Abstract
Intracellular bacterial pathogens manipulate host cell functions by producing enzymes that stimulate or antagonize signal transduction. The Listeria monocytogenes genome contains a gene, lmo1800, encoding a protein with a conserved motif of conventional tyrosine phosphatases. Here, we report that the lmo1800-encoded protein LipA is secreted by Listeria and displays tyrosine as well as lipid phosphatase activity in vitro. Bacteria lacking LipA are severely attenuated in virulence in vivo, thus revealing a so-far-undescribed enzymatic activity involved in Listeria infection.
INTRODUCTION
Listeria monocytogenes is a Gram-positive facultative intracellular pathogen and the causative agent of listeriosis. This food-borne disease is rare but can cause clinically serious symptoms with a high case fatality rate in immunocompromised individuals (37). After ingestion of contaminated food, bacteria cross the intestinal barrier and disseminate via blood and lymph nodes to the spleen and liver and the main target organs, the brain and the placenta. Listeria can either induce their uptake into nonphagocytic cells or infect macrophages via phagocytosis. Once ingested, bacteria evade the hostile environment of the endosomal or phagosomal compartment by lysis of the vacuole and escape to the cytoplasm, where they rapidly multiply and move within the cell by exploiting the actin cytoskeleton of the host cell. The actin polymerization also allows Listeria to spread into neighboring cells by forming a protrusion that is internalized in a double-membrane vacuole which is ultimately lysed, releasing bacteria in the cytosol (46, 56). These processes require the expression of a set of virulence genes, many of which are under the control of the transcription factor PrfA (positive regulatory factor A). Six of the PrfA-regulated genes are clustered on Listeria pathogenicity island 1 (13, 21, 50). Some genes, such as inlA, inlB, inlC, mpl, and hly, can be expressed from both PrfA-dependent and PrfA-independent promoters (20, 32–34). Other genes expressed independently of PrfA, such as iap and aut, also contribute to Listeria virulence (11, 32).
Phosphorylation of proteins as well as inositides is a common means of regulating cellular pathways in eukaryotic cells. However, it was not until the mid-1990s that bacterial interference with host protein and lipid metabolism by secretion of phosphatases was discovered (27). The secretion of kinases or phosphatases enables pathogens to directly interfere with host signal transduction and is employed by bacteria such as Mycobacterium, Salmonella, Shigella, and Yersinia. These bacteria target host pathways regulating survival, vesicle movement, or cytoskeletal rearrangements (10, 16, 24, 28, 39, 44, 45, 57).
The first phosphatase identified in L. monocytogenes is Stp, which regulates bacterial translation by acting on EF-Tu. Additionally, the enzyme controls the activity of L. monocytogenes superoxide dismutase. Accordingly, Stp adapts the bacteria to the stress imposed by their mammalian host, thus increasing intracellular multiplication and virulence (2, 3). In contrast, L. monocytogenes genes for tyrosine or lipid phosphatases have not been identified. Here we characterize the protein Listeria phosphatase A (LipA), encoded by the lmo1800 open reading frame of the Listeria monocytogenes genome. We show that this protein displays phosphatase activity on both protein and lipid substrates and plays a major role in virulence in mice.
MATERIALS AND METHODS
Bacterial strains, media, and growth conditions.
L. monocytogenes EGDe (serovar 1/2a, BUG1600) (26) and L. monocytogenes LO28 (29) were used as parental strains. L. monocytogenes EGDe ΔprfA (BUG2214), ΔsigB (BUG2215), ΔplcAB (17), and ΔlipA (BUG1959) strains as well as the L. monocytogenes LO28 ΔlipA strain were the isogenic deletion mutant strains used in this study. The last strain was used as a parental strain to construct the complemented L. monocytogenes LO28 ΔlipA::lipA strain. L. monocytogenes strains were routinely grown in brain heart infusion (BHI) broth (Difco) at 37°C and 200 rpm or on Oxford agar plates (Merck) at 37°C. When appropriate, L. monocytogenes strains were grown in a chemically defined improved minimal medium (IMM) described previously (43) or in LB medium. Growth conditions for L. monocytogenes gene expression analysis were described in our previous study (54). Escherichia coli DH5α, XL1-BlueMRF′, or Top10 was used as the host for plasmid constructions. E. coli strains were grown in LB medium at 37°C with shaking. When required, LB medium for growth of E. coli was supplemented with antibiotics to the following final concentrations: ampicillin (Amp), 100 μg/ml; chloramphenicol (Chl), 15 μg/ml; tetracycline (Tet), 15 μg/ml; erythromycin (Ery), 500 μg/ml; and kanamycin (Kan), 25 μg/ml. When required, BHI medium for growth of L. monocytogenes was supplemented with chloramphenicol at 3 μg/ml, erythromycin at 3 μg/ml, and kanamycin at 300 μg/ml.
Sensitivity to hydrogen peroxide, plumbagin, hydrochloric acid, or sodium chloride.
L. monocytogenes LO28 strains were grown overnight in BHI broth at 37°C and used to inoculate BHI at pH 5.5 (set with HCl) or BHI containing 5% NaCl at an optical density at 600 nm (OD600) of 0.05, and growth was monitored until stationary phase. L. monocytogenes EGDe strains were grown overnight in BHI broth at 37°C and used to inoculate BHI at an OD600 of 0.02. When the OD600 reached 0.5 to 1, 2 mM hydrogen peroxide or 15 μM plumbagin or hydrochloric acid was added to the cultures to set the pH at 6.5. Growth was monitored until stationary phase. Experiments were repeated at least twice independently.
Overexpression and purification of LipA.
For high-level expression of an N-terminally His6-tagged version, the lipA gene was cloned into pQE31 (Qiagen). PCR with genomic DNA of L. monocytogenes (LO28) was performed using primers lipA1800_5′ and lipA1800_3′, including the stop codon of lipA (see Table S1 in the supplemental material). After restriction with the appropriate enzymes, the PCR fragment was inserted into pQE31, resulting in plasmid pQE31-LipA. pQE31-LipA was transformed into E. coli XL1-BlueMRF′, creating the strain XL1-BlueMRF′(pQE31-LipA).
XL1-BlueMRF′(pQE31-LipA) bacteria were grown overnight in LB supplemented with Amp and Tet and reinoculated 1:100 in 1 ml LB with Amp and Tet the following day. At an OD600 of 0.5, LipA overexpression was induced by addition of 1 mM IPTG (isopropyl-β-d-thiogalactopyranoside). Bacteria were centrifuged for 15 min at 8,000 rpm and washed once with purification buffer. Purification with Ni-nitrilotriacetic acid (Ni-NTA) beads was performed according to the manufacturer's protocol.
Phosphatase activity assay.
To measure the amount of free phosphate released during dephosphorylation assays, a malachite green phosphate assay kit (Cayman Chemicals) was used with a range of substrates: diC8-PtdIns(3)P, diC8-PtdIns(4)P, diC8-PtdIns(5)P, diC8-PtdIns(3,4)P2, diC8-PtdIns(3,5)P2, diC8-PtdIns(4,5)P2, diC8-PtdIns(3,4,5)P3 (Echelon), O-phospho-dl-serine, O-phospho-dl-threonine, and O-phospho-l-tyrosine (Sigma). To measure phosphatase activity, 0.02 μg of purified His6-LipA or calf intestinal alkaline phosphatase (CIAP; Promega) was incubated with 20 mM phosphorylated amino acids or 25 μM phosphatidylinositol phosphate substrates for 10 min at 30°C.
For measurement of phosphatase activity in bacterial extracts, 10-ml stationary-phase cultures of L. monocytogenes LO28 wild-type (wt), ΔlipA, and ΔlipA::lipA strains were washed with 50 mM bis-Tris (pH 6.0) and incubated for 20 min at 4°C in the same buffer supplemented with 10 mg/ml of lysozyme and 1 unit of DNase I. Cells were sonicated and centrifuged for 30 min at 20,000 × g at 4°C. Supernatants (90 μl) were incubated with 185 μg of p-nitrophenylphosphate (pNPP) and incubated at 37°C for 90 min. The reaction was stopped by addition of 50 μl 4 N NaOH, and OD595 was measured. PNP release was normalized to the amount of total protein.
Expression of the GFP fusion protein.
