Abstract
Salmonella enterica serovar Typhi, the agent of typhoid fever in humans, expresses the surface Vi polysaccharide antigen that contributes to virulence. However, Vi expression can also be detrimental to some key steps of S. Typhi infectivity, for example, invasion, and Vi is the target of protective immune responses. We used a strain of S. Typhimurium carrying the whole Salmonella pathogenicity island 7 (SPI-7) to monitor in vivo Vi expression within phagocytic cells of mice at different times after systemic infection. We also tested whether it is possible to modulate Vi expression via the use of in vivo-inducible promoters and whether this would trigger anti-Vi antibodies through the use of Vi-expressing live bacteria. Our results show that Vi expression in the liver and spleen is downregulated with the progression of infection and that the Vi-negative population of bacteria becomes prevalent by day 4 postinfection. Furthermore, we showed that replacing the natural tviA promoter with the promoter of the SPI-2 gene ssaG resulted in sustained Vi expression in the tissues. Intravenous or oral infection of mice with a strain of S. Typhimurium expressing Vi under the control of the ssaG promoter triggered detectable levels of all IgG subclasses specific for Vi. Our work highlights that Vi is downregulated in vivo and provides proof of principle that it is possible to generate a live attenuated vaccine that induces Vi-specific antibodies after single oral administration.
INTRODUCTION
Typhoid fever is a systemic infection caused by the human-restricted pathogen Salmonella enterica serovar Typhi. Around 20 million cases of typhoid fever were estimated worldwide in 2000, including over 200,000 lethal cases (5). The few cases reported in industrialized countries are mainly associated with travel to South and Southeast Asia. The transmission of the disease occurs through the ingestion of water or food contaminated with feces from patients or chronic carriers and is a significant health burden in countries with very poor sanitation.
Three vaccines are currently available. The first is the orally administered, live attenuated vaccine Ty21a, derived by chemical mutagenesis, and the list of all the genes affected by the mutations was assessed only recently (19). Ty21a does not express the Vi antigen and is therefore incapable of eliciting any anti-Vi antibody response (6, 11). Overall, this vaccine is well tolerated and effective but nevertheless has several drawbacks, the main one being the need for several immunizations to develop effective protection (32). The second vaccine is a preparation of purified Vi antigen (11). Vi is a polymer of α-d-(1-4)-linked N-acetylgalactosaminuronate with 60 to 70% of the monomeric units O-acetylated at the C-3 position (7). Only S. Typhi, S. Paratyphi C, Citrobacter freundii, and some strains of S. Dublin are capable of producing Vi. The currently licensed purified Vi vaccine confers 55 to 75% protection against typhoid fever, mainly due to the induction of antibody responses (1, 16). However, this vaccine does not confer lasting immunological memory, is ineffective in children under the age of 2 years, and requires parenteral administration (20, 22). The third vaccine is a Vi conjugate delivered by parenteral administration, which has shown promise in field trials (22). Attempts to elicit strong anti-Vi responses by use of live vaccines have failed so far. One explanation for this is that Vi might not be expressed at immunogenic levels (i.e., may be downregulated in vivo) (30). In addition, efforts to express Vi constitutively in live S. Typhi failed to induce anti-Vi antibodies (31), suggesting that constitutive, unregulated expression of Vi may result in poor infectivity or persistence of the vaccine.
Vi is therefore a double-edged sword in the pathogenesis of infection. On the one hand, Vi can benefit bacteria by inhibiting complement deposition at the bacterial surface and the postphagocytic oxidative burst, thus resulting in reduced bacterial internalization and killing by phagocytes (2, 28). Vi may also modulate immune responses, perhaps by physically masking pathogen-associated molecular patterns (PAMPs) from innate immune receptors. For example, the Vi polysaccharide of S. Typhi reduces Toll-like receptor (TLR)-dependent interleukin-8 (IL-8) expression in the intestinal mucosa (26) and impairs the recognition of lipopolysaccharide (LPS) by TLR4 and prevents the secretion of tumor necrosis factor alpha (TNF-α) in macrophages (16, 37). However, inappropriate or constitutive expression of Vi on the bacterial surface could be detrimental to S. Typhi by hindering the secretion of proteins secreted by type 3 secretion systems (T3SS) and needed for invasion and intracellular survival of the bacterium (2). For example, the presence of Vi surrounding bacteria renders them less adherent and invasive to epithelial cells (36). Sustained expression of Vi could also trigger the production of anti-Vi antibodies, enhancing phagocytosis and killing of Vi+ S. Typhi. Thus, careful regulation of Vi biosynthesis at different anatomical sites and at different times after infection is needed for S. Typhi evasion of the host immune response without detriment to the infectivity of the bacterium due, for example, to the hindrance of Salmonella pathogenicity island (SPI)-encoded secretion systems.
Given the crucial impact of Vi on the pathogenesis of typhoidal infections, it is likely that Vi expression is tightly controlled throughout the infection process. The regulation of Vi is complex. Two widely separated loci are involved in Vi expression: viaA and viaB. The viaB operon resides on a 134-kb pathogenicity island known as SPI-7 and is specific to Vi-expressing strains. The viaB region in S. Typhi comprises 10 genes involved in either the biosynthesis (tviB, tviC, tviD, and tviE), export (vexA, vexB, vexC, and vexD), or membrane anchoring (vexE) of the Vi polysaccharide (35).
Vi expression is regulated by environmental stimuli, including osmolarity. Expression is enhanced at low or medium osmolarity (around 150 mM NaCl), whereas expression is downregulated at elevated osmolarity (above 300 mM NaCl) (2, 40). In the mammalian body, S. Typhi encounters several compartments with different osmolarities. In the gut lumen, where the osmolarity is approximately 300 mM, the bacterium expresses very little or no Vi antigen (2, 40). The low osmolarity of the blood and of the intracellular compartment is predicted to favor Vi expression, as also suggested by detectable Vi expression within bovine epithelial cells (33). This is in contrast to the very low or absent immune responses to Vi seen in the majority of typhoid patients, with anti-Vi antibodies found more frequently in the sera of chronic carriers and rarely in individuals with acute infection. S. Typhi resides mainly within phagocytic cells, and therefore it may be surprising that conditions within phagocytes are conducive to low or absent expression of Vi. However, it is reasonable to postulate that the regulation of Vi expression in S. Typhi varies throughout the course of infection, as previously identified for infection of human macrophages (10), and has evolved to favor the optimal infectivity of S. Typhi while avoiding immune detection.
