic agrB null mutant, and the agr complementing strain were each incubated in DS sporulation medium. By 8 h after incubation at 37°C in DS sporulation medium, wild-type F5603 had already formed refractile spores, with an efficiency of 60 to 70%. In contrast, the agrB null mutant formed only a trace amount of spores, barely detectable by phase-contrast microscopy (<1% sporulation efficiency). This effect was specifically due to inactivation of the agr locus, since the complementing strain showed similar sporulation efficiency as that of the wild-type parent.
After overnight culture, the sporulating abilities of F5603, the agrB null mutant, and the complementing strain were quantitatively compared by measuring their formation of heat-resistant spores. In this experiment, wild-type F5603 formed 5.8 × 104 ± 1.4 × 103 (±standard deviation) heat-resistant spores/ml. However, under the same incubation conditions, the isogenic agrB null mutant formed only 38 ± 5 heat-resistant spores/ml. This reduction in sporulation involved the inactivation of the agr locus rather than a nonspecific secondary mutation or polar effects, since the complementing strain formed 3.0 × 103 ± 2.0 × 102 heat-resistant spores/ml. Even when grown at 37°C in DS sporulation medium for 2, 3, or 7 days, sporulation of the agrB null mutant remained barely detectable (data not shown). Collectively, these results indicated that the agr locus is needed for efficient formation of heat-resistant mature spores.
The agr locus also regulates CPE, CPB2, and PFO expression by F5603 in DS sporulation medium.
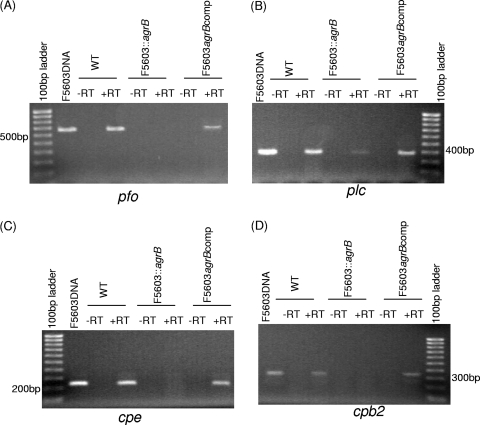
To detect whether AgrB regulates transcription of the cpe, cpb2, cpa, and pfo genes by F5603 growing in DS sporulating cultures, RT-PCR analyses were first performed. When wild-type F5603 was grown at 37°C in DS medium for 5 h, mRNA transcripts from all four toxin genes were detected (Fig. 4). In contrast, no cpe, cpb2, or pfo gene transcription was observed when the agrB null mutant was similarly grown for 5 h at 37°C in DS sporulation medium, although trace cpa transcription was detected under these growth conditions. This loss of toxin gene transcription in DS cultures of the agrB mutant was reversible by complementation.
Fig. 4.
RT-PCR analyses for toxin gene expression by F5603, the F5603::agrB mutant, and the complementing strain growing in DS medium. RNA was isolated from cultures grown for 5 h in DS medium. (A) Expression of the pfoA gene encoding PFO; (B) expression of the cpa gene encoding alpha-toxin; (C) expression of the cpe gene encoding enterotoxin; (D) expression of the cpb2 gene encoding beta2 toxin. The first lanes in panels A and C and last lanes in panels B and D show size markers. Lanes labeled with a plus sign (+) were from samples receiving reverse transcriptase, while lanes labeled with a minus sign (−) lacked reverse transcriptase to show the absence of DNA contamination.
Western blot analyses were then performed to confirm the importance of a functional agr locus for CPA, CPE, CPB2, and PFO production by DS sporulating cultures of F5603 (Fig. 3B). Using 16-h DS cultures that had been incubated at 37°C, Western blot studies detected CPB2 and PFO production by F5603 but not by the F5603::agrB mutant. However, production of both toxins was restored in the complementing strain under the same incubation conditions. No CPA production was detected in 16-h DS cultures, even for wild-type F5603.