For the expression of a LipA-green fluorescent protein (LipA-GFP) fusion, an in-frame fusion of lipA with the gfp gene was expressed under the control of the strong Listeria promoter Pdlt in the pAM401 backbone (23). To amplify the Pdlt promoter as described in reference 1, primers PDLT5 and PDLT3, adapted from oligonucleotides EA20 and EA21, respectively (see Table S1 in the supplemental material), were used. The lipA gene, including the ribosome binding site (RBS), was amplified by PCR from genomic L. monocytogenes LO28 DNA, using oligonucleotides RBS59-5 and RBSFULL-3 (see Table S1). The GFPmut2 gene was amplified using oligonucleotides GFP2-5 and GFP2-3 (see Table S1). Fragments were ligated and inserted into pAM401, resulting in plasmid pAM401-LipA-GFP. As a control, GFP was cloned into pAM401, controlled by the same promoter region. The resulting plasmid was designated pAM401-GFP.
Fractionation of bacterial cells.
Stationary-phase cultures of L. monocytogenes LO28(pAM401), LO28(pAM401-GFP), or LO28(pAM401-LipA-GFP) were grown overnight. The same numbers of bacteria of each strain (4 × 1010) were centrifuged at 6,000 × g for 15 min. On one hand, the supernatant was passed through a 0.22-μm filter, and protein was precipitated using 0.5% trichloroacetic acid (TCA) and 0.02% sodium deoxycholate (DOC) and washed with acetone. The precipitated proteins were resuspended in SDS sample buffer. On the other hand, the bacterial pellet was washed with phosphate-buffered saline (PBS), resuspended in PBS adjusted to a final concentration of 10 mM vanadate, 1 mM dithiothreitol (DTT), and 1× proteinase inhibitor mix (Roche Diagnostics), and lysed by sonication. The lysate was centrifuged at 15,000 × g for 30 min at 4°C. Both the supernatant, containing cytosolic protein, and the pellet, containing membrane proteins, were resuspended in SDS sample buffer and used for Western blot analysis.
Mutagenesis of L. monocytogenes.
The temperature-sensitive plasmid pEC64 was used to delete lipA from the L. monocytogenes LO28 genome. Plasmid pEC64 was kindly provided by Emmanuelle Charpentier (Umea University, Sweden). A 1,002-bp region located upstream of the lipA gene and a 1,001-bp region located downstream of the lipA gene were amplified by PCR using primers KOUP1 and KOUP2 and primers KODW1 and KODW2, respectively, from L. monocytogenes genomic DNA (see Table S1 in the supplemental material). After restriction with the appropriate enzymes, the two PCR fragments were ligated and cloned into the vector pEC64, resulting in plasmid pEC64-koLipA. The resulting plasmid was introduced into L. monocytogenes LO28 by electroporation (1,500 V, 400 Ω, and 25 μF), and chromosomal integration of the plasmid was achieved by growing the bacteria at 37°C in the presence of kanamycin. Excision of plasmid sequences occurred after repeated passage of bacteria at the replication-permissive temperature 28°C in the absence of kanamycin. The chromosomal deletion of the lipA gene in L. monocytogenes LO28, resulting in the L. monocytogenes LO28 ΔlipA strain, was verified by PCR. For mutagenesis of L. monocytogenes EGDe, 992-bp upstream and 971-bp downstream DNA regions flanking the lipA gene were amplified by PCR using genomic DNA from L. monocytogenes EGDe and oligonucleotides U155/L156 and U157/L158, respectively (see Table S1). The upstream BamHI-MluI and downstream MluI-BglII fragments were cloned sequentially into the thermosensitive vector pMAD (4), resulting in plasmid pOD57, which was maintained in E. coli TOP10, and creating the strain OD103. The pOD57 plasmid was electroporated into L. monocytogenes EGDe at 2,500 V, 250 Ω, and 25 μF. Deletion of the lipA gene by allelic exchange was carried out as previously described (4). Deletion mutants were identified by PCR with white colonies plated on BHI–X-Gal (BHI–5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside) agar using oligonucleotides U185 and L186 (see Table S1). The lipA deletion was analyzed by Southern blotting according to standard protocols. Briefly, genomic DNAs from L. monocytogenes EGDe wild-type and ΔlipA strains were digested by EcoRI. Fragments were separated on a 1% agarose gel and transferred onto a Hybond N+ nylon membrane (Amersham). The probe corresponding to the lipA gene was 5′ end labeled with [α-32P]dCTP using the Klenow fragment and a Megaprime DNA labeling system (Amersham). The membrane was incubated with the radiolabeled probe in Rapid-hyb buffer (Amersham) at 65°C overnight. Fragments that hybridized specifically with the radiolabeled probe were detected by autoradiography.
To complement the L. monocytogenes LO28 ΔlipA strain, the site-specific integrative bacteriophage vector pIMK2 (38) was used. The lipA gene was amplified from L. monocytogenes LO28 genomic DNA using primers LipA5′ and LipA3′ (see Table S1 in the supplemental material). After restriction with the appropriate enzymes, the PCR fragment was cloned into the overexpression vector pIMK2, resulting in plasmid pIMK2-LipA. Here, pIMK2-LipA was introduced into the L. monocytogenes LO28 ΔlipA strain by electroporation (1,500 V, 400 Ω, and 25 μF), followed by static incubation at 37°C for 6 h in BHI. Regenerated cells were plated on BHI agar containing kanamycin. The chromosomal integration of pIMK2, resulting in the Listeria monocytogenes LO28 ΔlipA::lipA strain, was verified by PCR with genomic DNA.
Mammalian cell culture.
RAW 264.7 macrophages, CMT-93 colon carcinoma cells, and L2 fibroblasts were cultured in Dulbecco's modified Eagle's medium (DMEM; Gibco, Invitrogen) supplemented with 10% fetal calf serum (FCS; Gibco, Invitrogen). Ptk-2 kangaroo rat kidney cells were cultured in MEM (Gibco, Invitrogen) supplemented with 20% FCS (Gibco, Invitrogen) and 1% l-glutamine–GlutaMAX-I (Gibco). Bone marrow-derived macrophages (BMMs) were obtained by culture of bone marrow of 7- to 10-week-old C57BL/6 mice and cultured in DMEM (Gibco, Invitrogen) supplemented with 10% FCS (Gibco, Invitrogen) and L929 fibroblast-derived colony-stimulating factor 1 (CSF-1) as described previously (5). The murine macrophage-like cell line J774 was cultured in DMEM (Gibco) supplemented with 4 mM l-glutamine (Gibco), 1 mM sodium pyruvate (Gibco), and 10% FCS (Biovalley). Cells were cultured at 37°C in a 5% CO2 atmosphere.
Infection of cells.
Cells were seeded in antibiotic-free medium and grown overnight. The OD600 of Listeria overnight cultures was measured, and the number of bacteria was determined by use of the following equation: OD600 of 1 = 2 × 109 bacteria/ml. Cells were infected with L. monocytogenes bacteria at the indicated multiplicity of infection (MOI). Using BMMs or J774 macrophage-like cells, extracellular bacteria were killed 1 h after infection by addition of gentamicin-containing medium (final concentration, 50 μg/ml). An additional hour later, the concentration of gentamicin in the medium was changed to 10 μg/ml. Two hours after infection of CMT-93, L2, or Ptk-2 cells, the medium was changed to medium containing 50 μg/ml gentamicin and 1 h later to medium containing 10 μg/ml gentamicin.
Determination of CFU in vitro.
Cells were infected as described above. At different time points, extracellular bacteria were removed by washing twice with PBS, and cells were lysed with distilled water. Serial dilutions of lysates were plated on BHI agar plates, and numbers of intracellular bacteria were determined by counting CFU.
Localization of Akt-PH-GFP in infected Ptk-2 cells.
Localization of Akt-pleckstrin homology-GFP (Akt-PH-GFP) in infected Ptk-2 cells was performed as described previously (51). Ptk-2 cells (3 × 106) were seeded on coverslips in six-well dishes and incubated for 40 to 48 h. Cells were transfected with 1.2 μg plasmid encoding Akt-PH-GFP, using 10 μg/ml Lipofectamine reagent (Invitrogen) and 11 μg/ml Plus reagent (Invitrogen) for 6 h. Forty-eight hours after transfection, cells were infected. Six hours after infection, cells were fixed with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100 in PBS. Bacteria were stained with rabbit antiserum to L. monocytogenes (Abcam) in antibody diluent (Dako, Glostrup, Denmark) for 1 h at room temperature. A secondary Alexa Fluor 594 chicken anti-rabbit IgG antibody (Molecular Probes) was applied for 1 h. Coverslips were mounted on slides using fluorescent mounting medium (Dako, Glostrup, Denmark).
