Abstract
The structural biology of Gram-positive cell surface adhesins is an emerging field of research, whereas Gram-negative pilus assembly and anchoring have been extensively investigated and are well understood. Gram-positive surface proteins known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) and individual proteins that assemble into long, hair-like organelles known as pili have similar features at the primary sequence level as well as at the tertiary structural level. Some of these conserved features are essential for their transportation from the cytoplasm and for cell wall anchoring. More importantly, the MSCRAMMs and the individual pilins are assembled with building blocks that are variants of structural modules used for human immunoglobulins. MSCRAMMs target the host's extracellular matrix proteins, such as collagen, fibrinogen, and fibronectin, and they have received considerable attention from structural biologists in the last decade, who have primarily been interested in understanding their interactions with host tissue. The recent focus is on the newly discovered pili of Gram-positive bacteria, and in this review, we highlight the advances in understanding of the individual pilus constituents and their associations and stress the similarities between the individual pilins and surface proteins.
Keywords: bacterial adhesions, Gram-positive pili, pilins, protein structure
Introduction
Adhesion to the host tissue is the first critical step for microbial colonization and infection, and it also helps the pathogen survive the pervasive fluid (shear) force present in the host. Both Gram-positive and Gram-negative bacteria use a multitude of proteins and protein assemblies that are anchored to their cell walls for adhesion. The chaperone-mediated assembly of hair-like, filamentous organelles known as pili in Gram-negative bacteria has been extensively investigated and is well understood,1–4 where the individual pilins are held together by an exquisite “donor strand complementation” technique. However, the pili in Gram-positive bacteria, a relatively recent discovery, 5–7 are assembled using covalent linkages and are mediated by a conserved and unique cysteine protease called sortase. 7 8 The main extended pilus segment, also known as the pilus shaft, is composed of several units of major (backbone) pilins that are covalently linked in a head-to-tail manner. In some cases, minor pilins (with or without the adhesive nature) are located at the tip and base.
In addition to the pili or fimbriae,9 a subfamily of surface proteins known as MSCRAMMs (microbial surface components recognizing adhesive matrix molecules) 10 are also covalently linked to their cell wall peptidoglycans by sortases and are used for targeting the host's extracellular matrix proteins (ECM), such as fibrinogen (Fg), 11 fibronectin (Fn), 12 and collagen 13 for adhesion.
In the last decade, the crystal structure analysis of MSCRAMMs revealed many interesting and unique features that dictate their adhesion to the host tissue (Table I), and the recent focus is on visualizing the individual pilin components and their associations. The MSCRAMMs and the individual pilin components have similar structural motifs [Fig. 1(a)] to facilitate their transport from the cytoplasm through the Sec system as well as for incorporation into the cell wall. The cell wall sorting motifs include a canonical LPXTG sequence at the C-terminus, where X is any of the 20 amino acids, followed by a hydrophobic stretch of residues and ending with a short, positively charged segment; these three motifs are sufficient and essential for anchoring to the cell wall.14 MSCRAMMs are modular proteins with individual domains for specific functions. Their N-terminal half primarily carries the ligand-binding domains (A-region), whereas the other structural domains that facilitate the extension of A-regions away from cell surface are in the C-terminal half [Fig. 1(b)]. In addition to having similar C-terminal sorting motifs, most major pilins contain a conserved segment called the pilin motif (WxxxVxVYPK) [Fig. 1(c)]. The side chain amide group of the Lys residue from this region is linked to a Thr residue carbonyl from the LPXTG motif of another pilin, and many repeats of the same linkage lead to the covalent polymerization of major pilins. This inter-molecular cross-linking catalyzed by a specific sortase, as demonstrated for Corynebacterium diphtheriae pilus assembly, 7 8 is unique to Gram-positives. To date, crystal structures are available for many full-length or fragments of individual pilins (Table I), 15–18 but, unlike the surface proteins, the type of ligands they target and their binding mode are still unclear.
Table I.
