Abstract
Streptococcus mutans is considered the primary etiologic agent of dental caries, a global health problem that affects 60 to 90% of the population, and a leading causative agent of infective endocarditis. It can be divided into four different serotypes (c, e, f, and k), with serotype c strains being the most common in the oral cavity. In this study, we demonstrate that in addition to OMZ175 and B14, three other strains (NCTC11060, LM7, and OM50E) of the less prevalent serotypes e and f are able to invade primary human coronary artery endothelial cells (HCAEC). Invasive strains were also significantly more virulent than noninvasive strains in the Galleria mellonella (greater wax worm) model of systemic disease. Interestingly, the invasive strains carried an additional gene, cnm, which was previously shown to bind to collagen and laminin in vitro. Inactivation of cnm rendered the organisms unable to invade HCAEC and attenuated their virulence in G. mellonella. Notably, the cnm knockout strains did not adhere to HCAEC as efficiently as the parental strains did, indicating that the loss of the invasion phenotype observed for the mutants was linked to an adhesion defect. Comparisons of the invasive strains and their respective cnm mutants did not support a correlation between biofilm formation and invasion. Thus, Cnm is required for S. mutans invasion of endothelial cells and possibly represents an important virulence factor of S. mutans that may contribute to cardiovascular infections and pathologies.
INTRODUCTION
The oral cavity is colonized by a large number of viridans group streptococci, including primarily soft tissue colonizers, such as Streptococcus salivarius and S. mitis, and predominantly hard tissue (tooth) colonizers, such as S. mutans and S. gordonii. Among tooth colonizers, S. mutans is considered the primary etiologic agent of dental caries, an infectious disease that affects 60 to 90% of the population worldwide (12). Strains of S. mutans can be grouped into four serotypes (c, e, f, and k) based on the composition and structure of the rhamnose glucose polysaccharide (RGP) associated with the cell wall. Epidemiological studies revealed that serotype c is the most common serotype isolated from dental plaque, being found in nearly 80% of S. mutans-positive samples. Serotypes e and f are found in about 20% and 2% of patients, respectively (22, 41, 54). Strains belonging to serotype k are the most infrequent, having been isolated thus far only from subjects from Japan, Thailand, and Finland (29, 42, 44).
In addition to colonizing the teeth in significant numbers, it is not unusual for S. mutans to gain access to the bloodstream during dental procedures (16, 21, 26). If a sufficient number of cells enter the circulation, transient bacteremia followed by adhesion to endothelial cells leads to infective endocarditis (IE) (26, 37), particularly in persons with predisposing cardiac conditions. In addition to IE, a significant association between dental infections and the occurrence of coronary atherosclerosis has been demonstrated (36). More specifically, oral streptococci and the periodontal pathogen Porphyromonas gingivalis have been associated with atherosclerotic/atheromatous plaques (15, 20, 34, 36, 60). Studies by Nakano and coworkers (38, 44, 45) reported that among bacterial species, S. mutans was the most frequently detected in diseased heart valve tissues and atheromatous plaque, suggesting that S. mutans may play an important and underestimated role in the onset of cardiovascular disease (CVD) (38). However, detection of bacteria in atheromas has been based on PCR amplification of S. mutans DNA, not on isolation of live bacteria. Recently, we demonstrated that two S. mutans strains, B14 and OMZ175, belonging to serotypes e and f, respectively, invade and persist in the cytoplasm of human coronary artery endothelial cells (HCAEC), revealing a possible new facet of the pathogenic potential of S. mutans and a mechanistic linkage of S. mutans to CVD (38).
In some cases, binding to the extracellular matrix (ECM) is the first step in the invasion of host cells (17, 58). The ECM is a macromolecular structure that becomes exposed when tissue integrity is damaged by lesions or traumas (63). Fibronectin, collagen, laminin, and elastin are considered the most common components of ECM (63) and can serve as receptors for bacteria during infection. Some S. mutans surface structures, such as the P1 protein (also known as antigen I/II or SpaP), the wall-anchored protein A (WapA), the biofilm regulatory protein A (BrpA), the autolysin AtlA, the glucosyltransferases (GtfB, GtfC, and GtfD), and the serotype-specific RGP, have been implicated in the pathogenesis of IE by promoting adherence to endothelial tissues and triggering inflammatory responses (17, 55, 61). More recently, a new surface protein with collagen- and laminin-binding activity, Cnm, which has an uneven distribution among the different serotypes of S. mutans, was identified (47, 51, 52). Interestingly, Cnm is found frequently in strains belonging to the uncommon serotype f but rarely in serotype c strains. An epidemiological survey revealed that Cnm is present in 10 to 20% of all isolated strains of S. mutans (43). Based on its capacity to bind to components of the ECM, it has been proposed that Cnm may play a role in the development of caries lesions in dentin and in colonization of heart valves (43). However, evidence validating this hypothesis was still lacking. In the present study, we investigated the role of Cnm in HCAEC invasion and virulence. Our findings revealed that Cnm plays a major role in the invasion and virulence properties of S. mutans and is therefore a virulence factor that could contribute to systemic infections by the organism.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
The S. mutans strains used in this study were isolated from either dental plaque or the blood of patients with bacteremia and/or endocarditis (Table 1). All strains had their serotype confirmed by PCR using serotype-specific primers described elsewhere (54). S. mutans strains were routinely cultured in brain heart infusion (BHI) medium at 37°C in a 5% CO2 atmosphere. When required, 1 mg ml−1 of kanamycin or 10 μg ml−1 of erythromycin was added to the growth medium. To induce expression of cnm in the complemented strain, cultures were grown overnight in BHI supplemented with a subinhibitory concentration of nisin (15 ng ml−1). To assess growth and survival in blood, overnight cultures of S. mutans UA159 and OMZ175 grown in TYG (3.5% tryptone, 0.5% yeast extract, 2.5% glucose) were diluted 1:20 in whole, pooled, heparinized human blood obtained from the University of Rochester Medical Center blood bank. Bacterial growth and survival were monitored by counting the CFU of serially diluted cultures collected every 3 h for the first 9 h and every 24 h thereafter for 6 days.
Table 1.