Western blot analyses also demonstrated the presence of CPE in 16-h DS cultures of F5603 (Fig. 3B). However, no CPE was detected in cultures of the agrB null mutant under similar growth conditions. Complementation partially restored CPE production, confirming that the loss of CPE expression by the agrB mutant had specifically involved disruption of the agr locus. The failure of complementation to fully restore the wild-type level of CPE production, which is sporulation specific, likely reflects the lower sporulation level noted for the complementing strain versus that of the wild-type parent (see preceding section).
Investigating how the agr locus regulates sporulation and CPE expression.
A previous study (8) demonstrated that a functional spo0A gene is essential for C. perfringens sporulation and for sporulation-specific CPE expression. Therefore, Western blotting was performed to compare Spo0A levels in 5-h DS cultures of wild-type F5603 to those of the isogenic agrB null mutant (Fig. 5 A). These analyses showed that the agrB null mutant contains less Spo0A than does the wild-type parent. This reduction in Spo0A levels in the mutant was specifically attributable to disruption of the agr locus, since Spo0A expression was restored to wild-type levels by complementation of the agrB null mutant with a plasmid carrying the agr locus (Fig. 5A).
Fig. 5.
Western blot analysis of DS cultures for Spo0A, SigF, and SigG production by wild-type F5603, the agrB null mutant, and the complementing strain. Cells from 5-h DS cultures were collected and lysed. Those lysates were then Western blotted for Spo0A expression (A), SigF expression (B), and SigG expression (C). The molecular weight of each band is indicated on the left.
In a previous study (12), we had demonstrated that C. perfringens sporulation and production of CPE requires a functional sigF gene. In addition, that study found that C. perfringens sporulation, but not CPE production, is also dependent upon a functional sigG gene. Therefore, a Western blot analysis was performed to assess SigF and SigG production by 5-h DS cultures of wild-type F5603, the isogenic agrB null mutant, and the complementing strain (Fig. 5B and C). Results of this analysis detected lower SigF levels, and an apparent absence of SigG production, in the agrB null mutant than in the wild-type F5603 strain. Furthermore, complementation increases both SigF and SigG levels compared to those of the agrB null mutant.
DISCUSSION
Sporulation plays a critical role in the pathogenesis of both Bacillus spp. and Clostridium spp. The importance of spores for transmission of anthrax, caused by Bacillus anthracis, is widely recognized (14). Similarly, spores contribute to the transmission of many clostridial diseases, including botulism, tetanus, gas gangrene, Clostridium difficile infection, and C. perfringens type A food poisoning (15). Sporulation can also be important for the regulation of toxin production. The prime example of this is the sporulation-specific production of CPE (12), which is the toxin responsible for causing the GI symptoms of C. perfringens type A food poisoning and some cases of human non-food-borne GI disease (22).
The sporulation process is relatively well understood in Bacillus spp. (25) but much less studied in the pathogenic clostridial spp. However, C. perfringens has emerged as the paradigm for studying sporulation and germination among the pathogenic clostridia. While more research is certainly needed, it has already become clear that both similarities and differences exist between the sporulation processes of Bacillus spp. versus C. perfringens. Similarities include observations that Spo0A and the same four alternative sigma factors are important for regulation of sporulation by both C. perfringens and Bacillus spp. (7–9, 12). Among noted differences, the sporulation-associated phosphorelay of Bacillus spp. has not been identified in Clostridium genomes (20).
In Bacillus spp., sporulation is a stress response to both nutrient starvation and high population density. QS systems are important for Bacillus spp. to sense their population density and then initiate sporulation. The importance of nutrient stress for initiating sporulation in pathogenic Clostridium spp. is less certain. For C. perfringens, the initiation of sporulation has been linked to such factors as pH, the presence of complex polysaccharides, and inorganic phosphate concentrations (10, 21, 29).