In this study, we tested the hypothesis that expression of Vi is downregulated within phagocytic cells in the host at different times after systemic infection. We also tested the hypothesis that Vi expression within phagocytic cells, directed by the use of in vivo-inducible promoters, triggers anti-Vi antibodies. We used S. Typhimurium experimental infections of mice as a model for typhoid fever. Since S. Typhimurium lacks the genes required for the expression of the Vi antigen, we used a strain of S. Typhimurium (C5.507) carrying the whole SPI-7, including the viaB operon, as a chromosomal integration. Although other elements involved in the regulation of Vi expression may be present in S. Typhi and may differ from those in S. Typhimurium, we show that Vi expression in strain C5.507 is regulated by osmolarity, similar to what is seen in S. Typhi. Our results show that Vi expression in the liver and spleen is downregulated with the progression of infection. Following these findings, we tested whether it was possible to achieve sustained expression of Vi in the liver and spleen by replacing the tviA promoter with the promoter of the ssaG gene from SPI-2, known to be upregulated when bacteria reach the Salmonella-containing vacuole (SCV) within macrophages (23). Our results show that the ssaG promoter allows sustained Vi expression in the tissues and results in the induction of anti-Vi antibody responses.
Our work highlights the observation that Vi is downregulated in vivo and provides proof of principle that it is possible to generate a live attenuated vaccine that induces Vi antibodies after single oral administration.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
Strains C5 and C5.507 of Salmonella enterica serovar Typhimurium were used in this study. The C5.507 strain is a Vi-expressing derivative of the virulent C5 strain. It contains the whole of SPI-7, including the viaB operon involved in the synthesis and export of the Vi antigen (12; our unpublished data). Preparation of electrocompetent Escherichia coli and S. enterica cells and transformations were performed as previously described (9). Media were supplemented with the appropriate antibiotic for selection (ampicillin, 100 μg/ml; kanamycin, 50 μg/ml; chloramphenicol, 20 μg/ml; or gentamicin, 40 μg/ml). The bacteria were cultured in classic Luria-Bertani (LB) broth and on LB agar (Sigma-Aldrich, United Kingdom) or in LB containing various concentration of NaCl. Low-pH (MM pH 5.8) or neutral-pH (MM pH 7.7) minimal medium was also used to check the expression of Vi under the regulation of the PssaG promoter in the Pssag::tviA GFP+ strain. The minimal medium contained 100 mM Bis-Tris buffer (Sigma-Aldrich, United Kingdom), 0.1% (wt/vol) Casamino Acids, 0.16% glycerol, and 10 μM MgCl2 (21). In vitro growth rates of Salmonella strains were determined by both optical density measurements and viability counts.
Animals.
All animals were handled in strict accordance with good animal practice as defined by the relevant international (Directive of the European Parliament and of the Council on the Protection of Animals Used for Scientific Purposes [Brussels 543/5]) and local (Department of Veterinary Medicine, University of Cambridge) animal welfare guidelines. All animal work was approved by the ethical review committee of the University of Cambridge and was covered by a project license granted by the Home Office under Animal (Scientific Procedures) Act 1986. BALB/c mice were purchased from Harlan Olac Ltd. (Blackthorn, United Kingdom). Female age-matched mice of 6 to 14 weeks old were used for experiments. Bacterial cultures were grown from single colonies in 10 ml LB broth incubated overnight without shaking at 37°C and then diluted in phosphate-buffered saline (PBS) to the appropriate concentration for inoculation. Inocula were enumerated by growth on LB agar pour plates. Mice were injected in a lateral tail vein or were infected by oral gavage with a volume of 0.2 ml.
Enumeration of viable salmonellae in mouse tissues.
Mice were killed by cervical dislocation, and livers and spleens were removed aseptically. Half of each organ was homogenized (separately) in a Seward Stomacher 80 Biomaster blender (Seward) and in 10 ml of sterile water in a Colworth Stomacher 80 blender. The resulting homogenates were diluted in a 10-fold series in PBS, and LB agar pour plates were used to enumerate viable bacteria.
Recombinant DNA techniques.
Standard methods were used for molecular cloning (29). Chromosomal and plasmid DNA purifications and routine DNA modifications, including restriction endonuclease digestion of DNA, modifications of DNA, and ligations, were carried out using commercial kits and supplies according to the manufacturers' instructions (Qiagen, United Kingdom; Promega, United Kingdom; Invitrogen, United Kingdom; Roche, United Kingdom; and New England BioLabs, United Kingdom). DNA concentration and purity were measured using a Nanodrop ND-1000 spectrophotometer. PCR primers were designed using Primer3 (http://frodo.wi.mit.edu/) and were purchased from Sigma (Sigma-Genosys, United Kingdom). PCRs were performed in 25-μl reaction volumes in 0.2-ml Eppendorf tubes in a Perkin Elmer Gene Amp 2400 thermal cycler. Reaction mixtures contained a 200 μM concentration of each deoxynucleoside triphosphate (dNTP), 2 mM Mg2+, 0.01 volume of Proof Start DNA polymerase (2.5 U μl−1; Qiagen), 0.1 volume polymerase buffer (10×), 1 μM (each) forward and reverse primers, and template DNA (∼50 ng plasmid DNA or ∼100 ng chromosomal DNA). Thermal cycler conditions were 94°C for 10 min; 35 cycles of 94°C for 1 min, 56°C for 1 min, and 72°C for 1 min/kb; and a final extension at 72°C for 10 min.
Genetic modification of bacterial strains.