Bacterial uptake and phagosomal escape assays.
Bacterial uptake and phagosomal escape assays were performed as described previously (47). For measuring bacterial uptake, Listeria bacteria were stained with 10 μM carboxyfluorescein diacetate succinimidyl ester (CFSE) in PBS for 20 min at 37°C. At the indicated time point, cells were harvested in prewarmed citrate buffer (135 mM KCl, 15 mM sodium citrate), washed with PBS, and resuspended in fluorescein isothiocyanate (FACS) buffer (0.2% bovine serum albumin [BSA], 0.1% sodium azide in PBS). Extracellular signals were quenched by addition of trypan blue solution (1 mg/ml in citrate buffer, pH 4).
For determining phagosomal escape, 3 × 105 RAW 264.7 cells were seeded on coverslips and infected with CFSE-labeled bacteria at an MOI of 10. Three hours after infection, cells were washed twice with PBS and fixed with 4% paraformaldehyde. After two PBS washing steps, cells were permeabilized with 0.2% Triton X-100 in PBS. Cells were again washed twice with PBS and incubated in 6.6 μM phalloidin-Alexa Fluor 594 (Molecular Probes, Invitrogen) in PBS for 40 min at room temperature. After two PBS washing steps, cells were counterstained with DAPI (4′,6-diamidino-2-phenylindole) (final concentration, 250 ng/ml).
Plaque assay.
A plaque assay with the L2 mouse fibroblast cell line was performed as described previously (36). In brief, 3 × 106 L2 cells were seeded in six-well plates and incubated for 40 to 48 h. Cells were infected at an MOI of 0.5, and 2 h after infection, the cells were washed three times with sterile PBS and overlaid with L2 cell overlay (2× DMEM containing 20% FCS and 20 μg/ml gentamicin, mixed 1:1 with 1.4% agarose). At 3 days postinfection, plaques were visualized by overlay with L2 staining overlay (L2 cell overlay, containing 0.35% 1 N HCl and 6% neutral red solution [Sigma]). Plates were scanned with AlphaImager (Bio-Rad) using white transillumination light. The plaque diameter (in pixels) was measured using the measure tool of GIMP 2.6 software. Due to the irregular shape of some plaques, the diameter was taken from left to right as well as from top to bottom, and the mean diameter was used for calculating the relative plaque area.
Western blot analysis.
Western blot analysis was performed by following the method described in reference 31, modified using fluorophore-linked secondary antibodies detected by the Odyssey infrared imaging system (Li-Cor Biosciences, Lincoln, NE). All of the following primary antibodies were stored in PBS supplemented with 2% BSA and 0.05% sodium azide: rabbit polyclonal listeriolysin O (LLO) antibody (1:1,500; Diatheva), mouse monoclonal GFP (B-2) antibody (1:200; Santa Cruz), mouse monoclonal pan-extracellular signal-regulated kinase (panERK) antibody (1:2,000; Becton Dickinson), rabbit polyclonal phospho-AKT (Ser473) antibody (1:1,000; Cell Signaling), and phospho-Stat1 (Tyr701) antibody (1:1,000; Cell Signaling). Antibody recognizing the bacterial ribosomal protein S9 was kindly provided by Isabella Moll (Max F. Perutz Laboratories, University of Vienna, Austria). The following secondary antibodies were diluted 1:20,000 in PBS: IRDye800-conjugated anti-rabbit IgG (Rockland), IRDye800-conjugated anti-mouse IgG (Rockland), IRDye700-conjugated anti-rabbit IgG (Rockland), and Alexa Fluor 680 anti-mouse IgG (Molecular Probes).
Animal studies.
Animal experiments were discussed and approved by the University of Veterinary Medicine Vienna institutional ethics committee and carried out in accordance with protocols approved by the Austrian law (GZ 680 205/67-BrGt/2003). Bacteria were prepared for infection as described previously (52). For infection with the L. monocytogenes LO28, ΔlipA, or ΔlipA::lipA strain, bacteria were washed with PBS (endotoxin free; Sigma) and injected into the peritoneum of 8- to 10-week-old C57BL/6 mice (Charles River). Alternatively, mice were infected with 1 × 109 CFU via intragastric gavage. The infectious dose was controlled by plating serial dilutions on Oxford agar plates. The survival of mice was monitored for 10 days, and data were displayed as Kaplan-Meier plots. For determination of bacterial loads of liver, spleen, and intestine, mice were killed at the indicated time points. The respective organs were isolated and homogenized in PBS (Polytron system PT 2100 and dispersing aggregate PT-DA 12/2 EC-D; Kinematica, Switzerland). Serial dilutions of the homogenates were plated on Oxford agar plates or BHI plates and incubated at 37°C for 48 h. Intestines were thoroughly flushed and washed with PBS before homogenization. L. monocytogenes EGDe growth in vivo was studied after intravenous injection of 8 × 103 CFU into 8-week-old female BALB/c mice (Charles River). At the indicated time points, the liver and spleen were dissected aseptically and the number of CFU was determined by plating serial dilutions of organ homogenates on BHI agar.
ELISA.
Blood was collected from the retro-orbital sinus of mice. Blood was incubated at room temperature for approximately 10 to 15 min to allow coagulation, followed by centrifugation for 10 min at 10,000 × g. Serum cytokine levels were measured by enzyme-linked immunosorbent assay (ELISA) using mouse tumor necrosis factor alpha (TNF-α) DuoSet, mouse interleukin-6 (IL-6) DuoSet, or mouse gamma interferon (IFN-γ) DuoSet by following the manufacturer's recommendations (R&D Systems).
RESULTS
LipA is homologous to known bacterial protein tyrosine phosphatases.
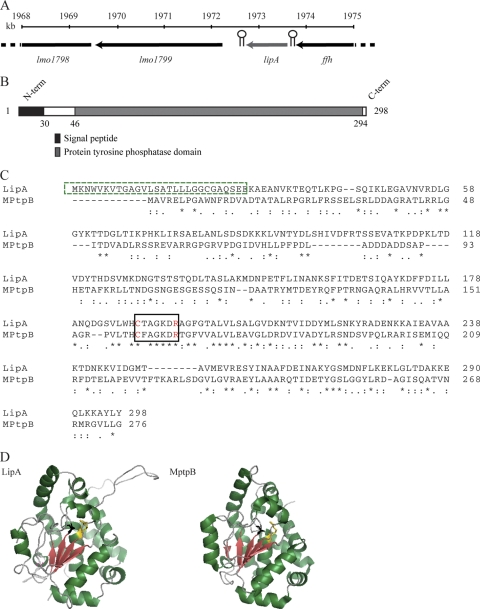
The genome of L. monocytogenes EGDe encodes the ORF lmo1800, with a putative protein tyrosine phosphatase (PTP) domain (COG2365) (53). The lmo1800 gene, from now on designated lipA (Listeria phosphatase A), is located at positions 1872724 to 1873620 of the chromosome and is surrounded by ffh, encoding the signal recognition particle protein, and lmo1799, a gene encoding a putative LPXTG protein (Fig. 1 A). Genes orthologous to lipA are present in the genomes of Listeria innocua (lin1914 encodes a protein with 92% homology to LipA), Listeria welshimeri (lwe1819 encodes a protein with 86% homology to LipA), Listeria seeligeri (YP_003465015 encodes a protein with 88% identity to LipA), and Listeria grayi (ZP_04443093 encodes a protein with 31% identity to LipA). Orthologs of LipA are present in the genomes of other bacteria, such as those of Lactobacillus brevis (42% identity), Bacillus cereus and Bacillus thuringiensis (36% identity), Enterococcus faecalis (33% identity), Clostridium difficile (25% identity), and Streptococcus pneumoniae (32% identity). Analysis of the LipA amino acid sequence using SignalP 3.0 software (http://www.cbs.dtu.dk/services/SignalP/) showed that the 298-amino-acid LipA protein contains a putative 30-amino-acid N-terminal signal sequence for secretion. Further examination revealed a C-terminal conserved domain (amino acids 46 to 294) belonging to the PTP superfamily (COG2365) (Fig. 1B and C). Amino acid sequence alignment with another homologous phosphatase known to be secreted, MPtpB from Mycobacterium tuberculosis, shows several conserved motifs (Fig. 1C) (8, 35). The C-terminal HCXXGKDRXG stretch corresponds to the distinctive active site CX5R signature motif of tyrosine phosphatases (53). The conformation of this motif, known as the P loop or PTP loop, is highly conserved and maintains close proximity between the catalytic cysteine and the arginine, which is important for phosphate binding. The predicted protein structure of LipA (http://toolkit.tuebingen.mpg.de/hhpred) shows strong homology to the M. tuberculosis phosphatase MPtpB (Fig. 1D) (35). This enzyme dephosphorylates phosphoserine (P-Ser), phosphothreonine (P-Thr), and phosphotyrosine (P-Tyr) as well as phosphoinositides (8). It promotes the ability of M. tuberculosis to grow in macrophages (9).