Whether high population density sensed by QS systems is also important for initiation of sporulation of pathogenic Clostridium spp. is only now coming under investigation. However, with genome sequencing studies revealing the presence of AgrB- and AgrD-encoding ORFs in many Clostridium spp., a recent study showed this QS system is needed for sporulation of C. sporogenes and C. botulinum (5). The current study now extends that work by showing that a functional Agr-like QS system is also important to obtain sporulation by C. perfringens. Specifically, inactivation of the agrB gene in C. perfringens strain F5603 decreased sporulation by ∼1,500-fold, which is similar to the ∼1,000-fold reduction in sporulation noted following inactivation of the agrD1 gene in C. botulinum (5). The reduction of sporulation noted for the C. perfringens agrB mutant can be specifically attributed to inactivation of the agr locus, since complementation increased sporulation by >100-fold over sporulation levels observed for the agrB mutant. The failure of the complementing strain to completely regain wild-type sporulation levels is interesting, since the same complementation fully restored expression of several toxins produced during vegetative growth. Those complementation differences might suggest that sporulating cultures are sensitive to Agr locus expression levels, i.e., complementation involving a multicopy plasmid carrying the cloned agr locus may have altered AgrB levels in the complementing strain versus the wild-type parent, which could then have affected the ability of the complementing strain to sporulate. This issue should be revisited in the future if AgrB or AgrD antibodies become available. It is not possible to compare the current sporulation complementation results obtained for C. perfringens agrB mutants against the ability of complementation to restore sporulation to agrD mutants of C. botulinum, because complementation was not performed in that earlier study (5).
The previous study by Cooksley et al. (5) also did not address how the Agr-like system regulates sporulation in C. botulinum. The current study now demonstrates that inactivation of the agrB gene in C. perfringens results in reduced and lost, respectively, expression of alternative sigma factors SigF and SigG, which are essential for sporulation of this species (12). In addition, the agrB mutant produced less Spo0A, which is also known to be required for C. perfringens sporulation (8). Assuming that, as for Bacillus spp., Spo0A is a master regulator of sporulation in C. perfringens that controls production of alternative sigma factors, this reduced Spo0A production could help to explain why the agrB mutant produces reduced amounts of alternative sigma factors and sporulates poorly. Collectively, these results strongly suggest that, in C. perfringens, the Agr-like QS system regulates sporulation at an early step, i.e., by controlling Spo0A and SigF synthesis.
It is now established that QS systems are important for sporulation in C. perfringens (this study), C. botulinum, and C. sporogenes (5), as well as Bacillus spp. and perhaps other pathogenic clostridial spp. However, even though Bacillus halodurans C-125 carries an ORF encoding an AgrB (GenBank accession number BH3475) with ∼21% homology to AgrB of C. perfringens F5603, it has not been examined (to our knowledge) whether AgrB is important for sporulation of that Bacillus spp. Additionally, it remains possible that other QS systems might be important for the regulation of C. perfringens sporulation.
Extensive studies have implicated the Agr QS system in regulating toxin production by the non-spore-forming bacterium Staphylococcus aureus (17). Similarly, previous studies had demonstrated that the Agr-like system of C. perfringens regulates the expression of two chromosomally encoded toxins, i.e., PFO and CPA, that are produced by type A strain 13 during vegetative growth (18, 27). The current study has now confirmed those findings for a second type A strain (F5603). More importantly, this work extends knowledge of AgrBD control of C. perfringens toxin production by showing that this QS system can also regulate the expression of a plasmid-encoded toxin, i.e., CPB2, during vegetative growth. This study found that supernatants from F5603 DS cultures also contain some CPB2 and PFO, likely due to the fact that 30% of the total bacteria in these sporulating cultures are vegetative cells. This agrees with observations of a previous study (6) that reported detection of CPB2 in sporulating DS cultures of F5603. The current results indicate that production of CPB2 and PFO by DS cultures of F5603 also requires the Agr-like QS system.
Finally, the current studies also demonstrate, for the first time, that a functional Agr-like QS system is necessary for sporulation-specific production of CPE by DS cultures of F5603. This regulation of CPE expression is attributable, at least in part, to the Arg-mediated effects on Spo0A and SigF expression demonstrated in this study, since Spo0A and SigF have both been shown to regulate CPE expression (8, 12). Although not yet demonstrated, it is likely that (as in Bacillus spp.) a functional spo0A gene is necessary for SigF production. The Agr-induced reduction of SigF production caused by reduced Spo0A levels should then control CPE expression, since SigF regulates expression of SigE and SigK, which have been shown to direct transcription from cpe promoters (7, 12).