S. mutans strains used in this study
| Strain | Origin | Serotype | Sourcea |
|---|---|---|---|
| UA159 | Dental plaque | c | University of Alabama |
| OMZ175 | Dental plaque | f | B. Guggenheim |
| B14 | Dental plaque | e | A. Bleiweis |
| MT4653 | Dental plaque | c | N. Jakubovicks |
| 13.1 | Dental plaque | e | N. Jakubovicks |
| LML4 | Dental plaque | e | N. Jakubovicks |
| LM7 | Dental plaque | e | P. Caufield |
| OM50E | Dental plaque | e | P. Caufield |
| OM96E | Dental plaque | f | P. Caufield |
| 19 | Dental plaque | f | N. Jakubovicks |
| 6139-99 | Blood | c | CDC |
| 1237-00 | Blood | c | CDC |
| 2955-00 | Blood | c | CDC |
| 190-01 | Blood/endocarditis | c | CDC |
| 2323-02 | Blood | e | CDC |
| 87-03 | Blood | c | CDC |
| 115-04 | Blood/endocarditis | c | CDC |
| 296-04 | Blood | c | CDC |
| 976-04 | Blood | c | CDC |
| 52-07 | Blood/endocarditis | c | CDC |
| 333-07 | Blood | c | CDC |
| OMZ175-cnm | cnm knockout | f | This study |
| B14-cnm | cnm knockout | e | This study |
| LM7-cnm | cnm knockout | e | This study |
| OM50E-cnm | cnm knockout | e | This study |
| 11060-cnm | cnm knockout | f | This study |
| OMZ175-cnm/pcnm | Complementation of cnm | f | This study |
| OMZ175-cnm/pMSP3535 | Control for complementation studies | f | This study |
CDC, Centers for Disease Control and Prevention.
Construction of cnm knockout strains and cnm complementation strain.
All strains listed in Table 1 and those from a previous study (2) were assessed for the presence of the cnm gene by PCR using the primers cnm-1F and cnm-1R (43, 54). Cnm+ strains had the cnm gene disrupted by insertion of a nonpolar kanamycin marker (28) 700 bp downstream of the ATG start codon, using a PCR-ligation mutagenesis strategy (30). Briefly, the 700-bp N-terminal portion of cnm was amplified from strain OMZ175 by use of primers cnm110-F (5′-CCGTTGCCATCATTTGC-3′) and cnm810BamHI-R (5′-CGGATCAGCGGATCCAGTTGCACC-3′), and the 750-bp C-terminal portion of the gene was amplified using primers cnm810BamHI-F (5′-GGTGCAACGGATCCGCTGATCCG-3′) and cnm1560R (5′-CAGGACCTTGTTTGGCT-3′). The underlined bases correspond to a BamHI restriction site that was included for cloning purposes. After amplification, the two PCR fragments were digested with BamHI and ligated to a nonpolar kanamycin resistance gene cassette that was obtained as a BamHI fragment. The ligation mixture was used to transform S. mutans OMZ175, followed by plating onto BHI containing kanamycin (1 mg ml−1). The insertional inactivation of cnm was confirmed by PCR sequencing. To generate cnm knockouts in the other Cnm+ strains (NCTC11060, LM7, and OM50E), a PCR product was generated from DNA of the OMZ175 cnm mutant strain by using primers cnm110F and cnm1560R, and 100 ng of this PCR product was used for transformation of competent cells.
To express the cnm gene in trans, the full-length cnm gene, including the ribosomal binding site, was amplified by PCR with primers containing BamHI (5′-GTAATATTCTGGATCCAAGAAAGGACTA-3′) and XbaI (5′-CCTGTTTTTAATCTAGATCAGCTATG-3′) restriction sites and ligated into pMSP3535 (11) which had been digested with BamHI and XbaI. A ligation mixture containing pMSP3535 expressing cnm (pcnm) was used to directly transform the S. mutans cnm knockout strain OMZ175-cnm to generate a complementation strain carrying the gene. Expression of cnm from pcnm was induced with 15 ng of nisin ml−1 as described elsewhere (32).
RNA isolation and real-time qRT-PCR.
To measure cnm expression levels in OMZ175, NCTC11060, B14, LM7, and OM50E, RNAs were extracted from cells grown to mid-exponential phase (optical density at 600 nm [OD600] = 0.5) in BHI broth as described elsewhere (1). A high-capacity cDNA reverse transcription kit containing random primers (Applied Biosystems, Foster, CA) was used to obtain cDNA from 1 μg each of three independent RNA samples. Quantitative reverse transcriptase PCR (qRT-PCR) was carried out using the cnm-specific primers cnm-CF (5′-CTGAGGTTACTGTCGTTAAA) and cnm-CR (5′-CACTGTCTACATAAGCATTC) (47) and protocols described elsewhere (1). Student's t test was performed to verify the significance of real-time RT-PCR quantification.
Adherence and invasion assays.
Antibiotic protection assays were performed to assess the capacity of S. mutans to invade HCAEC. Briefly, primary HCAEC (Lonza, Allendale, NJ) were cultured in endothelial cell basal medium 2 (EBM-2; Lonza) supplemented with EGM-2MV single-use aliquots (Lonza), as suggested by the supplier. The HCAEC were maintained at 37°C in a humidified 5% CO2 atmosphere. The cells were harvested by trypsinization and washed in EBM-2. One milliliter of suspension containing 105 endothelial cells was then seeded into each well of 24-well flat-bottomed tissue culture plates, followed by overnight incubation in the presence of gentamicin at 37°C in a 5% CO2 atmosphere. Prior to infection, the wells were washed three times with prewarmed EGM-2 without antibiotics. Overnight bacterial cultures were washed twice in phosphate-buffered saline (pH 7.2) and resuspended in supplemented EBM-2 without antibiotics to obtain bacterial suspensions containing 1 × 107 CFU ml−1 of S. mutans. One milliliter of bacterial cell suspension was used to infect HCAEC wells, in triplicate, for 2 h in the absence of antibiotics. Next, the wells were washed three times with 1 ml of EBM-2, followed by 3 h of incubation in 1 ml of EBM-2 containing 300 μg ml−1 gentamicin and 50 μg ml−1 penicillin G to kill extracellular bacteria. After the incubation period with antibiotics, the wells were washed three times with EBM-2. The HCAEC were then lysed for 20 min with 1 ml sterile water. The mixture of lysed HCAEC and S. mutans was plated onto BHI agar and incubated for 48 h at 37°C in a 5% CO2 atmosphere.