Fig. 1.
L. monocytogenes LipA. (A) Genomic organization of the lipA gene region. Data from the complete genome sequence of L. monocytogenes EGDe were used to draw the map. Hairpins depict putative terminators. The ffh gene encodes the signal recognition particle, lmo1799 encodes a putative LPXTG protein with 226 Ala-Asp tandem repeats, and lmo1798 encodes a protein of unknown function. (B) Amino acid sequence characteristics of LipA. The 30-amino-acid N terminus contains a signal sequence for secretion or membrane insertion (black). A tyrosine phosphatase domain (COG2365) extends from amino acids 46 to 294 (dark gray). (C) Multiple alignment of L. monocytogenes LipA with the Mycobacterium tuberculosis phosphatase MPtpB (31). Asterisks, colons, or dots indicate amino acid residues that are identical, highly related, or only weakly related, respectively. The N-terminal box indicates the signal peptide (green dashed line). The C-terminal box indicates the conserved active site of tyrosine phosphatases (solid black line). Catalytic cysteine and phosphate binding arginine are shown in red. Protein sequences are for L. monocytogenes EGDe Lmo1800 (MP_465325.1) and M. tuberculosis MPtpB (P96830). (D) Predicted protein structure of LipA and published 3D structure of M. tuberculosis MPtpB (31). The P-loop cysteine is marked in black and aspartic acid in yellow.
LipA contains a functional export signal for secretion to the extracellular environment.
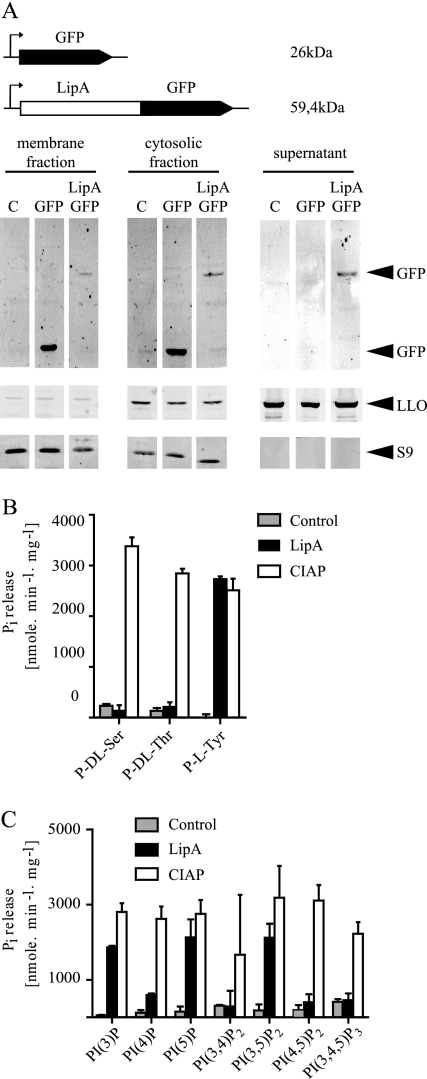
To investigate the relevance of the predicted signal sequence of LipA in vivo, we constructed a pAM401-based expression vector to fuse GFP to the C terminus of LipA. The same plasmid was also used to express GFP alone as a control. L. monocytogenes was transformed with plasmid pAM401(LipA-GFP) or pAM401(pAM40-GFP). The growth medium from overnight cultures was precipitated, and the cytoplasmic as well as the membrane fractions were prepared from the cell pellet. Anti-GFP Western blot analysis showed that the fusion protein was detectable in the cytoplasm, the cell membrane, and cellular supernatants, suggesting that LipA contains a functional export signal (Fig. 2 A). To control the purity of the cell fractions, antibodies to the secreted virulence factor listeriolysin O (LLO) and to the cytoplasmic ribosomal protein S9 were used. LLO showed a distribution highly similar to that of LipA-GFP, whereas S9 could not be detected in the culture supernatant.
Fig. 2.
LipA secretion and phosphatase activity of recombinant LipA. (A) The supernatant, membrane, and cytosolic fraction were collected from overnight cultures of L. monocytogenes LO28 clones, containing an empty plasmid (C), a plasmid expressing only GFP (GFP), or a plasmid expressing the LipA protein fused with GFP (LipA-GFP). Western blot analysis was performed using antibodies to GFP, the secreted protein listeriolysin O (LLO), and the ribosomal protein S9. (B and C) The specific activity of LipA compared to the activity of calf intestine alkaline phosphatase (CIAP) toward phosphorylated amino acids (B) and phosphatidylinositol phosphates (C) was assessed by measuring the release of inorganic phosphate (Pi) using a malachite green assay. Controls represent the catalyst-independent Pi release for each substrate.
LipA exhibits phosphotyrosine as well as lipid phosphatase activity.
We characterized the enzymatic activity profile and substrate specificity of the putative phosphatase LipA in vitro using a recombinant His6-LipA fusion protein. Using P-Ser, P-Thr, and P-Tyr as substrates, we found that LipA specifically dephosphorylates P-Tyr but not P-Ser or P-Thr (Fig. 2B). The similarity between the predicted protein structure of LipA and the triple-specificity phosphatase MPtpB suggested that LipA might dephosphorylate phosphoinositides. To test this hypothesis, we tested LipA activity against seven phosphoinositide species. Indeed, LipA readily dephosphorylated phosphatidylinositol 3-phosphate [PI(3)P], PI(5)P, and PI(3,5)P2 (Fig. 2C). The catalytic activity of recombinant LipA toward p-nitrophenylphosphate (pNPP), a chromogenic substrate used for detection of phosphatase activity, was optimal at pH 6.5 and at temperatures between 25 and 40°C (see Fig. S1 in the supplemental material).
Growth and stress resistance remain unchanged in the L. monocytogenes ΔlipA strain.
To analyze LipA function, we inactivated the lipA gene by homologous recombination in both the EGDe and LO28 strains of L. monocytogenes. First, we compared the growth profiles of the mutants and the corresponding wild-type strains at 37°C in BHI and LB or minimal medium. Both knockout strains had identical growth profiles in BHI compared to the respective wild-type strains (see Fig. S2 in the supplemental material). The lag phases and doubling times of the mutant and wild-type strains were identical. Both strains reached stationary phase at the same density. In minimal or LB medium, the mutant and wild-type strains had similar growth profiles (see Fig. S2). Thus, inactivation of lipA did not significantly affect Listeria replication in broth media.
In order to investigate the possible roles of LipA under conditions mimicking those faced by Listeria in the infected host, the growth profiles of the mutant and wild-type strains in the presence of subinhibitory concentrations of acid, salt, or reactive oxygen species were compared. Growth of L. monocytogenes LO28 was tested in BHI supplemented with 5% NaCl and BHI at pH 5.5. The LO28 ΔlipA strain did not differ from the parental strain in the in vitro growth rate under those conditions. Similarly, addition of hydrochloric acid to cultures of the L. monocytogenes EGDe wild-type or ΔlipA strain at mid-log phase did not reveal any growth defect of the ΔlipA strain (see Fig. S2 in the supplemental material). Also, growth characteristics of the mutant EGDe strain were similar to those of the wild type in the presence of hydrogen peroxide or the superoxide generator plumbagin (see Fig. S2). These results indicate that LipA is not required for resistance against acid, osmotic, or oxidative stress in vitro.
The LipA deletion does not affect intracellular replication of L. monocytogenes.