Strains C5 and C5.507 were genetically modified to express the gfp gene constitutively. The primers MalXT1 (5′-CCGCAGGTTCAGTCGGTAAAAGATGAAATGGTTGGCCTGATGAATACCGTTCAGGCATAACCTGGGGTAATGACTCTCTAGC-3′) and MalYCam (5′-CTACGTACACCATGTCCCGCGTCGGTCAACTTCCTGTGAAAAATCGAACATATCCCTTCCGACGTCATTTCTGCCATTCATCC-3′) were used to amplify the chromosomal region of S. Typhimurium strain JH3016 containing the fusion rpsM′-gfp and the cat gene (15). The PCR fragment was integrated into the chromosomes of S. Typhimurium strains C5 and C5.507 (between two pseudogenes—malX and malY) by the Lambda Red method as previously detailed (8). The resultant strains were confirmed by PCR, sequencing, and Southern blot analysis. The expression of green fluorescent protein (GFP) was verified by fluorescence microscopy. The insertion of the gfp gene did not affect the growth rate of the two strains in vivo and in vitro (data not shown). The primers gentaFRTFwd (5′-ATTATCTAGAAAGTATAGGAACTTCGACTGACCTTTACGCATGGG-3′) and gentaFRTRev (5′-GCGCTCTAGAGAATAGGAACTTCGGAATAGGAACTTCCACAAAACGGTGCAAATAAGTAT-3′) were used to amplify the chromosomal region containing the aacC1 gene from S. Typhimurium SL1344 degP::Gm (24). The PCR product was cloned into the XbaI site of pBADTOPOKanFRT as an XbaI fragment, replacing the kanamycin resistance cassette and providing the final plasmid pBADTOPOgentaFRT, containing the gentamicin resistance cassette flanked by two FRT sites. The promoter PssaG, containing the ribosome binding site, was PCR amplified from S. Typhimurium C5.507 genomic DNA by use of the primers PssaGFwd (5′-ATATGGATCCTACGAAGCTTCCTGGCAGGGATTGGCC-3′) and PssaGRev (5′-AATGCTTTTCCTTAAAATAAATACATCG-3′) and then cloned into pGEM-T Easy (Promega, United Kingdom) to give the pGEMTeasy/PssaG plasmid. The fragment containing the gentamicin resistance cassette flanked by the full FRT sites was cloned into the BamHI sites of pGEMTeasy/PssaG as a BamHI fragment generated by PCR amplification from pBADTOPOgentaFRT, using the primers HNgentaFRTFwdBamHI (5′-ATTAGGATCCATGACGATGACAAGCTCGCCCTTTCG-3′) and HNgentaFRTRev (5′-GAGAGGATCCAGGGATAGGCTTACCTTCAAGCTC-3′) (BamHI sites incorporated into primers), giving the pGEMTeasy/GentaFRT-PssaG plasmid. The gentaFRT-PssaG cassette was PCR amplified from pGEMTeasy/GentaFRT-PssaG with the primers ODMssaGFwd (5′-CATTCGATTTTCTAGACTAAATAAGATTTTTTGATAGGTACAAACAATGAATTGTGCAGGTAGGGATAGGCTTACCTTCAAGCTC-3′) and ODMssaGRev (5′-ATGCCAGCAGCTCCAACCCCGAAATAGATATCATTCGGAGGCCAGAAATGATGAAACCTCATAATGCTTTTCCTTAAAATAAATACATCG-3′). The product was integrated into the chromosome of C5.507 GFP+ (replacing the tviA promoter) by the Lambda Red method as previously detailed (8). This intermediate strain, still carrying the gentamicin resistance cassette, was then transformed with pCP20 as previously described (4) to excise the aacC1 gene between the FRT sites and to give the final strain S. Typhimurium Pssag::tviA GFP+, which was confirmed by PCR, sequencing, and Southern blot analysis.
Agglutination of in vitro culture.
The C5.507 GFP+ strain was cultured overnight in LB or LB containing 150, 300, 400, or 600 mM NaCl. For agglutination, 10 μl of the overnight culture was mixed with 10 μl of rabbit anti-Vi polyclonal antibody (Remel) or rabbit anti-LPS O4 agglutinating serum (Remel) and then mounted onto a glass slide. Agglutination was visualized by phase-contrast microscopy using a Leica DM6000B fluorescence microscope (magnification, ×630).
Immunostaining of in vitro cultures by fluorescence microscopy.
For immunostaining, 100 μl of an overnight bacterial culture was centrifuged at 13,000 × g for 5 min; the bacteria were then fixed for 5 min at room temperature in 10% buffered formalin. Subsequently, pellets were washed in PBS (2 5-min washes) and then incubated for 1 h at room temperature or overnight at 4°C with 1:500 rabbit anti-Vi polyclonal antibody (Remel) or 1:500 normal rabbit serum (Dako). Subsequently, pellets were washed in PBS (2 5-min washes) and then incubated for 1 h at room temperature in the dark with 1:200 Alexa Fluor 568-conjugated goat anti-rabbit antibody (Invitrogen-Molecular Probes, United Kingdom). All pellets were washed in PBS (2 5-min washes) and mounted onto glass slides with Vectashield (Vector Laboratories Ltd.). The analysis of tissue sections was done by multicolor fluorescence microscopy (MCFM) using a Leica DM6000B fluorescence microscope running FW4000 acquisition software.
Immunostaining of tissue sections for fluorescence microscopy.
Half of each organ was fixed overnight in 4% paraformaldehyde diluted in PBS, washed for a total of 90 min in three changes of PBS, and then immersed in 20% sucrose (in PBS) for 16 h at 4°C before being embedded in optimal cutting temperature (OCT) reagent (Raymond A Lamb Ltd., United Kingdom) in cryomolds (Park Scientific, Northampton, United Kingdom). Samples were frozen and stored at −80°C. Thirty-micrometer sections were cut, blocked, and permeabilized for 10 min in a permeabilizing solution containing 10% normal goat serum and 0.002 mg/ml saponin in PBS (Sigma, Poole, United Kingdom). Sections were stained for 16 h at 4°C with a 1:500 dilution of rabbit anti-Vi polyclonal antibody (Remel). A 1:500 dilution of rabbit anti-LPS O4 agglutinating serum (Remel), diluted in permeabilizing solution, and a 1:500 dilution of normal rabbit serum (Dako) were used as negative controls. Subsequently, sections were washed in PBS (3 30-min washes) and then incubated for 1 h at room temperature with 1:200 Alexa Fluor 568-conjugated goat anti-rabbit antibody (Invitrogen-Molecular Probes, United Kingdom). All sections were washed in PBS (3 30-min washes) and mounted onto Vectabond-treated glass slides (Vector Laboratories Ltd.), using Vectashield containing DAPI (4′,6-diamidino-2-phenylindole) (Vector Laboratories Ltd.). The analysis of tissue sections was done by MCFM using a Leica DM6000B fluorescence microscope running FW4000 acquisition software. Between 100 and 200 GFP+ bacteria were counted per organ, and the expression of Vi was recorded for each bacterium counted.
ELISA for detection of anti-Vi IgG.