The capacity of S. mutans strains to adhere to the surfaces of HCAEC was assessed in the presence of cytochalasin D (Sigma) as described elsewhere (13), with minor modifications. Briefly, the HCAEC were cultured, seeded, and maintained in the same way as described above. Prior to infection, HCAEC-containing wells were washed three times with Hanks' balanced salt solution (Lonza) and then exposed to EBM-2 containing 5 μg ml−1 cytochalasin D without antibiotics for 30 min at 37°C in a 5% CO2 atmosphere. Overnight bacterial cultures were washed twice with phosphate-buffered saline (pH 7.2) and diluted in EBM-2 containing 5 μg ml−1 cytochalasin D without antibiotics to obtain suspensions containing 1 × 107 CFU ml−1. One milliliter of bacterial suspension was used to infect HCAEC cultures, followed by 30 min of incubation at 37°C in a 5% CO2 atmosphere. The HCAEC wells were then washed three times with Hanks' balanced salt solution to remove unbound bacteria, followed by HCEAC lysis with 1 ml of ice-cold sterile water for 20 min. Lysates containing dead HCAEC and intact S. mutans were serially diluted and plated onto BHI agar. All agar plates were incubated for 48 h at 37°C in a 5% CO2 atmosphere.
Biofilm assay.
The capacity of the invasive strains and their respective cnm knockouts to form biofilms in the presence of sucrose or glucose in saliva-coated 96-well microtiter plates was assessed. Briefly, the wells were coated for 1 h at 37°C with 100 μl of sterile, clarified, pooled human saliva (49). Strains grown in BHI medium to an OD600 of 0.5 were diluted 1:100 in low-molecular-weight medium (LMW) (27) supplemented with 1% glucose or 1% sucrose, using a total of six wells per culture. The plates were incubated for 24 h at 37°C in a 5% CO2 atmosphere. One well per strain was assessed for total growth yield by removing planktonic and sessile cells and measuring the OD600. The remaining 5 wells were blotted, rinsed, and stained with 0.1% crystal violet as described elsewhere (3). The incorporated crystal violet was recovered by performing two extractions with 200 μl of 33% acetic acid, and biofilm formation was quantified by measuring the optical density of the solution at 575 nm. Experiments were performed in triplicate.
Galleria mellonella infection.
For G. mellonella killing assays, insects in the final instar larval stage were purchased from Vanderhorst Inc. (St. Marys, OH), stored at 4°C in the dark, and used within 7 days of shipment. Groups of 15 larvae, ranging from 200 to 300 mg and with no signs of melanization, were chosen randomly and used for subsequent infection. A 10-μl syringe (Hamilton, Reno, NV) was used to inject 5-μl aliquots containing 1 × 106 CFU of S. mutans, grown overnight in BHI containing 5% serum and washed twice with sterile saline, into the hemocoel of each larva via the last left proleg. Bacterial colony counts on BHI plates were used to confirm initial inocula. Groups injected with saline solution or with heat-inactivated S. mutans OMZ175 (30 min at 75°C) were used as controls in each experiment. After injection, larvae were incubated at 37°C, and appearance (signs of melanization) and survival were recorded at selected intervals. Larvae were scored as dead when they displayed no movement in response to touch. Kaplan-Meier killing curves were plotted, and estimations of differences in survival were compared using the log rank test. P values of ≤0.05 were considered significant. All data were analyzed with GraphPad Prism 4.0 software.
RESULTS
Identification of new invasive strains supports the link between serotypes e and f and invasive behavior.
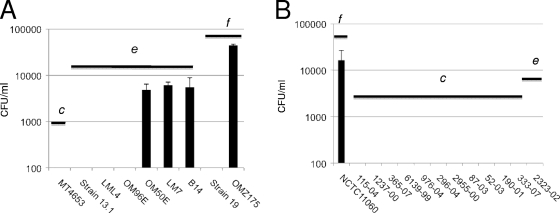
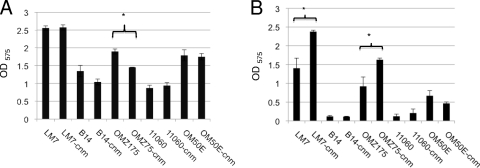
In a previous study, strains B14 and OMZ175, belonging to serotypes e and f, respectively, were able to invade HCAEC, whereas none of the 8 serotype c strains tested displayed invasive behavior (2). To increase the number of serotype e and f strains screened, we assessed the capacity of 19 additional strains to invade HCAEC: 12 isolated from the blood of patients with bacteremia and/or IE (10 serotype c, 1 serotype e, and 1 serotype f strain) and 7 isolated from dental plaque (1 serotype c, 4 serotype e, and 2 serotype f strains). OMZ175 and B14 were used as controls for high and low invasive rates in our experiments. Of the strains isolated from dental plaque, two serotype e strains, LM7 and OM50E, were found to be invasive (Fig. 1A), with 4.8 × 103 and 6.1 × 103 CFU, respectively, recovered from the cytoplasm of HCAEC. Among the blood isolates, serotype f strain NCTC11060 was found to be highly invasive, with 1.6 × 104 CFU recovered from the cytoplasm of HCAEC (Fig. 1B). Notably, while most of the tested strains belonged to the more prevalent serotype c, no serotype c strains were capable of invading HCAEC. On the other hand, 3 of the 5 serotype e and 2 of the 3 serotype f strains tested were capable of invading HCAEC. Note that strains belonging to serotype f were consistently more invasive than serotype e strains, with approximately 5-fold more cells able to reach the cytoplasm of HCAEC.
Fig. 1.
Invasive properties of Streptococcus mutans strains isolated from dental plaque (A) and from blood of patients with bacteremia and/or endocarditis (B). The number of S. mutans CFU recovered from the intracellular compartment of HCAEC after 5 h of infection is shown. The serotypes of the strains are indicated (c, e, or f). The data represent the averages and standard deviations (SD) for at least three independent experiments.