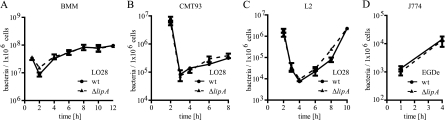
The intracellular growth of Listeria was assessed using a classical gentamicin assay. Infected cells were lysed at the indicated time points, and serial dilutions of intracellular bacteria were plated on BHI agar. To exclude cell-type-specific effects, we performed our analysis in bone marrow-derived macrophages, the J774 macrophage-like cell line, the rectal epithelial cell line CMT-93, and L2 fibroblast cells (Fig. 3). The intracellular growth profiles of wt and ΔlipA bacteria did not differ significantly in any of the infected cell types. In line with this assay, we did not observe a difference in the uptake of wt or ΔlipA Listeria into BMM and CMT-93 cells or a difference in phagosomal escape in RAW 264.7 cells (see Fig. S3 in the supplemental material). The intracellular presence of LipA might affect lipid phosphorylation as well as tyrosine phosphorylation signaling. The PIP3-dependent activation of PI 3-kinase causes phosphorylation and activation of the downstream Ser/Thr kinase Akt. The phosphorylation levels of Akt at the onset of infection of either macrophages or epithelial cells were similar after infection with wt and ΔlipA bacteria (see Fig. S4 in the supplemental material). Likewise, activation of Stat1 by tyrosine phosphorylation in macrophages, an event stimulated after the release of type I interferons (18), was not affected by the presence or absence of LipA (see Fig. S4). The lactate dehydrogenase release levels used to assess host cell lysis/death were the same in the presence and absence of LipA, as seen in Fig. S5 in the supplemental material.
Fig. 3.
Effect of L. monocytogenes LipA on intracellular growth. Growth of L. monocytogenes LO28 or EGDe was assessed by plating serial dilutions of cellular lysates at the indicated time points. Numbers of CFU are displayed as log CFU per 1 × 106 cells. BMM (A), CMT-93 (B), and L2 (C) cells were infected with L. monocytogenes LO28 wt and ΔlipA strains at MOIs of 10, 50, and 20, respectively. (D) J774 cells were infected with L. monocytogenes EGDe wt and ΔlipA strains at an MOI of 50.
LipA is not required for cell-to-cell spread.
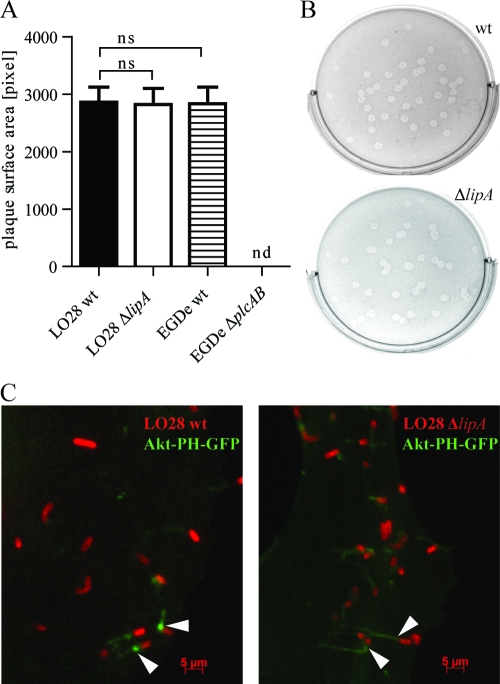
To test the impact of LipA deletion on the ability of Listeria to spread from cell to cell, a plaque assay was performed. Semiconfluent layers of L2 fibroblast cells were infected at a low MOI and overlaid with a mixture of tissue culture medium, agarose, and gentamicin. After an incubation time of 3 days, plaques were visualized by addition of another overlay containing neutral red, a stain for live host cells. L. monocytogenes wt and phospholipase ΔplcAB bacteria were included as controls. ΔplcAB strains are not able to escape from secondary vacuoles and do not form plaques. Comparison of the average surface areas of plaques formed by wt and ΔlipA bacteria showed no significant difference, whereas the ΔplcAB strain showed the expected inability to form plaques (Fig. 4 A and B).
Fig. 4.
Effect of L. monocytogenes LipA on movement and cell-to-cell spread. (A) Detection of L. monocytogenes cell-to-cell spread by a plaque assay. L2 cells were infected with L. monocytogenes LO28 wt and ΔlipA strains at an MOI of 0.5. Three days after infection, relative plaque surface area was calculated for 50 plaques per bacterial strain, and the mean for two independent experiments was calculated. ns, not significant; nd, not detected. (B) Representative pictures of plaques formed by L. monocytogenes LO28 wt (upper panel) and ΔlipA (lower panel) strains. (C) Localization of Akt-PH-GFP (green) in Ptk-2 cells infected with L. monocytogenes LO28 wt and ΔlipA strains. Cells were transfected with Akt-PH-GFP and infected with Listeria for 6 h. Bacteria were visualized using an antibody to Listeria and a secondary Alexa Fluor 594-conjugated antibody (red).
During ActA-dependent movement within the cytoplasm, Listeria bacteria accumulate PI(3,4,5)P3 around their surface (51). The host Akt kinase contains a PH domain that binds PI(3,4)P2 and PI(3,4,5)P3, and a fusion protein of GFP linked to the PH domain of Akt has been reported to concentrate on moving Listeria and the actin-rich tails (51). We used this construct to investigate Akt-PH recruitment to L. monocytogenes wt and ΔlipA bacteria. Ptk-2 cells were transfected with Akt-PH-GFP and infected with L. monocytogenes for 6 h. Bacteria were stained with a primary anti-Listeria antibody and a red secondary Alexa Fluor 594-conjugated antibody. Recruitment of the Akt-PH-GFP fusion protein to one pole of L. monocytogenes as well as to polarized tails, presumably representing the actin-rich tails, was detected upon microscopic examination of cytoplasmic L. monocytogenes wt and ΔlipA strains (Fig. 4C).
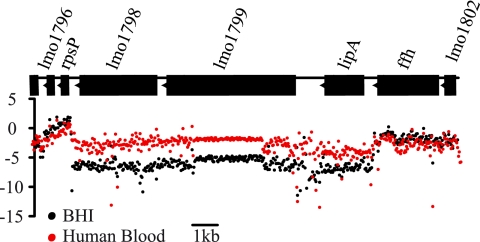
LipA is highly expressed in blood.
To get insight into the contribution of LipA to infection, we examined lipA transcription after growth of Listeria in vitro and in vivo, using our L. monocytogenes-specific tiling arrays and gene expression arrays (54). The level of the lipA transcript in wild-type L. monocytogenes grown to exponential phase in BHI broth at 37°C, the reference condition, was compared to the level of lipA transcribed in bacteria grown to stationary phase in BHI broth at 37°C following growth in BHI broth at 37°C and 6% oxygen, growth in BHI broth at 30°C, growth in the intestinal lumen after oral inoculation of axenic mice, and growth in human blood for 60 min. Strikingly, lipA transcription was induced 13.4-fold when bacteria were grown in human blood (Fig. 5). The expression levels of lipA were similar to those under the reference condition for all the other growth conditions and were independent of PrfA, the main activator of Listeria virulence genes. Likewise, lipA expression was not influenced by the alternative sigma factor SigB.
Fig. 5.
Expression of L. monocytogenes lipA in human blood. Transcriptional tiling maps of L. monocytogenes EGDe grown in BHI to exponential phase at 37°C (black dots) or in human blood at 37°C for 60 min (red dots) show normalized hybridization intensities (y axis, in arbitrary units) and genomic coordinates at the lipA locus (x axis, in bp). Each dot represents the average intensity signal for one probe from three independent biological replicates. Annotated ORFs are indicated as black boxes along the x axis.
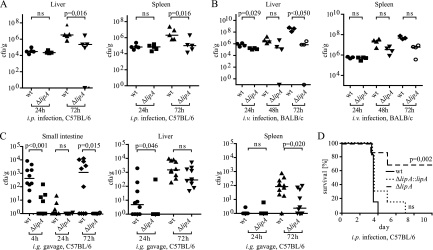
LipA is required for effective host colonization and Listeria virulence in vivo.
To examine the contribution of LipA to Listeria virulence in mice, the spread of ΔlipA bacteria to internal organs was measured. To exclude the possibility that the consequences of lipA deletion were restricted to a particular genetic background of either L. monocytogenes or its murine host, C57BL/6 and BALB/c mice were infected with both the EGD ΔlipA and LO28 ΔlipA strains and their respective parental strains. Intraperitoneal (i.p.), intravenous (i.v.), and intragastric (i.g.) routes were chosen for infection and subsequent measurement of spread to internal organs. In addition, LO28 wt, LO28 ΔlipA, and LO28 ΔlipA::lipA bacteria were used to determine the survival of infected C57BL/6 mice.