Flat-bottomed 96-well plates were coated at 4°C overnight with 100 μl of 2 μg/ml of purified Vi from Citrobacter strain 3056 (obtained from the Novartis Vaccine Institute for Global Health [NVGH]) in carbonate buffer (0.05 M; pH 9.6). The wells were blocked with 200 μl of PBS containing 0.05% Tween 20 (PBST) and 5% milk for 1 h at room temperature. One-hundred-microliter serum dilutions in PBST–0.1% bovine serum albumin (BSA) were then added and incubated for 2 h at room temperature. To detect total immunoglobulins, the plates were incubated for 1 h at 37°C with 1:2,000 horseradish peroxidase (HRP)-conjugated goat anti-mouse (Dako). After three washes with PBST, the reaction was revealed by adding 100 μl of o-phenylenediamine (OPD; Sigma-Aldrich, United Kingdom) for 15 min and stopped with 100 μl of 3 M H2SO4. Optical density (OD) was measured at 492 nm using a FluoStar Galaxy microplate reader. To detect IgG subclasses, 100 μl of isotype-specific antibody (Sigma, United Kingdom) (diluted 1:1,000 in PBST–0.1% BSA) was added to the plates and then incubated for 30 min at room temperature. After three washes in PBST, 100 μl of 1:5,000 HRP-conjugated rabbit anti-goat, diluted in PBST–0.1% BSA, was added, and the plates were incubated for 15 min. The reaction was revealed by adding 100 μl of OPD. A negative control (naïve mouse sera, obtained from naïve mice from the same batch as that used in the experiment and then pooled) and a positive control (standard murine anti-Vi serum obtained from NVGH) were used in each experiment. We also tested pooled sera from mice immunized with live attenuated aroA mutant S. Typhimurium (strain SL3261) and found this serum to be negative by enzyme-linked immunosorbent assay (ELISA), at a level similar to that observed for naïve serum. The results are shown as arbitrary units. The arbitrary unit value for each serum represents the reciprocal of the serum dilution that would give an OD reading of 0.1 using a best-fit linear regression curve.
Statistical analysis.
All data analysis was produced using the open-source R statistical language (27). For growth curve analyses, two replicate measurements were obtained for each organ from each mouse, and the data represent the means for these replicates. To calculate the proportions of cells expressing Vi, between 100 and 200 GFP+ bacteria were counted per organ, and Vi expression was recorded for each bacterium counted. Data for the liver and spleen were analyzed separately.
To assess whether there was evidence that net growth rates between successive time points varied between C5 GFP+- and C5.507 GFP+-infected mice, a regression model including time (categorical) and genotype main effects and a time × genotype interaction effect was fitted to the log10 CFU counts. Each effect was added in turn, and the statistical significance of the change in model fit was assessed using likelihood ratio tests (LRT; in this case, a P value of <0.05 suggests that the new model gives a statistically significantly better fit to the data than the previous one). If necessary, pairwise comparisons could be conducted using the multcomp package in R (17), and 95% adjusted confidence intervals (95% CIs) were produced for these differences.
A similar method was used to explore the relationships between the proportion of Vi+ bacteria and time (see Fig. 2 and 5) and the log10 CFU counts and genotype (see Fig. 6).
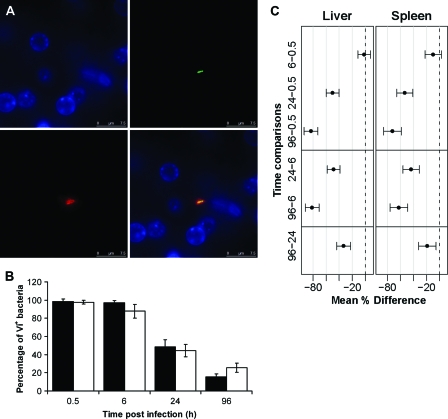
Fig. 2.
(A) Detection of Vi antigen expression in vivo by immunofluorescence in the liver at 30 min postinfection. The panels represent different channels: green, GFP+ Salmonella; red, Vi; blue, nucleic acid (indicated by DAPI staining). A merged image is shown at bottom right. Bars, 7.5 μm. (B) Percentages of C5.507 GFP+ bacteria expressing Vi antigen in vivo in livers (black bars) and spleens (white bars) at 0.5 h, 6 h, 24 h, and 96 h p.i. Results are expressed as mean percentages of C5.507 GFP+ bacteria expressing Vi antigen ± standard deviations (n = 4 mice per group; a total of 200 bacteria were counted per organ and per time point). (C) Means and adjusted 95% CIs for the differences between the percentages of Vi+ bacteria at consecutive time points (shown in panel B). If the CIs do not cross zero, then they are equivalent to ascertaining statistical significance. Gray lines correspond to increments of 20%.
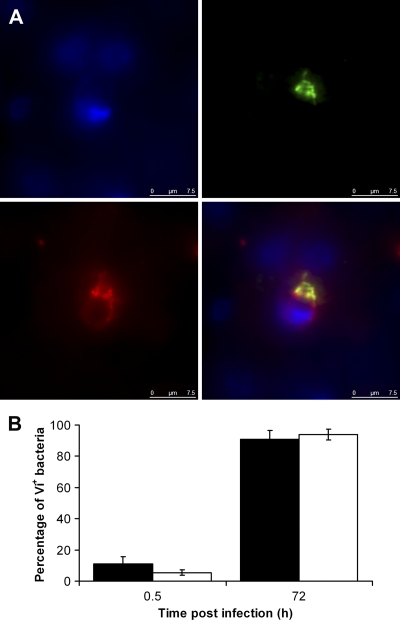
Fig. 5.
(A) Detection of Vi antigen expression in vivo by immunofluorescence in the livers of mice infected with PssaG::tviA GFP+. The panels represent different channels: green, GFP+ Salmonella; red, Vi; blue, nucleic acid (indicated by DAPI staining). A merged image is shown at bottom right. Bars, 7.5 μm. (B) Percentages of PssaG::tviA GFP+ bacteria expressing Vi antigen in vivo in livers (black bars) and spleens (white bars) at 0.5 h and 72 h p.i. Results are expressed as mean percentages of C5.507 GFP+ bacteria expressing Vi antigen ± standard deviations (n = 4 mice per group; a total of 200 bacteria were counted per organ and per time point).
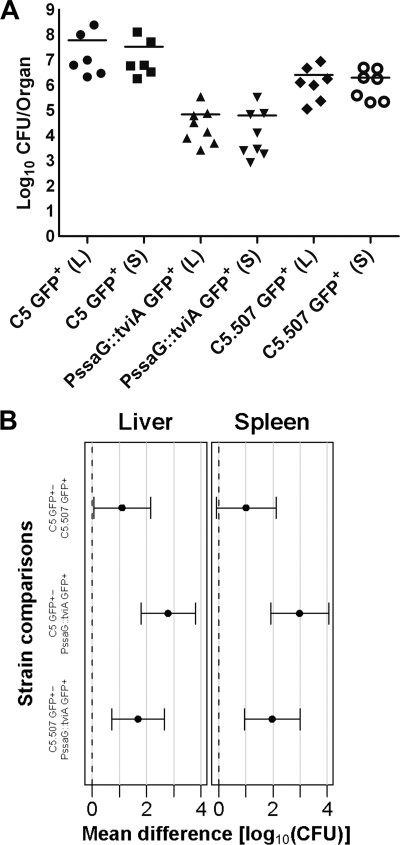
Fig. 6.