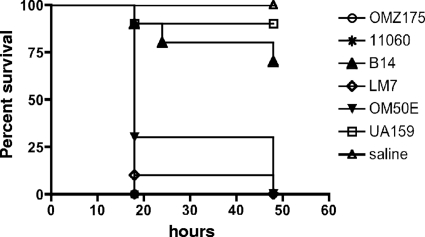
Invasive strains are more virulent than noninvasive strains in the greater wax worm model.
We asked whether G. mellonella could be used to identify differences in the virulence potential of invasive and noninvasive strains. In fact, with the exception of the serotype e strain B14, we found significantly higher mortality rates (P < 0.01) for the groups of worms infected with invasive strains (70 to 100% mortality) within the first 48 h. In contrast, only 10% of the larvae infected with noninvasive serotype c strain UA159 died over the same period (Fig. 2). Six additional noninvasive strains, belonging to serotypes c (52-03, 2955-00, 190-01, and GS-5), e (2323-02), and f (strain 19), were tested in this model, and all behaved similarly to UA159 (data not shown).
Fig. 2.
Killing of G. mellonella larvae infected with the noninvasive strain UA159 and the invasive strains OMZ175, B14, OM50E, LM7, and NCTC11060 of S. mutans. The experiments were repeated three times, and the results are representative of a typical experiment.
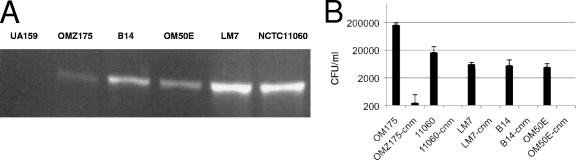
Invasive strains carry the cnm gene.
Recently, it was demonstrated that 10 to 20% of S. mutans strains isolated in Asia and Europe carry the cnm gene, which encodes a collagen- and laminin-binding protein (43, 47). Notably, this gene is associated predominantly with serotypes f and k and is rarely found in serotype c and serotype e strains (43, 47, 51). Because there was a strong correlation between non-serotype c strains and HCAEC invasion, we hypothesized that cnm plays a role in cellular invasion by S. mutans. Indeed, we found that of the 33 S. mutans strains tested in the present study and a previous study (2), only the 5 invasive strains harbored a copy of the cnm gene (Fig. 3A).
Fig. 3.
(A) Detection of the cnm gene by PCR reveals that cnm is found only in invasive strains. (B) Invasion of HCAEC by cnm knockout strains. The number of S. mutans CFU recovered from the intracellular compartment of HCAEC after 5 h of infection is shown. The data represent the averages and SD for at least three independent experiments.
All invasive strains express cnm.
To verify cnm expression levels, the mRNA levels of cnm in exponential-phase cultures of all five invasive strains were measured by qRT-PCR. Strain OMZ175 displayed the highest expression level of cnm (1.2 × 107 copies) among all strains. Strain NCTC11060 showed the lowest expression level of cnm (1.7 × 106 copies), followed by B14 (3.3 × 106 copies), OM50E (4.6 × 106 copies), and LM7 (5.3 × 106 copies). These results confirmed the expression of cnm in all invasive strains but failed to correlate levels of invasion with higher expression levels of cnm mRNA.
cnm knockout strains have a diminished capacity to adhere to and invade HCAEC.
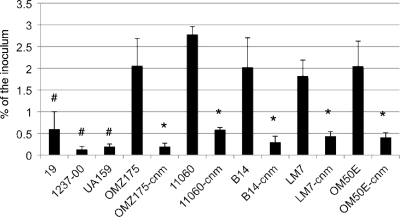
Based on the published sequence of cnm (51), we used a PCR-ligation mutagenesis approach (30) to inactivate the cnm gene in strain OMZ175, using a nonpolar marker. In comparison to the parental strain OMZ175, the cnm mutant strain, designated OMZ175-cnm, did not display any growth defect under standard laboratory growth conditions (data not shown). First, HCAEC were infected separately with UA159 (noninvasive), OMZ175 (invasive), and OMZ175-cnm. The results clearly revealed that inactivation of cnm completely abolished the ability of OMZ175 to invade HCAEC. Based on this finding, we inactivated cnm in the 4 additional invasive strains and observed that, in all cases, a functional cnm gene was required for invasion of HCAEC (Fig. 3B). Adherence assays revealed that both invasive and noninvasive strains could adhere to the surfaces of HCAEC (Fig. 4). However, all invasive strains were recovered in significantly (P < 0.005) larger numbers than the noninvasive strains UA159, 19, and 1237-00. In addition, inactivation of cnm in all of the invasive strains rendered a significant (P < 0.005) 10-fold decrease in the adherence rate (Fig. 4). These results strongly support the hypothesis that Cnm plays an essential role in the invasion process by enhancing bacterial adherence to HCAEC.
Fig. 4.
Adherence of S. mutans to HCAEC. Data shown represent percentages of adherent cells in relation to the initial bacterial inoculum. #, statistically significant difference between the noninvasive strains UA159, 19, and 1237-00 and the invasive strain OMZ175 (P < 0.01); *, statistically significant difference (P < 0.01) between the cnm knockout strains and their respective parental strains. The data represent the averages and SD for at least three independent experiments.
cnm knockout strains are less virulent than the parental strains.
We tested the cnm mutant strains for the ability to kill larvae of G. mellonella. The virulence of the cnm knockout strain in the OMZ175 background was dramatically attenuated, with killing rates that were identical to those found for noninvasive strains (Fig. 5). An identical pattern was observed for the other four cnm knockout strains (data not shown). This result adds further support for an association between intracellular invasion and systemic virulence in S. mutans and corroborates the usefulness of the G. mellonella infection model to assess bacterial pathogenesis.
Fig. 5.
(A) Killing of G. mellonella larvae infected with the noninvasive strain UA159 (□), the invasive strain OMZ175 (⧫), and the cnm knockout strain OMZ175-cnm (•). The experiments were repeated three times, and the results are representative of a typical experiment. (B) Wax worms infected with UA159, OMZ175, and OMZ175-cnm in a typical experiment at 24 h postinjection.
The role of Cnm in biofilm formation is strain dependent.