The loss of CPE production by the agrB mutant was partially reversible by complementation, confirming that the observed reduction in CPE production by this mutant specifically involved inactivation of its agr locus. The inability of complementation to fully restore CPE production to wild-type levels is likely attributable to the lower sporulation levels observed for the complementing strain relative to wild-type parent sporulation levels (see earlier discussion). The amount of CPE produced by a C. perfringens strain generally correlates with sporulation levels (4).
In summary, this study now demonstrates that, as for C. botulinum and C. sporogenes, the Agr-like QS system regulates sporulation in C. perfringens. This regulation is shown, for the first time in any pathogenic clostridial species, to involve (at least in part) reduced production of Spo0A, a protein necessary for initiation of sporulation in C. perfringens. Agr control of sporulation mediates production of CPE, whose sporulation-dependent expression requires Spo0A and sporulation-associated sigma factors. Future studies should explore how the Agr-like system controls Spo0A production, whether this pathway is similar in other pathogenic clostridial species, and whether this Agr-like system interacts with other signaling mechanisms to initiate sporulation in pathogenic Clostridium spp.
ACKNOWLEDGMENTS
This work was generously supported by grant R37-AI19844-29 from the National Institute of Allergy and Infectious Diseases.
We thank Richard Losick, Masaya Fujita, Paul Hauer, and Mafuzer Sarker for supplying antibodies used in this study.
Footnotes
Published ahead of print on 4 April 2011.
REFERENCES
- 1. Bischofs I., Hug J., Liu A., Wolf D., Arkin A. 2009. Complexity in bacterial cell-cell communication: quorum signal integration and subpopulation signaling in the Bacillus subtilis phosphorelay. Proc. Natl. Acad. Sci. U. S. A. 106:6459–6464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Carman R. J. 1997. Clostridium perfringens in spontaneous and antibiotic-associated diarrhoea of man and other animals. Rev. Med. Microbiol. 8(Suppl. 1):S43–S45 [Google Scholar]
- 3. Chen Y., McClane B. A., Fisher D. J., Rood J. I., Gupta P. 2005. Construction of an alpha toxin gene knockout mutant of Clostridium perfringens type A by use of a mobile group II intron. Appl. Environ. Microbiol. 71:7542–7547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Collie R. E., Kokai-Kun J. F., McClane B. A. 1998. Phenotypic characterization of enterotoxigenic Clostridium perfringens isolates from non-food-borne human gastrointestinal diseases. Anaerobe 4:69–79 [DOI] [PubMed] [Google Scholar]
- 5. Cooksley C., et al. 2010. Regulation of neurotoxin production and sporulation by a putative agrBD signaling system in proteolytic Clostridium botulinum. Appl. Environ. Microbiol. 76:4448–4460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Fisher D. J., et al. 2005. Association of beta2 toxin production with Clostridium perfringens type A human gastrointestinal disease isolates carrying a plasmid enterotoxin gene. Mol. Microbiol. 56:747–762 [DOI] [PubMed] [Google Scholar]
- 7. Harry K. H., Zhou R., Kroos L., Melville S. B. 2009. Sporulation and enterotoxin (CPE) synthesis are controlled by the sporulation-specific factors SigE and SigK in Clostridium perfringens. J. Bacteriol. 191:2728–2742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huang I. H., Waters M., Grau R. R., Sarker M. R. 2004. Disruption of the gene (spoOA) encoding sporulation transcription factor blocks endospore formation and enterotoxin production in enterotoxigenic Clostridium perfringens type A. FEMS Microbiol. Lett. 233:233–240 [DOI] [PubMed] [Google Scholar]
- 9. Kroos L. 2007. The Bacillus and Myxococcus developmental networks and their transcriptional regulators. Annu. Rev. Genet. 41:13–39 [DOI] [PubMed] [Google Scholar]
- 10. Labbe R. G. 1989. Clostridium perfringens, p. 192–234 In Doyle M. P. (ed.), Foodborne bacterial pathogens. Marcel Decker Press, New York, NY [Google Scholar]