(A) Viable counts in the livers (L) and spleens (S) of BALB/c mice at day 6 p.i. for oral infection with ∼9 log10 CFU of the C5 GFP+, C5.507 GFP+, or PssaG::tviA GFP+ strain. Results are expressed as log10 viable counts for individual mice (the mean for each group is indicated by a horizontal bar; n = 6 to 8 mice per group). (B) Means and adjusted 95% CIs for the differences between the log10 CFU counts between strains (shown in panel A). Gray lines correspond to 1 log10 unit.
RESULTS
Expression of Vi antigen in C5.507 GFP+ strain in vitro.
The expression of Vi has been demonstrated to be osmoregulated in vitro in S. Typhi (2, 25, 38). Before proceeding to the analysis of Vi expression in S. Typhimurium C5.507 GFP+ in vivo, we ascertained that Vi is regulated in this strain in vitro by a known stimulus (osmolarity), using LB broth containing various concentrations of NaCl. After overnight growth in LB containing either 150 mM or 300 mM NaCl, cultures of S. Typhimurium C5.507 GFP+ showed significant expression of Vi, as indicated by strong agglutination with anti-Vi serum (data not shown). The Vi agglutination was weaker for cultures grown overnight at an osmolarity of 400 mM and disappeared almost completely at 600 mM NaCl. The parental C5 strain was used as a negative control, and no agglutination was seen with this strain. Conversely, agglutination with anti-O4 serum was observed when the C5.507 GFP+ bacteria were grown at 600 mM NaCl and when the C5 strain was used as a positive control. The expression of Vi in the C5.507 GFP+ strain is thus sensitive to variation of the osmolarity in the broth and appears to be upregulated under low-osmolarity conditions and downregulated in high-osmolarity environments.
Growth of C5 GFP+ and C5.507 GFP+ strains in the mouse spleen and liver.
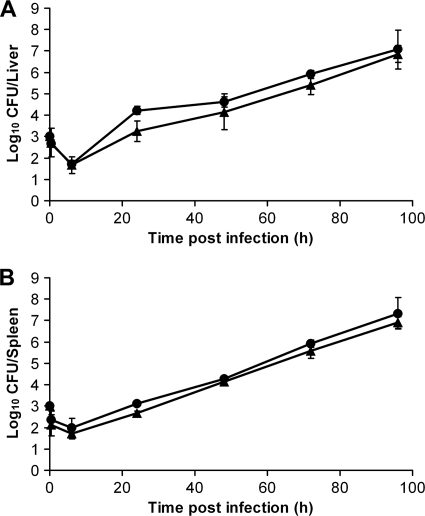
BALB/c mice were injected intravenously (i.v.) with ∼3 log10 CFU of the C5 GFP+ or C5.507 GFP+ strain, and the growth rates of these strains in the liver and spleen were followed for up to 96 h postinfection (p.i.) (Fig. 1). The growth profiles of the C5.507 GFP+ strain and the C5 GFP+ strain were similar, with increases of ∼1 log10 CFU per day in both organs and the appearance of moderate signs of infection at 96 h p.i. Though there was a strong relationship between the log10 CFU counts and time (LRT P value of ≪0.01 in both cases), beyond this there was no evidence of a strong genotype effect in the liver (LRT P value, 0.11), and there was only a small effect in the spleen (LRT P value, 0.01; C5 GFP+ counts were, on average, 0.28 log10 CFU higher [95% CI, 0.09 to 0.48 log10 CFU]). This is consistent with the fact that the inoculum was slightly higher for C5 GFP+ in the spleen and is probably a direct result of this fact. In neither case was there any evidence of a time-genotype interaction effect (LRT P values of 0.40 and 0.94, respectively); thus, it appears that the presence of SPI-7 and expression of viaB under the control of PtviA do not greatly change the growth of C5.507 GFP+ in vivo in comparison to the parental Vi-nonexpressing C5 GFP+ strain.
Fig. 1.
In vivo growth curves for S. Typhimurium C5 GFP+ (circles) and C5.507 GFP+ (triangles) in the livers (A) and spleens (B) of BALB/c mice infected intravenously with ∼3 log10 CFU. Results are expressed as mean log10 viable counts ± standard deviations (n = 4 mice per group).
Expression of Vi antigen in C5.507 GFP+ strain in vivo.
We visualized by immunofluorescence the expression of Vi in the spleens and livers of mice infected with C5.507 GFP+ at 0.5 h, 6 h, 24 h, and 96 h post-i.v. infection. Figure 2 A shows an example of Vi-expressing bacteria in the liver at 0.5 h p.i.: the GFP+ bacteria are surrounded by Vi staining. We then proceeded to quantify the percentage of bacteria that expressed detectable amounts of Vi in the tissues. At early time points (0.5 h and 6 h p.i.), the majority of bacteria (between 88 and 99%) were observed to express Vi, whereas at 96 h p.i., most of the bacteria were observed to be Vi negative (between 74.5 and 84.4%) (Fig. 2B). Thus, downregulation of Vi expression was detectable from 24 h p.i. and was more dramatic at 96 h p.i., when most of the bacteria were not expressing detectable amounts of Vi. The results from analysis of variance (ANOVA) backed up these conclusions, suggesting that time dependence was strongly associated with the proportion of Vi-expressing bacteria in both the liver and the spleen (LRT P values of ≪0.01 in both cases). Adjusted 95% confidence intervals for the mean differences between the comparison groups are shown in Fig. 2C.
Vi expression of PssaG::tviA GFP+ strain in vitro and in vivo.