The capacity of the invasive strains and their respective cnm knockouts (designated B14-cnm, LM7-cnm, OM50E-cnm, 11060-cnm, and OMZ175-cnm) to form biofilms in saliva-coated microtiter plates was assessed. Differences among strains were clear, with LM7 displaying an increased capacity to form biofilms compared to the other strains, regardless of the sugar source (Fig. 6). On the other hand, strains B14 and NCTC11060 showed a diminished capacity to form biofilms, particularly in the presence of glucose (Fig. 6). In sucrose, there was a statistically significant reduction in the amount of biofilm formed by OMZ175-cnm compared to the amount formed by the parent strain OMZ175 (P = 0.008) (Fig. 6A). Conversely, biofilm formation in the presence of glucose was significantly enhanced in the OMZ175-cnm and LM7-cnm mutants (P ≤ 0.0035) compared to the parent strains (Fig. 6B). Overall, the inactivation of cnm in the five invasive strains rendered variable biofilm phenotypes among the different strains. Therefore, the role of cnm in biofilm formation appears to be strain specific.
Fig. 6.
Biofilm assays of cells grown in LMW supplemented with 1% sucrose (A) and 1% glucose (B) on the surfaces of 96-well microtiter plates. Results shown are averages for three separate experiments plus SD. Statistically significant differences between strains were assessed by Student's t test and are indicated by asterisks.
Expression of cnm in trans in OMZ175-cnm partially restores the capacity to invade HCAEC and the capacity for virulence.
The capacity of OMZ175, OMZ175-cnm, and OMZ175-cnm harboring pcnm (OMZ175-cnm/pcnm; complemented strain) to invade HCAEC was assessed. Although the levels of invasion were not completely restored to wild-type levels, the complemented cnm mutant strain displayed an invasive phenotype in the presence of nisin (Fig. 7A). Notably, uninduced OMZ175-cnm/pcnm displayed much lower invasion levels than those under nisin-induced conditions (Fig. 7A), suggesting that optimal levels of cnm mRNA (or Cnm) could not be achieved in trans.
Fig. 7.
Expression of cnm in trans partially restored the invasive phenotype (A) and virulence (B) of OMZ175-cnm.
The virulence of the complemented strain in the G. mellonella wax worm model was also assessed. As observed in the invasion assay, the attenuated virulence of the OMZ-cnm strain was partially restored in the complemented strain grown in the presence of nisin (Fig. 7B). The survival rates at 86 h were 10%, 50%, and 90% for OMZ175, OMZ175-cnm/pcnm, and OMZ175-cnm, respectively (Fig. 7B).
Cnm does not contribute to growth and survival in human blood.
To assess whether Cnm can contribute to survival in blood, the ability of the cnm knockout strain in OMZ175 (OMZ175-cnm) and of the invasive (OMZ175, NCTC11060, LM7, B14, and OM50E) and noninvasive (UA159, 19, OM96E, 2955-00, and 2323-02) strains to grow and survive in blood was assessed. Compared to the parent strain OMZ175, growth and survival of the cnm knockout strain were not affected when cells were cultivated in whole blood, with both strains displaying similar patterns over a 6-day period (data not shown). In addition, comparing invasive strains to noninvasive strains, we did not find a correlation between the invasive phenotype and the ability to grow and survive in blood (data not shown). Finally, there were no obvious differences in survival in blood between strains isolated from dental plaque and those from blood of patients with bacteremia/IE (data not shown).
DISCUSSION
Non-serotype c strains of S. mutans, comprising serotypes e, f, and k, have been detected at high frequency in specimens from patients who underwent surgery for removal of atheromatous plaque and heart valve replacement (38, 40, 44). It has been speculated that non-serotype c strains were isolated at higher frequency because they were highly persistent in blood (44). However, the results presented herein show no correlation between the different serotypes and survival in blood. Therefore, it is likely that the high recovery of non-serotype c strains from cardiovascular specimens is due to other virulence factors specific to non-serotype c strains. Our observation that, to date, only non-serotype c strains could invade heart endothelial cells suggests that these strains have the potential to persist in the heart tissue by hiding in the intracellular niche, leading to antibiotic treatment failure, chronic inflammation, and increased morbidity. In this study, we identified three new invasive S. mutans strains, two belonging to serotype e (LM7 and OM50E) and one belonging to serotype f (NCTC11060), that, in addition to our previously identified serotype e (B14) and f (OMZ175) invasive strains (2), strongly associate invasive behavior with non-serotype c strains. Collectively, our previous and present studies have evaluated the capacity of 33 strains to invade HCAEC and identified a total of 5 invasive strains.
Recently, a number of laboratories (4–6, 8, 9) have demonstrated that the larvae of the greater wax worm G. mellonella can be used to model systemic bacterial infections, showing a strong correlation with results obtained for mammals (18, 35, 50). Insects possess a complex, multicomponent innate immune system that kills pathogens by using mechanisms similar to those used by mammals, including the production of enzymes (lysozymes), reactive oxygen species, and antimicrobial peptides (25). In particular, there are significant similarities between the oxidative burst pathways of insect hemocytes and mammalian neutrophils (10). Recently, we demonstrated the usefulness of systemic infection of G. mellonella as an adjunct model to study the virulence of S. mutans (24). Notably, the invasive strains were more virulent in the G. mellonella model than noninvasive strains, establishing for the first time a correlation between specific serotypes and cellular invasion and virulence. In addition, we showed that cnm, a gene encoding a collagen- and laminin-binding protein, was present only in invasive strains and that inactivation of cnm abolished the capacity of these strains to invade HCAEC and attenuated virulence in G. mellonella. It has been suggested that Cnm contributes to caries development and to CVD due to its ability to avidly bind to collagen, a major component of dentin and heart valves (43, 47). Studies are under way to disclose the role of Cnm in S. mutans virulence in an animal model of IE.