Mice infected with 2 × 106 L. monocytogenes LO28 ΔlipA bacteria exhibited a significant reduction of bacterial loads in both the liver and spleen at day 3 postinfection compared to the wt strain (Fig. 6 A). Likewise, BALB/c mice injected i.v. with 8 × 103 viable units of the EGDe or isogenic ΔlipA mutant strain showed reduced organ loads of mutant bacteria. The differences were most pronounced in both organs at 72 h postinfection, although statistical significance was not reached in the spleen (Fig. 6B). Importantly, multiplication of the mutant bacteria was controlled at 72 h postinfection in both the liver and spleen, while the number of wild-type bacteria continued to increase. To assess whether the decrease in virulence of ΔlipA strains observed upon systemic infection also occurred upon infection via the gastrointestinal tract, C57BL/6 mice were infected with L. monocytogenes LO28 wt and ΔlipA strains via intragastric gavage (Fig. 6C). wt bacteria were taken up by or tightly associated with intestinal tissue 4 h after infection, whereas significantly fewer bacteria were found after infection with the L. monocytogenes LO28 ΔlipA strain. This is unlikely to result from a decreased resistance to acid conditions in the gastrointestinal tract because the acid resistance of L. monocytogenes was unchanged upon deletion of the lipA gene (see Fig. S2 in the supplemental material; data not shown). Bacterial counts were below the detection limit after 24 h and increased approximately to the values found at 4 h 3 days after infection. Again, numbers of ΔlipA bacteria were significantly lower. Likewise, a reduction in ΔlipA counts 3 days after infection was observed in the spleen and liver. Whereas the difference was pronounced in the spleen, it did not reach statistical significance in the liver. The reduction of the pathogen burden was in line with an increased survival of mice (Fig. 6D). After i.p. infection of C57BL/6 mice with 1 × 106 L. monocytogenes LO28 wt bacteria, all animals died by day 4, whereas 70% of the population injected with the ΔlipA strain survived. To ascertain that the reduction of virulence was indeed due to the lack of LipA, the ΔlipA strain was complemented with an integrative plasmid encoding the lipA gene to generate the L. monocytogenes LO28 ΔlipA::lipA strain. The presence of this plasmid restored PTP activity in Listeria extracts (see Fig. S6 in the supplemental material) and lethal infection of mice upon intraperitoneal infection (Fig. 6D).
Fig. 6.
Effect of L. monocytogenes LipA on virulence in mice. (A) Groups of 4 to 5 C57BL/6 wt mice were infected intraperitoneally with 2 × 106 L. monocytogenes LO28 wt and ΔlipA bacteria. At days 1 and 3, the spleen and liver were homogenized and the bacterial titer was determined. Medians were plotted, and statistical significance was calculated using the Mann-Whitney U test. (B) L. monocytogenes EGDe wt and ΔlipA strains (8 × 103 CFU) were inoculated intravenously in groups of 4 BALB/c mice. Bacterial growth was followed in the spleen and liver at 24, 48, and 72 h. Medians were plotted, and statistical significance was calculated using the Mann-Whitney U test. (C) Groups of 10 C57BL/6 wt mice were infected via intragastric gavage of 1 × 109 L. monocytogenes LO28 wt and ΔlipA bacteria. At the indicated time points, numbers of CFU were determined in the small intestine, liver, and spleen by plating serial dilutions on Oxford agar plates. Medians were plotted, and statistical significance was calculated using the Mann-Whitney U test. (D) Survival assay. Groups of 6 to 7 C57BL/6 wt mice were infected intraperitoneally with 1 × 106 L. monocytogenes LO28 wt, ΔlipA, or ΔlipA::lipA bacteria, and survival was monitored for 10 days. Data are depicted as Kaplan-Meier plots. The survival of mice infected with the L. monocytogenes LO28 ΔlipA strain was strongly increased compared to that of mice infected with the wt strain (P = 0.002). This difference was no longer observed when the mice were infected with the complemented strains (not significant for the wt versus the ΔlipA::lipA strain and P = 0.009 for the ΔlipA strain versus the ΔlipA::lipA strain). Statistical significance was calculated using the Gehan-Breslow-Wilcoxon test.
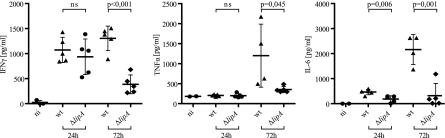
Serum cytokine levels are reduced after infection with ΔlipA bacteria.
To test whether proinflammatory cytokine production reflected bacterial loads, serum levels of IFN-γ, TNF-α, and IL-6 were measured by ELISA. IFN-γ was robustly produced after i.p. infection of C57BL/6 mice with wt L. monocytogenes LO28 on days 1 and 3 after infection. A significant reduction was detected after infection with ΔlipA bacteria on the third day of infection. Production of TNF-α was detected at low levels 3 days after infection with wt bacteria but with quite a high variability between mice. After infection with the ΔlipA strain, TNF-α levels were only marginally elevated compared to those in uninfected mice and were significantly lower than those in mice infected with wt bacteria. Likewise, serum IL-6 levels were highly reduced during infection with ΔlipA bacteria compared to those during infection with wt bacteria (Fig. 7). During infection with L. monocytogenes, leukocyte populations in blood or lymphoid organs were subject to changes in number and composition (see Fig. S7 in the supplemental material). The extents to which these changes occurred were similar for wt and ΔlipA bacteria.
Fig. 7.
Effect of L. monocytogenes LipA on serum cytokine levels. Levels of IFN-γ, TNF-α, and IL-6 in sera of C57BL/6 wt mice after intraperitoneal infection with L. monocytogenes LO28 wt and ΔlipA strains was measured by ELISA. Cytokine levels in individual mice are depicted, and means with standard deviation are shown. Statistical significance was calculated using the Student t test. ni, noninfected.
DISCUSSION
L. monocytogenes is a well-established model organism in infection biology. Extensive studies on the infectious cycle of Listeria have provided profound knowledge of the virulence factors that bacteria require for successfully using host cells as a replication niche. Posttranslational modification of host proteins and pathways by bacterial enzymes is increasingly recognized as an important facet of the host-pathogen relationship (48, 49). An enhancement of virulence through secretion of phosphatases and their interference with host metabolism had originally been associated with the pathogenicity of enteric Gram-negative bacteria and the use of type III secretion systems (10, 25, 39). Whereas S/T dephosphorylation and the ensuing adaptation to the host environment have previously been found to contribute to L. monocytogenes virulence (2, 3), we now provide the first report showing that tyrosine dephosphorylation of hitherto-unidentified host proteins similarly contributes to the virulence of this bacterium. We characterize lmo1800 as a gene encoding the phosphatase LipA, a member of the classical tyrosine phosphatase family. Among phospho-amino acid substrates, recombinant LipA specifically dephosphorylated P-Tyr. Typically, tyrosine-specific phosphatases display a bell-shaped pH rate profile with a low pH optimum (30). In line with this, LipA had its highest activity at pH 6.5. Besides P-Tyr, LipA dephosphorylated the phosphoinositides PI(3)P, PI(5)P, and PI(3,5)P2. This dual activity as both phospho-amino acid and phospholipid phosphatase is in line with the high similarity between the predicted LipA structure and that of phosphatase MPtpB, a virulence factor required for intracellular growth of Mycobacterium tuberculosis (6, 8, 35). MPtpB dephosphorylates P-Tyr, P-Ser, and P-Thr and, additionally, a broad range of phosphoinositide substrates (8). MPtpB contains a flexible lid formed by two helices that protects the active site from oxidative inactivation. This is mediated by the conformational dynamics of the lid and might represent an adaptation to oxidative challenges in vivo (22). The predicted three-dimensional (3D) model of LipA suggests that it also contains this flexible loop structure. Important for the contribution of LipA to L. monocytogenes virulence, our data show the functionality of the predicted signal peptide for secretion or membrane insertion. Using a LipA-GFP fusion construct, we found that LipA-GFP was present in the supernatants of Listeria cultured in broth, indicating that LipA is secreted. A recent study suggested that LipA was among the proteins lipidated by the prolipoprotein diacylglyceryl transferase (Lgt), leading to its anchorage to the bacterial surface (7). Although the 30 N-terminal amino acids of LipA contain a potential lipobox for acylation (L19-G-G-C-G23), the membrane association of LipA could be only transient before its release into the extracellular environment.