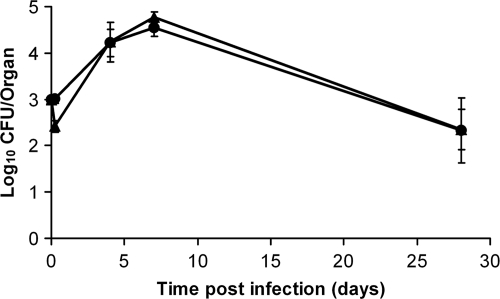
To explore the possibility of maintaining sustained expression of Vi in the tissues and to determine whether this would lead to the induction of anti-Vi immune responses, we replaced the tviA promoter with the SPI-2 ssaG promoter, so the whole viaB operon is under the control of this promoter in the PssaG::tviA GFP+ strain. To test that regulation of Vi expression would be consistent with the conditions known to activate or suppress the ssaG promoter (21), the strain was grown in minimal medium (MM) at pH 5.8 (ssaG-inducing conditions) or pH 7.7 (ssaG-suppressing conditions). Strong Vi expression was observed when the bacterium was grown in MM pH 5.8 (Fig. 3 A), but the expression of Vi was not detectable when the bacterium was grown in MM pH 7.7 (Fig. 3B). Thus, Vi expression in the PssaG::tviA GFP+ strain was pH sensitive, as expected. We next proceeded to monitor the growth characteristics of the PssaG::tviA GFP+ strain in the tissues of infected mice. Mice were injected i.v. with ∼3 log10 CFU of the PssaG::tviA GFP+ strain. Bacterial numbers in the tissues increased until day 8, when they reached ∼5 log10 CFU per organ and then steadily declined in both organs (Fig. 4). Therefore, the PssaG::tviA GFP+ strain showed an attenuated phenotype compared to the C5 GFP+ and C5.507GFP+ strains after i.v. infection. We then monitored the expression of Vi in the livers and spleens of mice infected i.v. with the PssaG::tviA GFP+ strain at 0.5 h and 72 h p.i. (Fig. 5 A and B). The majority of bacteria (89%) found in livers and spleens did not express detectable levels of Vi at 0.5 h p.i., but 91% expressed detectable levels of Vi at 72 h p.i. (Fig. 5B). This was validated by ANOVA (data not shown). Thus, replacing the natural PtviA promoter with PssaG resulted in the in vivo upregulation of Vi rather than in the normal downregulation of this polysaccharide antigen. We also infected mice orally with ∼9 log10 CFU of the C5 GFP+, C5.507 GFP+, and PssaG::tviA GFP+ strains. Viable counts performed on day 6 for livers and spleens of the infected mice showed that the PssaG::tviA GFP+ strain was able to efficiently colonize the spleens and livers of mice infected orally, although the bacterial counts were lower than those seen with C5 GFP+ and C5.507 GFP+ (Fig. 6 A). The strain effect was highly significant (LRT P values of ≪0.01) for both the liver and the spleen, and the mean differences (with 95% CIs) are shown in Fig. 6B.
Fig. 3.
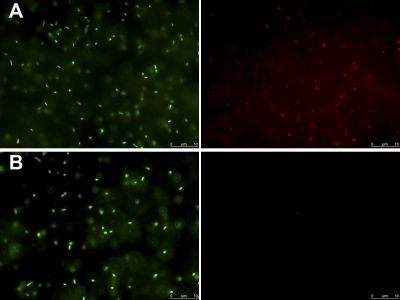
Vi expression in the PssaG::tviA GFP+ strain grown overnight in MM pH 5.8 (A) or MM pH 7.7 (B). The panels represent the same fields of view in different channels: green, GFP+ Salmonella; red, Vi.
Fig. 4.
In vivo growth curve for S. Typhimurium PssaG::tviA in the livers (circles) and spleens (triangles) of BALB/c mice infected intravenously with ∼3 log10 CFU. Results are expressed as mean log10 viable counts ± standard deviations (n = 4 mice per group).
Production of anti-Vi antibodies.
We established that expression of Vi from PssaG results in in vivo upregulation of this polysaccharide antigen rather than in the normal downregulation seen when Vi is controlled by its natural tviA promoter. We then determined whether infection with the PssaG::tviA GFP+ strain would be able to induce an anti-Vi antibody response after oral infection. The sera of mice infected either i.v. (4 mice infected with ∼3 log10 CFU and 4 mice infected with ∼4 log10 CFU) or orally (8 mice infected with ∼9 log10 CFU from an inoculum grown in LB and 6 mice infected with ∼9 log10 CFU from an inoculum grown in MM pH 7.7) were tested by ELISA for the presence of anti-Vi antibodies at 28 days p.i. Both i.v. and oral immunization with PssaG::tviA GFP+ induced anti-Vi serum antibody (Fig. 7). We then proceeded to test the IgG isotype profile of the anti-Vi response. Sera from the mice infected via the same route were pooled together and were found to contain anti-Vi antibodies of all subclasses (IgG1, IgG2a, IgG2b, and IgG3) (data not shown) as an indication of the presence of a T-cell-dependent IgG seroconversion of the humoral response (14). Thus, using an appropriate inducible promoter such as PssaG, it is possible to induce an anti-Vi antibody response after single i.v. or oral administration of live S. enterica.
Fig. 7.
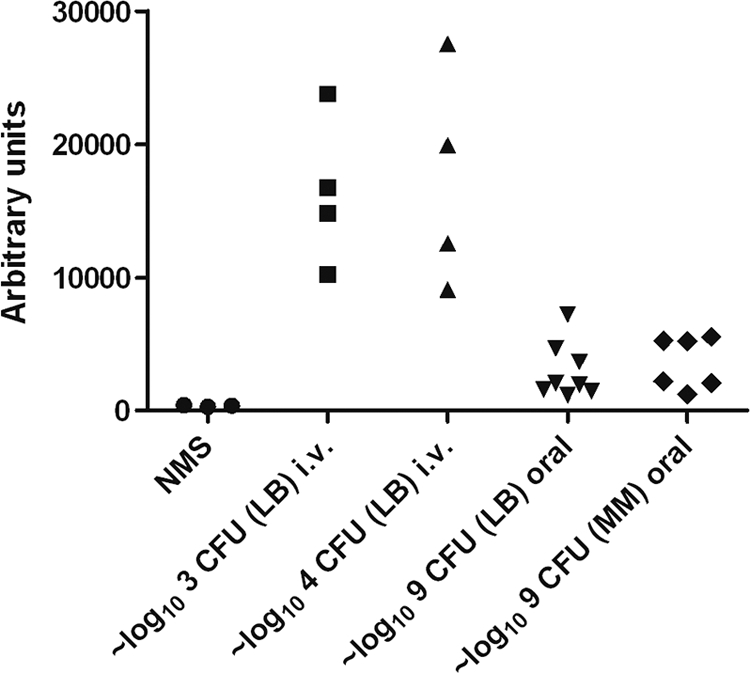
Determination of anti-Vi antibodies by ELISA at 28 days p.i. following i.v. infection of groups of 4 mice with either ∼3 log10 CFU or ∼4 log10 CFU of PssaG::tviA GFP+ and at 28 days p.i. following oral infection of 8 mice with ∼9 log10 CFU of PssaG::tviA GFP+ grown in LB and of 6 mice with ∼9 log10 CFU of PssaG::tviA GFP+ grown in MM pH 7.7. Three batches of naïve mouse serum (NMS) were used as negative controls. The results are shown in arbitrary units. The arbitrary unit value for each serum represents the reciprocal of the serum dilution that would give an OD reading of 0.1 using a best-fit linear regression curve.