The distribution of the cnm gene in S. mutans is on par with the frequency of invasive strains identified in our present study and a previous study (2). While cnm is detected in approximately 20% of S. mutans populations (40, 47, 51), this gene is overrepresented in the minor serotype f (approximately 80% of strains) (39, 47), suggesting that Cnm-dependent cellular invasion constitutes an important virulence factor of non-serotype c strains. Subsequent adherence experiments confirmed that Cnm acts as an adhesin, as noninvasive and cnm knockout strains adhered less efficiently to HCAEC than cnm+ strains did. Streptococcal adherence to host cells is mediated by surface proteins and is considered an essential step in the intracellular invasion process (46). Once bacterial adhesins recognize a host receptor and attach to the host surface, host cell signaling cascades can be triggered, leading to various outcomes, such as bacterial internalization, proinflammatory responses, and host cell apoptosis (46). While it is clear that Cnm plays an essential role in the invasion process by enhancing the ability of S. mutans cells to adhere to the surfaces of HCAEC, it remains to be determined whether Cnm also participates in the subsequent steps associated with the invasion process.
The ability to bind to surfaces and to form biofilms is considered an important virulence attribute of S. mutans (7, 31, 33). In addition, the capacity to adhere to ECM proteins has been suggested to be an important factor in the colonization of the heart valves by oral bacteria (48, 53, 56). Similar to the case for S. gallolyticus (62), our data revealed that the capacity to invade does not seem to be associated with the ability to form biofilms and that Cnm plays a strain-specific role in biofilm formation. In S. gordonii, inactivation of glucosyltranferase (gtf), which is responsible for biosynthesis of the extracellular polysaccharide glucan that contributes to the adhesion of streptococci to cultured human umbilical vein endothelial cells (59), led to a significant reduction in the ability of the strain to invade these cells. In S. mutans, three glucosyltransferases, GtfB, GtfC, and GtfD, are responsible for the production of the water-insoluble and water-soluble glucans and play a major role in sucrose-dependent biofilm formation (7, 59). The S. mutans glucosyltransferases, in particular GtfB and GtfC, contribute to virulence in animal models of caries and endocarditis (55, 64). In addition to the Gtfs, the wall-associated protein A (WapA), which has collagen-binding activity, is thought to participate in the pathogenesis of IE (19). However, inactivation of gtfB, gtfC, and wapA in OMZ175 did not affect the capacity of the mutant strains to invade HCAEC (J. Abranches et al., unpublished data), suggesting that the S. mutans invasion process is strongly dependent upon the presence of Cnm.
Among our invasive strains, we observed different invasion efficiency rates, with serotype f strains displaying higher invasion rates than serotype e strains. Differences in invasion rates have been shown for other oral bacteria, such as P. gingivalis and S. gordonii (13, 23, 57), as well as for an inhabitant of the gastrointestinal flora, Streptococcus gallolyticus subsp. gallolyticus (62). Furthermore, certain clinical strains of S. mutans display low expression levels of cnm mRNA (47). Although some variability in the level of cnm mRNA was observed among strains, it was not possible to establish a correlation between invasion rates and the expression levels of Cnm.
In conclusion, we showed that S. mutans adhesion to and invasion of HCAEC are intimately linked with the presence of a matrix adhesion-dependent virulence factor, revealing a previously unrecognized mechanism of S. mutans pathogenesis. Our current working hypothesis is that the ability to invade HCAEC helps S. mutans to evade immune surveillance and antibiotic treatment, thereby increasing the morbidity of IE as well as stimulating chronic inflammatory responses that could contribute to CVD. Furthermore, we propose that the Cnm molecule could serve as a biomarker for screening patients who need to receive preventive treatment prior to dental procedures, as well as being a target for the development of novel therapeutic approaches to treat streptococcal infections.
ACKNOWLEDGMENTS
We thank P. Caulfield, N. Jakubovics, and B. Beall for kindly providing some of the strains. We also thank A. Progulske-Fox and Edith Sampson for helpful discussions.
This work was supported by American Heart Association grant 10GRNT4210049 and by NIH-NIDCR Training Program in Oral Sciences grant T32 DE007202 to J.A. and by American Heart Association grant 0655897T to P.J.S.-H.
Footnotes
Published ahead of print on 21 March 2011.
REFERENCES
- 1. Abranches J., Candella M. M., Wen Z. T., Baker H. V., Burne R. A. 2006. Different roles of EIIABMan and EIIGlc in regulation of energy metabolism, biofilm development, and competence in Streptococcus mutans. J. Bacteriol. 188:3748–3756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Abranches J., et al. 2009. Invasion of human coronary artery endothelial cells by Streptococcus mutans OMZ175. Oral Microbiol. Immunol. 24:141–145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ahn S. J., Lemos J. A., Burne R. A. 2005. Role of HtrA in growth and competence of Streptococcus mutans UA159. J. Bacteriol. 187:3028–3038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andrejko M., Mizerska-Dudka M., Jakubowicz T. 2008. Changes in Galleria mellonella apolipophorin III level during Pseudomonas aeruginosa infection. J. Invertebr. Pathol. 97:14–19 [DOI] [PubMed] [Google Scholar]
- 5. Andrejko M., Mizerska-Dudka M., Jakubowicz T. 2008. Changes in Galleria mellonella lysozyme level and activity during Pseudomonas aeruginosa infection. Folia Microbiol. (Prague) 53:147–151 [DOI] [PubMed] [Google Scholar]
- 6. Aperis G., et al. 2007. Galleria mellonella as a model host to study infection by the Francisella tularensis live vaccine strain. Microbes Infect. 9:729–734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Banas J. A. 2004. Virulence properties of Streptococcus mutans. Front. Biosci. 9:1267–1277 [DOI] [PubMed] [Google Scholar]