The important contribution of LipA to the virulence of L. monocytogenes was revealed by a comprehensive study in mice. LipA deficiency decreased pathogen load and inflammatory cytokine production, irrespective of the genetic background of the infected mice (C57BL/6 or BALB/c), the Listeria strain (LO28 or EGDe), or the route of infection (i.v., i.p., or i.g.). This strongly suggests that LipA targets host cell components rather than bacterial components. ΔlipA bacteria grew equally well in broth and host cells, and they were equally resistant to adverse conditions such as low pH, osmotic stress, and an oxidizing environment. Importantly, the L. monocytogenes genome does not contain proven tyrosine kinases (26). Therefore, it appears likely that the effect of LipA on virulence results from dephosphorylation of host cell proteins or lipids. Our data further suggest that LipA targets do not affect the L. monocytogenes life cycle in either the phagocytes or the nonphagocytic colonic epithelial cells investigated in this study. Macrophages are relevant as both replication niches and effector cells, depending on activation status, which in turn requires signals by pattern recognition receptors and IFN-γ-mediated tyrosine phosphorylation of Stat transcription factors (19). The intestinal epithelium represents the initial contact between L. monocytogenes and its mammalian host. A plethora of signals, including both tyrosine phosphorylation and phospholipid-dependent events, derives from InlA and InlB ligands, pattern recognition and cytokine receptors, and the invasion process per se to accompany bacterial uptake (15, 18, 40). Surprisingly, we could not detect an impact of LipA on these pathways. Assuming that lipid phosphatase activity is relevant for virulence, our future experiments to identify events and pathways affected by LipA will have to take into account that the enzyme shows pronounced preference for some phosphoinositides over others. Strikingly, a phosphate at C-4 strongly reduces dephosphorylation. Since many lipid signaling pathways require PI(3,4)P2 or PI(3,4,5)P3 (12), our search will be directed toward events such as endosome or phagosome maturation that require PI(3)P (14, 28), PI(5)P monophosphates (41, 42), or PI(3,5)P2 diphosphates (45, 57).
The decrease in the number of intestinal ΔlipA bacteria already 4 h after intragastric inoculation points toward a role for LipA during the immediate innate phase of the anti-Listeria immune response. Speculating about the possible function of LipA, we hypothesize that it targets a pathway in a cell type not analyzed so far or a cooperation of different cell types that may not be readily amenable to studies ex vivo. Alternatively, LipA might interfere with a biological process undetected using the assays performed in our study. The reduction in serum cytokines observed in infected mice would fit both explanations. The identical compositions of blood and splenic leukocyte populations rather argue that reduced serum levels are a consequence of reduced bacterial numbers and not due to a change in the quality of the immune response. Important effector cells of early anti-Listeria immunity are neutrophils (55), a cell type notoriously difficult to study due to its short life span in culture. As LipA is highly upregulated in human blood, our future aims must be directed at investigating the effect of LipA deletion during infections of blood monocytes and neutrophils.
Taken together, we characterized a novel virulence determinant of L. monocytogenes, namely, the secreted phosphatase LipA. We demonstrated that LipA is essential for promoting Listeria infections in vivo irrespective of the route of infection. The importance of this newly described factor is highlighted by the drastic defect in early host colonization after intragastric delivery of ΔlipA bacteria. Further investigation of LipA function and the identification of physiological LipA substrates will be interesting future tasks for a better understanding of Listeria interactions with its mammalian host.
Supplementary Material
ACKNOWLEDGMENTS
Work in the laboratory of T.D. was supported by the University of Vienna through the research focus “Symbiosis Research and Molecular Principles of Recognition” and the Austrian Science Fund (FWF) through grant P 20522-B05. Work in the laboratory of P.C. was supported by the Institut Pasteur (GPH9), Inserm, INRA, ANR (ANR-06-PATHO-011-01), and ERC (Advanced Grant 233348).
P.C. is an International Research Scholar of the Howard Hughes Medical Institute.
Footnotes
Supplemental material for this article may be found at http://iai.asm.org/.
Published ahead of print on 28 March 2011.
ADDENDUM IN PROOF
While our paper was under review, Beresford and colleagues (N. J. Beresford, C. Saville, H. J. Bennett, I. S. Roberts, and L. Tabernero, BMC Genomics 11:457–469, 2010) reported lipid phosphatase activity of the protein encoded by lmo1800, in agreement with the data reported in our study.
REFERENCES
- 1. Abachin E., et al. 2002. Formation of d-alanyl-lipoteichoic acid is required for adhesion and virulence of Listeria monocytogenes. Mol. Microbiol. 43:1–14 [DOI] [PubMed] [Google Scholar]
- 2. Archambaud C., Gouin E., Pizarro-Cerda J., Cossart P., Dussurget O. 2005. Translation elongation factor EF-Tu is a target for Stp, a serine-threonine phosphatase involved in virulence of Listeria monocytogenes. Mol. Microbiol. 56:383–396 [DOI] [PubMed] [Google Scholar]
- 3. Archambaud C., Nahori M. A., Pizarro-Cerda J., Cossart P., Dussurget O. 2006. Control of Listeria superoxide dismutase by phosphorylation. J. Biol. Chem. 281:31812–31822 [DOI] [PubMed] [Google Scholar]
- 4. Arnaud M., Chastanet A., Debarbouille M. 2004. New vector for efficient allelic replacement in naturally nontransformable, low-GC-content, gram-positive bacteria. Appl. Environ. Microbiol. 70:6887–6891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Baccarini M., Bistoni F., Lohmann Matthes M. L. 1985. In vitro natural cell-mediated cytotoxicity against Candida albicans: macrophage precursors as effector cells. J. Immunol. 134:2658–2665 [PubMed] [Google Scholar]
- 6. Bach H., Papavinasasundaram K. G., Wong D., Hmama Z., Av-Gay Y. 2008. Mycobacterium tuberculosis virulence is mediated by PtpA dephosphorylation of human vacuolar protein sorting 33B. Cell Host Microbe 3:316–322 [DOI] [PubMed] [Google Scholar]
- 7. Baumgartner M., et al. 2007. Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189:313–324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Beresford N., et al. 2007. MptpB, a virulence factor from Mycobacterium tuberculosis, exhibits triple-specificity phosphatase activity. Biochem. J. 406:13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beresford N. J., et al. 2009. Inhibition of MptpB phosphatase from Mycobacterium tuberculosis impairs mycobacterial survival in macrophages. J. Antimicrob. Chemother. 63:928–936 [DOI] [PubMed] [Google Scholar]
- 10. Bliska J. B., Falkow S. 1993. The role of host tyrosine phosphorylation in bacterial pathogenesis. Trends Genet. 9:85–89 [DOI] [PubMed] [Google Scholar]
- 11. Cabanes D., Dussurget O., Dehoux P., Cossart P. 2004. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol. Microbiol. 51:1601–1614 [DOI] [PubMed] [Google Scholar]
- 12. Cantley L. C. 2002. The phosphoinositide 3-kinase pathway. Science 296:1655–1657 [DOI] [PubMed] [Google Scholar]
- 13. Chakraborty T., et al. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Coronas S., et al. 2007. PtdIns5P: a little phosphoinositide with big functions? Biochem. Soc. Symp. 74:117–128 [DOI] [PubMed] [Google Scholar]
- 15. Cossart P., Bierne H. 2001. The use of host cell machinery in the pathogenesis of Listeria monocytogenes. Curr. Opin. Immunol. 13:96–103 [DOI] [PubMed] [Google Scholar]
- 16. Cozzone A. J., Grangeasse C., Doublet P., Duclos B. 2004. Protein phosphorylation on tyrosine in bacteria. Arch. Microbiol. 181:171–181 [DOI] [PubMed] [Google Scholar]