DISCUSSION
Our results show that Vi expression is regulated in vivo and is suppressed in the liver and spleen with the progression of infection. We have also shown that by replacing the tviA promoter with the promoter of the ssaG gene, it is possible to achieve sustained expression of Vi in the tissues and to trigger anti-Vi IgG responses in the sera of mice infected intravenously or orally with this strain.
Vi regulation in vivo is likely to be complex, and bacteria need to maintain an optimal level of Vi expression in each phase of the infection in order to maximize infectivity and to avoid immune responses. Osmolarity is one of the recognized regulators of Vi. Vi is downregulated in the high-osmolarity gut environment, and this is likely to be a mechanism used by bacteria to minimize hindrance of the T3SS 1 that contributes to invasion via M cells in the gut (13). After the invasion process has been completed, the bacteria reach the lymph and bloodstream, where they would normally be opsonized by complement and directed to complement receptors on the surfaces of spleen and liver phagocytes. At this stage, Vi would be beneficial to the bacteria due to its known ability to limit complement deposition at the bacterial surface. Indeed, Vi expression is upregulated by exposure to/growth in serum (our unpublished data). Once the bacteria are cleared from the blood, they reach an intracellular location within phagocytes. The low osmolarity of the intracellular compartment is permissive to Vi expression, and thus one would expect detectable levels of Vi on the surfaces of intracellular bacteria if osmolarity was the prevailing signal in the regulation of Vi within phagocytes. Conversely, we found that Vi expression was suppressed in the intracellular compartment of the liver and spleen with the progression of the infection and that by day 4 of a parenteral infection, the majority of bacteria did not express detectable levels of Vi. This indicates that the regulation of Vi expression within phagocytes is complex and is likely to be affected by signals other than osmolarity. The lack of sustained expression of Vi in vivo is also consistent with the observation that Vi antibody titers are usually low or absent in the majority of acutely infected typhoid patients (3), suggesting that downregulation of Vi contributes to Salmonella immunoevasion of the antibody response.
We have shown that the natural downregulation of Vi can be overcome by using the ssaG promoter, which is inducible inside phagocytes. This allows sustained expression in the tissues and does not affect the ability of bacteria to colonize the spleen and liver after oral infection. However, the PssaG::tviA GFP+ strain that we used in this study (and other clones with similar chromosomal insertions) appeared to be attenuated in its growth in the spleen and liver. It is plausible to attribute this to the fact that intracellular Vi expression may hinder secretion of SPI-2 effectors that play a role in Salmonella growth within the phagosome (13). Expression of TviA has been shown to increase systemic infectivity of S. Typhimurium in a chicken model (39). Conversely, in the current study, we did not observe increased systemic colonization of C5.507. This is likely due to differences in the animal species used and, more specifically, to the fact that S. Typhimurium can overcome the mucosal barriers of mice and chickens with different dynamics.
We found that intravenous or oral administration of the PssaG::tviA GFP+ strain to mice could induce an IgG antibody response. The response was smaller in animals immunized orally than in those immunized i.v. This is not unusual for immunization with live salmonellae (18) and could be due to the lower systemic bacterial loads that are usually reached after oral vaccination. The response contained all IgG subclasses, including IgG2a, which we have shown in the past to be the most efficient at mediating uptake and intracellular killing of Salmonella by phagocytes via FcRI (34). The presence of a class-switched IgG response to Vi is similar to what we previously observed in analyzing the immunoglobulin response to LPS in mice immunized with live and killed vaccines, whereby a T-cell-dependent response to the LPS polysaccharide antigen was seen when the animals received live vaccination (14). This work therefore provides proof of principle that it is possible to elicit class-switched IgG responses to Vi via single oral immunization with live bacteria that express Vi under the control of an in vivo-inducible promoter.
ACKNOWLEDGMENTS
We thank Isabelle Hautefort and Jay Hinton for JH3010, Sarah Peters for SL1344 degP::Gm, Helen Northern for pBADTOPOKanFRT and pCP20, and Novartis Vaccine Institute for Global Health (NVGH) for supply of purified Vi.
This work was supported by a Wellcome Trust grant (081743/Z/06/Z) awarded to P.M. and A.J.G. A.J.G. and F.J.E.M. are funded by the Medical Research Council (grant G0801161). T.J.M. is supported by the Department for the Environment, Food and Rural Affairs/Higher Education Funding Council of England (grant VT0105).
The authors have no conflicting financial interests.
Footnotes
Published ahead of print on 14 March 2011.
REFERENCES
- 1. Acharya I. L., et al. 1987. Prevention of typhoid fever in Nepal with the Vi capsular polysaccharide of Salmonella Typhi. A preliminary report. N. Engl. J. Med. 317:1101–1104 [DOI] [PubMed] [Google Scholar]
- 2. Arricau N., et al. 1998. The RcsB-RcsC regulatory system of Salmonella Typhi differentially modulates the expression of invasion proteins, flagellin and Vi antigen in response to osmolarity. Mol. Microbiol. 29:835–850 [DOI] [PubMed] [Google Scholar]
- 3. Baker S., Favorov M., Dougan G. 2010. Searching for the elusive typhoid diagnostic. BMC Infect. Dis. 10:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cherepanov P. P., Wackernagel W. 1995. Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158:9–14 [DOI] [PubMed] [Google Scholar]
- 5. Crump J. A., Mintz E. Z. 2010. Global trends in typhoid and paratyphoid fever. Clin. Infect. Dis. 50:241–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cryz S. J., Jr., et al. 1989. Construction and characterization of a Vi-positive variant of the Salmonella Typhi live oral vaccine strain Ty21a. Infect. Immun. 57:3863–3868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Daniels D. L., Schneerson R., Egan W. M., Szu S. C., Robbins J. B. 1989. Characterization of the Salmonella Paratyphi C Vi polysaccharide. Infect. Immun. 57:3159–3164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Datsenko K. A., Wanner B. L. 2000. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc. Natl. Acad. Sci. U. S. A. 97:6640–6645 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Dower W. J., Miller J. F., Ragsdale C. W. 1988. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 16:6127–6145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Faucher S. P., Porwollick S., Dozois C. M., McClelland M., Daigle F. 2006. Transcriptome of Salmonella enterica serovar Typhi within macrophages revealed through the selective capture of transcribed sequences. Proc. Natl. Acad. Sci. U. S. A. 103:1906–1911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Guzman C. A., et al. 2006. Vaccines against typhoid fever. Vaccine 24:3804–3811 [DOI] [PubMed] [Google Scholar]