- 8. Bergin D., Brennan M., Kavanagh K. 2003. Fluctuations in haemocyte density and microbial load may be used as indicators of fungal pathogenicity in larvae of Galleria mellonella. Microbes Infect. 5:1389–1395 [DOI] [PubMed] [Google Scholar]
- 9. Bergin D., Murphy L., Keenan J., Clynes M., Kavanagh K. 2006. Pre-exposure to yeast protects larvae of Galleria mellonella from a subsequent lethal infection by Candida albicans and is mediated by the increased expression of antimicrobial peptides. Microbes Infect. 8:2105–2112 [DOI] [PubMed] [Google Scholar]
- 10. Bergin D., Reeves E. P., Renwick J., Wientjes F. B., Kavanagh K. 2005. Superoxide production in Galleria mellonella hemocytes: identification of proteins homologous to the NADPH oxidase complex of human neutrophils. Infect. Immun. 73:4161–4170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bryan E. M., Bae T., Kleerebezem M., Dunny G. M. 2000. Improved vectors for nisin-controlled expression in gram-positive bacteria. Plasmid 44:183–190 [DOI] [PubMed] [Google Scholar]
- 12. Centers for Disease Control Prevention 2001. Promoting oral health: interventions for preventing dental caries, oral and pharyngeal cancers, and sports-related craniofacial injuries. MMWR Recomm. Rep. 50(RR-21):1–13 [PubMed] [Google Scholar]
- 13. Dorn B. R., Burks J. N., Seifert K. N., Progulske-Fox A. 2000. Invasion of endothelial and epithelial cells by strains of Porphyromonas gingivalis. FEMS Microbiol. Lett. 187:139–144 [DOI] [PubMed] [Google Scholar]
- 14. Dorn B. R., Dunn W. A., Jr., Progulske-Fox A. 1999. Invasion of human coronary artery cells by periodontal pathogens. Infect. Immun. 67:5792–5798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Douglas C. W., Heath J., Hampton K. K., Preston F. E. 1993. Identity of viridans streptococci isolated from cases of infective endocarditis. J. Med. Microbiol. 39:179–182 [DOI] [PubMed] [Google Scholar]
- 16. Drangsholt M. T. 1998. A new causal model of dental diseases associated with endocarditis. Ann. Periodontol. 3:184–196 [DOI] [PubMed] [Google Scholar]
- 17. Engels-Deutsch M., et al. 2003. Insertional inactivation of pac and rmlB genes reduces the release of tumor necrosis factor alpha, interleukin-6, and interleukin-8 induced by Streptococcus mutans in monocytic, dental pulp, and periodontal ligament cells. Infect. Immun. 71:5169–5177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Fedhila S., et al. 2009. Comparative analysis of the virulence of invertebrate and mammalian pathogenic bacteria in the oral insect infection model Galleria mellonella. J. Invertebr. Pathol. 103:24–29 [DOI] [PubMed] [Google Scholar]
- 19. Han T. K., Zhang C., Dao M. L. 2006. Identification and characterization of collagen-binding activity in Streptococcus mutans wall-associated protein: a possible implication in dental root caries and endocarditis. Biochem. Biophys. Res. Commun. 343:787–792 [DOI] [PubMed] [Google Scholar]
- 20. Haraszthy V. I., Zambon J. J., Trevisan M., Zeid M., Genco R. J. 2000. Identification of periodontal pathogens in atheromatous plaques. J. Periodontol. 71:1554–1560 [DOI] [PubMed] [Google Scholar]
- 21. Hill E. E., Herijgers P., Herregods M. C., Peetermans W. E. 2006. Evolving trends in infective endocarditis. Clin. Microbiol. Infect. 12:5–12 [DOI] [PubMed] [Google Scholar]
- 22. Hirasawa M., Takada K. 2003. A new selective medium for Streptococcus mutans and the distribution of S. mutans and S. sobrinus and their serotypes in dental plaque. Caries Res. 37:212–217 [DOI] [PubMed] [Google Scholar]
- 23. Jandik K. A., et al. 2008. Invasive differences among Porphyromonas gingivalis strains from healthy and diseased periodontal sites. J. Periodontal Res. 43:524–530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kajfasz J. K., et al. Two Spx proteins modulate stress tolerance, survival, and virulence in Streptococcus mutans. J. Bacteriol. 192:2546–2556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kavanagh K., Reeves E. P. 2004. Exploiting the potential of insects for in vivo pathogenicity testing of microbial pathogens. FEMS Microbiol. Rev. 28:101–112 [DOI] [PubMed] [Google Scholar]
- 26. Kilian M. 1982. Systemic disease: manifestations of oral bacteria. Dent. Microbiol. 1982:832–838 [Google Scholar]
- 27. Koo H., et al. 2003. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J. Antimicrob. Chemother. 52:782–789 [DOI] [PubMed] [Google Scholar]
- 28. Kremer B. H., et al. 2001. Characterization of the sat operon in Streptococcus mutans: evidence for a role of Ffh in acid tolerance. J. Bacteriol. 183:2543–2552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lapirattanakul J., et al. 2009. Detection of serotype k Streptococcus mutans in Thai subjects. Oral Microbiol. Immunol. 24:431–433 [DOI] [PubMed] [Google Scholar]
- 30. Lau P. C., Sung C. K., Lee J. H., Morrison D. A., Cvitkovitch D. G. 2002. PCR ligation mutagenesis in transformable streptococci: application and efficiency. J. Microbiol. Methods 49:193–205 [DOI] [PubMed] [Google Scholar]
- 31. Lemos J. A., Abranches J., Burne R. A. 2005. Responses of cariogenic streptococci to environmental stresses. Curr. Issues Mol. Biol. 7:95–107 [PubMed] [Google Scholar]
- 32. Lemos J. A., Brown T. A., Jr., Burne R. A. 2004. Effects of RelA on key virulence properties of planktonic and biofilm populations of Streptococcus mutans. Infect. Immun. 72:1431–1440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lemos J. A., Burne R. A. 2008. A model of efficiency: stress tolerance by Streptococcus mutans. Microbiology 154:3247–3255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Li L., Messas E., Batista E. L., Jr., Levine R. A., Amar S. 2002. Porphyromonas gingivalis infection accelerates the progression of atherosclerosis in a heterozygous apolipoprotein E-deficient murine model. Circulation 105:861–867 [DOI] [PubMed] [Google Scholar]
- 35. Mahajan-Miklos S., Rahme L. G., Ausubel F. M. 2000. Elucidating the molecular mechanisms of bacterial virulence using non-mammalian hosts. Mol. Microbiol. 37:981–988 [DOI] [PubMed] [Google Scholar]