- 17. Darji A., Mohamed W., Domann E., Chakraborty T. 2003. Induction of immune responses by attenuated isogenic mutant strains of Listeria monocytogenes. Vaccine 21(Suppl. 2):S102–S109 [DOI] [PubMed] [Google Scholar]
- 18. Decker T., Muller M., Stockinger S. 2005. The yin and yang of type I interferon activity in bacterial infection. Nat. Rev. Immunol. 5:675–687 [DOI] [PubMed] [Google Scholar]
- 19. Decker T., Stockinger S., Karaghiosoff M., Muller M., Kovarik P. 2002. IFNs and STATs in innate immunity to microorganisms. J. Clin. Invest. 109:1271–1277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Domann E., et al. 1993. Detection of a prfA-independent promoter responsible for listeriolysin gene expression in mutant Listeria monocytogenes strains lacking the PrfA regulator. Infect. Immun. 61:3073–3075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dussurget O., Pizarro-Cerda J., Cossart P. 2004. Molecular determinants of Listeria monocytogenes virulence. Annu. Rev. Microbiol. 58:587–610 [DOI] [PubMed] [Google Scholar]
- 22. Flynn E. M., Hanson J. A., Alber T., Yang H. 2010. Dynamic active-site protection by the M. tuberculosis protein tyrosine phosphatase PtpB lid domain. J. Am. Chem. Soc. 132:4772–4780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Fujimoto S., Ike Y. 2001. pAM401-based shuttle vectors that enable overexpression of promoterless genes and one-step purification of tag fusion proteins directly from Enterococcus faecalis. Appl. Environ. Microbiol. 67:1262–1267 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Galan J. E. 2001. Salmonella interactions with host cells: type III secretion at work. Annu. Rev. Cell Dev. Biol. 17:53–86 [DOI] [PubMed] [Google Scholar]
- 25. Galan J. E., Collmer A. 1999. Type III secretion machines: bacterial devices for protein delivery into host cells. Science 284:1322–1328 [DOI] [PubMed] [Google Scholar]
- 26. Glaser P., et al. 2001. Comparative genomics of Listeria species. Science 294:849–852 [DOI] [PubMed] [Google Scholar]
- 27. Grangeasse C., Cozzone A. J., Deutscher J., Mijakovic I. 2007. Tyrosine phosphorylation: an emerging regulatory device of bacterial physiology. Trends Biochem. Sci. 32:86–94 [DOI] [PubMed] [Google Scholar]
- 28. Hilbi H. 2006. Modulation of phosphoinositide metabolism by pathogenic bacteria. Cell. Microbiol. 8:1697–1706 [DOI] [PubMed] [Google Scholar]
- 29. Kocks C., et al. 1992. L. monocytogenes-induced actin assembly requires the actA gene product, a surface protein. Cell 68:521–531 [DOI] [PubMed] [Google Scholar]
- 30. Kolmodin K., Aqvist J. 2001. The catalytic mechanism of protein tyrosine phosphatases revisited. FEBS Lett. 498:208–213 [DOI] [PubMed] [Google Scholar]
- 31. Kovarik P., Stoiber D., Novy M., Decker T. 1998. Stat1 combines signals derived from IFN-gamma and LPS receptors during macrophage activation. EMBO J. 17:3660–3668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lalic-Multhaler M., Bohne J., Goebel W. 2001. In vitro transcription of PrfA-dependent and -independent genes of Listeria monocytogenes. Mol. Microbiol. 42:111–120 [DOI] [PubMed] [Google Scholar]
- 33. Lingnau A., et al. 1995. Expression of the Listeria monocytogenes EGD inlA and inlB genes, whose products mediate bacterial entry into tissue culture cell lines, by PrfA-dependent and -independent mechanisms. Infect. Immun. 63:3896–3903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Luo Q., Rauch M., Marr A. K., Muller-Altrock S., Goebel W. 2004. In vitro transcription of the Listeria monocytogenes virulence genes inlC and mpl reveals overlapping PrfA-dependent and -independent promoters that are differentially activated by GTP. Mol. Microbiol. 52:39–52 [DOI] [PubMed] [Google Scholar]
- 35. Madhurantakam C., et al. 2005. Crystal structure of low-molecular-weight protein tyrosine phosphatase from Mycobacterium tuberculosis at 1.9-Å resolution. J. Bacteriol. 187:2175–2181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Marquis H. 2006. Tissue culture cell assays used to analyze Listeria monocytogenes. Curr. Protoc. Microbiol. 9:9B.4. [DOI] [PubMed] [Google Scholar]
- 37. Mead P. S., et al. 1999. Food-related illness and death in the United States. Emerg. Infect. Dis. 5:607–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Monk I. R., Gahan C. G., Hill C. 2008. Tools for functional postgenomic analysis of Listeria monocytogenes. Appl. Environ. Microbiol. 74:3921–3934 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nhieu G. T., Enninga J., Sansonetti P., Grompone G. 2005. Tyrosine kinase signaling and type III effectors orchestrating Shigella invasion. Curr. Opin. Microbiol. 8:16–20 [DOI] [PubMed] [Google Scholar]
- 40. O'Riordan M., Portnoy D. A. 2002. The host cytosol: front-line or home front? Trends Microbiol. 10:361–364 [DOI] [PubMed] [Google Scholar]
- 41. Pendaries C., et al. 2006. PtdIns5P activates the host cell PI3-kinase/Akt pathway during Shigella flexneri infection. EMBO J. 25:1024–1034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Pendaries C., et al. 2005. Emerging roles of phosphatidylinositol monophosphates in cellular signaling and trafficking. Adv. Enzyme Regul. 45:201–214 [DOI] [PubMed] [Google Scholar]
- 43. Phan-Thanh L., Gormon T. 1997. A chemically defined minimal medium for the optimal culture of Listeria. Int. J. Food Microbiol. 35:91–95 [DOI] [PubMed] [Google Scholar]
- 44. Philpott D. J., Girardin S. E., Sansonetti P. J. 2001. Innate immune responses of epithelial cells following infection with bacterial pathogens. Curr. Opin. Immunol. 13:410–416 [DOI] [PubMed] [Google Scholar]
- 45. Pizarro-Cerda J., Cossart P. 2004. Subversion of phosphoinositide metabolism by intracellular bacterial pathogens. Nat. Cell Biol. 6:1026–1033 [DOI] [PubMed] [Google Scholar]
- 46. Portnoy D. A., Auerbuch V., Glomski I. J. 2002. The cell biology of Listeria monocytogenes infection: the intersection of bacterial pathogenesis and cell-mediated immunity. J. Cell Biol. 158:409–414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Reutterer B., et al. 2008. Type I IFN are host modulators of strain-specific Listeria monocytogenes virulence. Cell. Microbiol. 10:1116–1129 [DOI] [PubMed] [Google Scholar]
- 48. Ribet D., Cossart P. 2010. Pathogen-mediated posttranslational modifications: a re-emerging field. Cell 143:694–702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ribet D., Cossart P. 2010. Post-translational modifications in host cells during bacterial infection. FEBS Lett. 584:2748–2758 [DOI] [PubMed] [Google Scholar]
- 50. Scortti M., Monzo H. J., Lacharme-Lora L., Lewis D. A., Vazquez-Boland J. A. 2007. The PrfA virulence regulon. Microbes Infect. 9:1196–1207 [DOI] [PubMed] [Google Scholar]
- 51. Sidhu G., et al. 2005. Phosphoinositide 3-kinase is required for intracellular Listeria monocytogenes actin-based motility and filopod formation. J. Biol. Chem. 280:11379–11386 [DOI] [PubMed] [Google Scholar]
- 52. Stockinger S., et al. 2009. Characterization of the interferon-producing cell in mice infected with Listeria monocytogenes. PLoS Pathog. 5:e1000355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Tabernero L., Aricescu A. R., Jones E. Y., Szedlacsek S. E. 2008. Protein tyrosine phosphatases: structure-function relationships. FEBS J. 275:867–882 [DOI] [PubMed] [Google Scholar]
- 54. Toledo-Arana A., et al. 2009. The Listeria transcriptional landscape from saprophytism to virulence. Nature 459:950–956 [DOI] [PubMed] [Google Scholar]
- 55. Unanue E. R. 1997. Inter-relationship among macrophages, natural killer cells and neutrophils in early stages of Listeria resistance. Curr. Opin. Immunol. 9:35–43 [DOI] [PubMed] [Google Scholar]
- 56. Vazquez-Boland J. A., et al. 2001. Listeria pathogenesis and molecular virulence determinants. Clin. Microbiol. Rev. 14:584–640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Weber S. S., Ragaz C., Hilbi H. 2009. Pathogen trafficking pathways and host phosphoinositide metabolism. Mol. Microbiol. 71:1341–1352 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.