- 11. Li J., McClane B. A. 2008. A novel small acid soluble protein variant is important for spore resistance of most Clostridium perfringens food poisoning isolates. PLoS Pathog. 4:e1000056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Li J., McClane B. A. 2010. Evaluating the involvement of alternative sigma factors SigF and SigG in Clostridium perfringens sporulation and enterotoxin synthesis. Infect. Immun. 78:4286–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li J., McClane B. A. 2006. Further comparison of temperature effects on growth and survival of Clostridium perfringens type A isolates carrying a chromosomal or plasmid-borne enterotoxin gene. Appl. Environ. Microbiol. 72:4561–4568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Mallozzi M., Viswanathan V., Vedantam G. 2010. Spore-forming Bacilli and Clostridia in human disease. Future Microbiol. 5:1109–1123 [DOI] [PubMed] [Google Scholar]
- 15. McClane B. A. 2007. Clostridium perfringens, p. 423–444 In Doyle M. P., Beuchat L. R. (ed.), Food microbiology, 3rd ed. ASM Press, Washington, DC [Google Scholar]
- 16. Myers G. S., et al. 2006. Skewed genomic variability in strains of the toxigenic bacterial pathogen, Clostridium perfringens. Genome Res. 16:1031–1040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Novick R. P., Geisinger E. 2008. Quorum-sensing in staphylococci. Annu. Rev. Genet. 42:541–564 [DOI] [PubMed] [Google Scholar]
- 18. Ohtani K., et al. 2010. Identification of a two-component VirR/VirS regulon in Clostridium perfringens. Anaerobe 16:258–264 [DOI] [PubMed] [Google Scholar]
- 19. Ohtani K., et al. 2009. Virulence gene regulation by the agr system in Clostridium perfringens. J. Bacteriol. 191:3919–3927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Paredes C. J., Alsaker K. V., Papoutsakis E. T. 2005. A comparative genomic view of clostridial sporulation and physiology. Nat. Rev. Microbiol. 3:969–978 [DOI] [PubMed] [Google Scholar]
- 21. Philippe V., et al. 2006. Inorganic phosphate induces spore morphogenesis and enterotoxin production in the intestinal pathogen Clostridium perfringens. Infect. Immun. 74:3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarker M. R., Carman R. J., McClane B. A. 1999. Inactivation of the gene (cpe) encoding Clostridium perfringens enterotoxin eliminates the ability of two cpe-positive C. perfringens type A human gastrointestinal disease isolates to affect rabbit ileal loops. Mol. Microbiol. 33:946–958 [DOI] [PubMed] [Google Scholar]
- 23. Sayeed S., et al. 2008. Beta toxin is essential for the intestinal virulence of Clostridium perfringens type C disease isolate CN3685 in a rabbit ileal loop model. Mol. Microbiol. 67:15–30 [DOI] [PubMed] [Google Scholar]
- 24. Scallan E., et al. 2011. Foodborne illness acquired in the United States—major pathogens. Emerg. Infect. Dis. 17:7–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Schultz D., Wolynes P., Ben J. E., Onuchic J. 2009. Deciding fate in adverse times: sporulation and competence in Bacillus subtilis. Proc. Natl. Acad. Sci. U. S. A. 106:21027–21034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Shimizu T., et al. 2002. Complete genome sequence of Clostridium perfringens, an anaerobic flesh-eater. Proc. Natl. Acad. Sci. U. S. A. 99:996–1001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Vidal J. E., Chen J., Li J., McClane B. A. 2009. Use of an EZ-Tn5-based random mutagenesis system to identify a novel toxin regulatory locus in Clostridium perfringens strain 13. PLoS One 4:e6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Vidal J. E., McClane B. A., Saputo J., Parker J., Uzal F. A. 2008. Effects of Clostridium perfringens beta-toxin on the rabbit small intestine and colon. Infect. Immun. 76:4396–4404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wrigley D., Hanwella H., Thon B. 1995. Acid exposure enhances sporulation of certain strains of Clostridium perfringens. Anaerobe 1:163–169 [DOI] [PubMed] [Google Scholar]