- 12. Hale C., et al. 2006. Evaluation of a novel Vi conjugate vaccine in a murine model of salmonellosis. Vaccine 24:4312–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Haragga A., Ohlson M. B., Miller S. I. 2008. Salmonellae interplay with host cells. Nat. Rev. Microbiol. 6:53–66 [DOI] [PubMed] [Google Scholar]
- 14. Harrison J. A., Villarreal-Ramos B., Mastroeni P., Cemarco de Hormaeche R., Hormaeche C. E. 1997. Correlates of protection induced by live Aro− Salmonella typhimurium vaccines in the murine typhoid model. Immunology 90:618–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hautefort I., Proença M. J., Hinton J. C. D. 2003. Single-copy green fluorescent protein gene fusions allow accurate measurement of Salmonella gene expression in vitro and during infection of mammalian cells. Appl. Environ. Microbiol. 69:7480–7491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hirose K., et al. 1997. Survival of Vi-capsulated and Vi-deleted Salmonella typhi strains in cultured macrophage expressing different levels of CD14 antigen. FEMS Microbiol. Lett. 147:259–265 [DOI] [PubMed] [Google Scholar]
- 17. Hothorn T., Bretz F., Westfall P. 2008. Simultaneous inference in general parametric models. Biom. J. 50:346–363 [DOI] [PubMed] [Google Scholar]
- 18. Khan C. M., et al. 1994. Construction, expression, and immunogenicity of the Schistosoma mansoni P28 glutathione S-transferase as a genetic fusion to tetanus toxin fragment C in a live Aro attenuated vaccine strain of Salmonella. Proc. Natl. Acad. Sci. U. S. A. 91:11261–11265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Kopecko D. J., et al. 2009. Genetic stability of vaccine strain Salmonella Typhi Ty21a over 25 years. Int. J. Med. Microbiol. 299:233–246 [DOI] [PubMed] [Google Scholar]
- 20. Kossaczka Z., et al. 1999. Safety and immunogenicity of Vi conjugate vaccines for typhoid fever in adults, teenagers, and 2- to 4-year-old children in Vietnam. Infect. Immun. 67:5806–5810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kox L. F., Wosten M. M., Groisman E. A. 2000. A small protein that mediates the activation of a two-component system by another two-component system. EMBO J. 19:1861–1872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Lin F. Y., et al. 2001. The efficacy of a Salmonella typhi Vi conjugate vaccine in two-to-five-year-old children. N. Engl. J. Med. 344:1263–1269 [DOI] [PubMed] [Google Scholar]
- 23. McKelvie N. D., et al. 2004. Expression of heterologous antigens in Salmonella Typhimurium vaccine vectors using the in vivo-inducible, SPI-2 promoter, ssaG. Vaccine 22:3243–3255 [DOI] [PubMed] [Google Scholar]
- 24. Mo E., Peters S. E., Willers C., Maskell D. J., Charles I. G. 2006. Single, double and triple mutants of Salmonella enterica serovar Typhimurium degP (htrA), degQ (hhoA) and degS (hhoB) have diverse phenotypes on exposure to elevated temperature and their growth in vivo is attenuated to different extents. Microb. Pathog. 41:174–182 [DOI] [PubMed] [Google Scholar]
- 25. Pickard D., et al. 1994. Characterization of defined ompR mutants of Salmonella Typhi: ompR is involved in the regulation of Vi polysaccharide expression. Infect. Immun. 62:3984–3993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rafatellu M., et al. 2005. The Vi capsule antigen of Salmonella enterica serotype Typhi reduces Toll-like receptor-dependent interleukin-8 expression in the intestinal mucosa. Infect. Immun. 73:3367–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. R Development Core Team 2008. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria: http://www.R-project.org [Google Scholar]
- 28. Robbins J. D., Robbins J. B. 1984. Reexamination of the protective role of the capsular polysaccharide (Vi antigen) of Salmonella typhi. J. Infect. Dis. 150:436–449 [DOI] [PubMed] [Google Scholar]
- 29. Sambrook J., Russell D. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 30. Tacket C. O., et al. 1991. Lack of immune response to the Vi component of a Vi-positive variant of the Salmonella typhi live oral vaccine strain Ty21a in human studies. J. Infect. Dis. 163:901–904 [DOI] [PubMed] [Google Scholar]
- 31. Tacket C. O., Pasetti M. F., Sztein M. B., Livio S., Levine M. M. 2004. Immune responses to an oral typhoid vaccine strain that is modified to constitutively express Vi capsular polysaccharide. J. Infect. Dis. 190:565–570 [DOI] [PubMed] [Google Scholar]
- 32. Tacket C. O., Levine M. M. 2007. CVD 908, CVD 908-htrA, and CVD 909 live oral typhoid vaccines: a logical progression. Clin. Infect. Dis. 45:S20–S23 [DOI] [PubMed] [Google Scholar]
- 33. Tran Q. T., et al. 2010. The Salmonella enterica serotype Typhi Vi capsular antigen is expressed after the bacterium enters the ileal mucosa. Infect. Immun. 78:527–535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Uppington H., et al. 2006. Effect of immune serum and role of individual Fcgamma receptors on the intracellular distribution and survival of Salmonella enterica serovar Typhimurium in murine macrophages. Immunology 119:147–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Virlogeux I., Waxin H., Ecobichon C., Popoff M. 1995. Role of the viaB locus in synthesis, transport and expression of Salmonella Typhi Vi antigen. Microbiology 141:3039–3047 [DOI] [PubMed] [Google Scholar]
- 36. Waxin H., Virlogeux I., Kolyva S., Popoff M. Y. 1993. Identification of six open reading frames in the Salmonella enterica subsp. enterica ser. Typhi viaB locus involved in Vi antigen production. Res. Microbiol. 144:363–371 [DOI] [PubMed] [Google Scholar]
- 37. Wilson R. P., et al. 2008. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell. Microbiol. 10:876–890 [DOI] [PubMed] [Google Scholar]
- 38. Winter S. E., et al. 2009. The TviA auxiliary protein renders the Salmonella enterica serotype Typhi RcsB regulon responsive to changes in osmolarity. Mol. Microbiol. 74:175–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Winter S. E., et al. 2010. A rapid change in virulence gene expression during the transition from the intestinal lumen into tissue promotes systemic dissemination of Salmonella. PLoS Pathog. 6:e1001060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhao L., et al. 2001. Vi-suppressed wild strain Salmonella Typhi cultured in high osmolarity is hyperinvasive toward epithelial cells and destructive of Peyer's patches. Microbiol. Immunol. 45:149–158 [DOI] [PubMed] [Google Scholar]