- 36. Meurman J. H., Sanz M., Janket S. J. 2004. Oral health, atherosclerosis, and cardiovascular disease. Crit. Rev. Oral Biol. Med. 15:403–413 [DOI] [PubMed] [Google Scholar]
- 37. Moreillon P., Que Y. A. 2004. Infective endocarditis. Lancet 363:139–149 [DOI] [PubMed] [Google Scholar]
- 38. Nakano K., et al. 2006. Detection of cariogenic Streptococcus mutans in extirpated heart valve and atheromatous plaque specimens. J. Clin. Microbiol. 44:3313–3317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nakano K., et al. 2007. Streptococcus mutans clonal variation revealed by multilocus sequence typing. J. Clin. Microbiol. 45:2616–2625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nakano K., et al. 2007. Serotype distribution of Streptococcus mutans a pathogen of dental caries in cardiovascular specimens from Japanese patients. J. Med. Microbiol. 56:551–556 [DOI] [PubMed] [Google Scholar]
- 41. Nakano K., Nomura R., Nakagawa I., Hamada S., Ooshima T. 2004. Demonstration of Streptococcus mutans with a cell wall polysaccharide specific to a new serotype, k, in the human oral cavity. J. Clin. Microbiol. 42:198–202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nakano K., et al. 2007. Detection of novel serotype k Streptococcus mutans in infective endocarditis patients. J. Med. Microbiol. 56:1413–1415 [DOI] [PubMed] [Google Scholar]
- 43. Nakano K., et al. 2010. Molecular characterization of Streptococcus mutans strains containing the cnm gene encoding a collagen-binding adhesin. Arch. Oral Biol. 55:34–39 [DOI] [PubMed] [Google Scholar]
- 44. Nakano K., Ooshima T. 2009. Serotype classification of Streptococcus mutans and its detection outside the oral cavity. Future Microbiol. 4:891–902 [DOI] [PubMed] [Google Scholar]
- 45. Nemoto H., Nakano K., Nomura R., Ooshima T. 2008. Molecular characterization of Streptococcus mutans strains isolated from the heart valve of an infective endocarditis patient. J. Med. Microbiol. 57:891–895 [DOI] [PubMed] [Google Scholar]
- 46. Nobbs A. H., Lamont R. J., Jenkinson H. F. 2009. Streptococcus adherence and colonization. Microbiol. Mol. Biol. Rev. 73:407–450 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Nomura R., et al. 2009. Molecular and clinical analyses of the gene encoding the collagen-binding adhesin of Streptococcus mutans. J. Med. Microbiol. 58:469–475 [DOI] [PubMed] [Google Scholar]
- 48. Okahashi N., et al. Pili of oral Streptococcus sanguinis bind to fibronectin and contribute to cell adhesion. Biochem. Biophys. Res. Commun. 391:1192–1196 [DOI] [PubMed] [Google Scholar]
- 49. Phan T. N., Reidmiller J. S., Marquis R. E. 2000. Sensitization of Actinomyces naeslundii and Streptococcus sanguis in biofilms and suspensions to acid damage by fluoride and other weak acids. Arch. Microbiol. 174:248–255 [DOI] [PubMed] [Google Scholar]
- 50. Rahme L. G., et al. 2000. Plants and animals share functionally common bacterial virulence factors. Proc. Natl. Acad. Sci. U. S. A. 97:8815–8821 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Sato Y., et al. 2004. Streptococcus mutans strains harboring collagen-binding adhesin. J. Dent. Res. 83:534–539 [DOI] [PubMed] [Google Scholar]
- 52. Sato Y., et al. 2004. Application of in vitro mutagenesis to identify the gene responsible for cold agglutination phenotype of Streptococcus mutans. Microbiol. Immunol. 48:449–456 [DOI] [PubMed] [Google Scholar]
- 53. Scheld W. M., Strunk R. W., Balian G., Calderone R. A. 1985. Microbial adhesion to fibronectin in vitro correlates with production of endocarditis in rabbits. Proc. Soc. Exp. Biol. Med. 180:474–482 [DOI] [PubMed] [Google Scholar]
- 54. Shibata Y., et al. 2003. Analysis of loci required for determination of serotype antigenicity in Streptococcus mutans and its clinical utilization. J. Clin. Microbiol. 41:4107–4112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Shun C. T., et al. 2005. Glucosyltransferases of viridans streptococci are modulins of interleukin-6 induction in infective endocarditis. Infect. Immun. 73:3261–3270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sommer P., Gleyzal C., Guerret S., Etienne J., Grimaud J. A. 1992. Induction of a putative laminin-binding protein of Streptococcus gordonii in human infective endocarditis. Infect. Immun. 60:360–365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Stinson M. W., Alder S., Kumar S. 2003. Invasion and killing of human endothelial cells by viridans group streptococci. Infect. Immun. 71:2365–2372 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Tamura G. S., Kuypers J. M., Smith S., Raff H., Rubens C. E. 1994. Adherence of group B streptococci to cultured epithelial cells: roles of environmental factors and bacterial surface components. Infect. Immun. 62:2450–2458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Vacca-Smith A. M., Jones C. A., Levine M. J., Stinson M. W. 1994. Glucosyltransferase mediates adhesion of Streptococcus gordonii to human endothelial cells in vitro. Infect. Immun. 62:2187–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Van der Meer J. T., et al. 1992. Efficacy of antibiotic prophylaxis for prevention of native-valve endocarditis. Lancet 339:135–139 [DOI] [PubMed] [Google Scholar]
- 61. Vernier-Georgenthum A., al-Okla S., Gourieux B., Klein J. P., Wachsmann D. 1998. Protein I/II of oral viridans streptococci increases expression of adhesion molecules on endothelial cells and promotes transendothelial migration of neutrophils in vitro. Cell. Immunol. 187:145–150 [DOI] [PubMed] [Google Scholar]
- 62. Vollmer T., Hinse D., Kleesiek K., Dreier J. 2010. Interactions between endocarditis-derived Streptococcus gallolyticus subsp. gallolyticus isolates and human endothelial cells. BMC Microbiol. 10:78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Westerlund B., Korhonen T. K. 1993. Bacterial proteins binding to the mammalian extracellular matrix. Mol. Microbiol. 9:687–694 [DOI] [PubMed] [Google Scholar]
- 64. Yamashita Y., Bowen W. H., Burne R. A., Kuramitsu H. K. 1993. Role of the Streptococcus mutans gtf genes in caries induction in the specific-pathogen-free rat model. Infect. Immun. 61:3811–3817 [DOI] [PMC free article] [PubMed] [Google Scholar]