Available Crystal Structures of Gram-Positive Adhesins
| Organism | Surface/pilins | Receptors | Fragment | PDB code | References |
|---|---|---|---|---|---|
| S. aureus | MSCRAMM | Collagen | Cna A-region (N2 domain) | 1AMX | 21 |
| S. aureus | MSCRAMM | Collagen | Cna A-region | 2F68 (apo form) | 50 |
| (N1 & N2 domains) | 2F6A (complex) | ||||
| S. aureus | MSCRAMM | – | Cna B-repeats | 1D2O (B1) | 22 |
| 1D2P (B1B2) | |||||
| E. faecalis | MSCRAMM | Collagen | ACE (N2 domain) | 2OKM | 48 |
| E. faecalis | MSCRAMM | Collagen | ACE (N1 and N2 domains) | 2Z1P | 49 |
| S. aureus | MSCRAMM | Fibrinogen | ClfA (N2 and N3 domains) | 1N67 | 51 |
| S. aureus | MSCRAMM | Fibrinogen | ClfA (N2 and N3 domains) | 2VR3 (complex) | 61 |
| S. epidermidis | MSCRAMM | Fibrinogen | SdrG (N2 and N3 domains) | 1R19 (apo form) | 60 |
| 1R17 (complex) | |||||
| S. epidermidis | MSCRAMM | Fibrinogen | SdrG (N2 and N3 mutants) | 2RAL (E381C/P595C) | 68 |
| S. aureus | MSCRAMM | Fibronectin | FnBPA-1/2–3F1 | 2RKZ (FnBPA-1/2–3F1) | 66 |
| 2RKY (FnBPA-1/4–5F1) | |||||
| 3CAL (FnBPA-5/2–3F1) | |||||
| 2RL0 (FnBPA-1/2–3F1) | |||||
| S. agalactiae | Minor pilin | – | GBS52 | 3PHS | 15 |
| S. pyogenes | Major pilin | – | Spy0128 | 3B2M | 26 |
| S. pyogenes | Major pilin | – | Spy0128 mutant | 3GLD (E177A) | 27 |
| 3GLE (N168A) | |||||
| C. diphtheriae | Major pilin | – | SpaA | 3HR6/3HTL | 16 |
| S. pneumoniae | Minor pilin | – | RrgA | 2WW8 | 17 |
| B. cereus | Major pilin | – | BcPA* | 3KPT | 18 |
| S. pyogens | Minor pilin | – | FctB | 3KLQ | 30 |
| S. pneumoniae | Major pilin | – | RrgB* | 2X9W/2X9X/2X9Y/2X9Z | 40 |
| A. Naeslundii | Major pilin | – | FimA* | 3QDH | To be published |
| S. agalactiae | Major pilin | – | GBS80* | 3PF2, 3PG2 | 69 |
–Obtained by limited proteolysis.
Figure 1.

The domain organization for Gram-positive adhesins. (a) Common structural motifs present in the MSCRAMMs and pilin components. An N-terminal signal sequence (S) followed by a varying number of non-repetitive and repetitive regions suitable for various functions (ligand binding, projecting binding region/stalking, etc.) and C-terminal region for cell wall anchoring that includes a cell wall sorting region (W) containing the LPXTG motif, the membrane-spanning hydrophobic domain (M) and the cytoplasmic positively charged C-terminal tail (C). (b) The organization of three distinct MSCRAMMs. Collagen-binding (CNA/ACE), fibrinogen-binding (ClfA/B, SdrG), and fibronectin-binding (FnbpA/B) MSCRAMMs. In addition to common structural motifs (S, W, M, and C), most adhesins carry ligand-binding domains (N1, N2, N3, …) in the N-terminal half (A-region) and other structural domains that facilitate the extension of A-regions away from the cell surface and unknown functions in the C-terminal half (B-region). Exceptions to this rule are the bifunctional FnbpA/B MSCRAMMs, which display Fn-binding activity at the C-terminal repeats (D1–D4) and fibrinogen-binding activity in the N-terminal half. CNA and ACE are highly homologous; however, the ACE is composed of only two subdomains (N1 and N2) but exhibits equal affinity to the three domain (N1, N2, and N3) CNA. (c) The organization of pilins. In addition to common structural motifs (S, W, M, and C), the pilins carry a varying number of IgG-like domains (often) that makeup pilus shaft and for adhesion. Several major pilins contain conserved pilin motif (WxxxVxVYPK) for covalent polymerization that is catalyzed by an enzyme sortase. The tip pilins contain a vWFA domain, which is flanked by IgG-like domains for adhesion. The base pilin likely contains a lysine in the C-terminal half or in the domain linker for their incorporation into the pilus shaft at the base, and a proline-rich C-terminal tail for cell wall anchoring.
One important common building block for both types of Gram-positive adhesins is the IgG-constant domain, which appears in two or more different forms that are different from the widely observed V-, C-, H-, and I-type folds of the immunoglobulin superfamily.19 20 The IgG-constant domain has a four-stranded β-sheet (I), ABED on one side of the barrel and a three-stranded β-sheet (II) CFG on the other side [Fig. 2(a)]. Two variants of the IgG-C fold are frequently seen in Gram-positive adhesins (pilins and MSCRAMMs) and are named DEv-IgG and IgG-rev folds. Similar to the IgG-C fold, the DEv-IgG fold has two β-sheets, in which strands ABED form β-sheet I and CFG form β-sheet II. However, the DEv-IgG fold also exhibits a few extra strands (often two) between strands E and D, which align with strands CFG at the interface of sheets I and II [Fig. 2(b)]. We first observed this variant in the ligand-binding A-region of collagen-binding Staphylococcus aureus CNA, 21 sometimes referred to as a cnaA-type fold. The IgG-rev fold has three- (CBE) and four- (DAGF) stranded β-sheets or four- (CBEF) and three- (DAG) stranded sheets, but the strands are arranged in the reverse order [Fig. 2(c)]. We first observed the IgG-rev fold in the domains of the CNA B-region, 22 and it is thus sometimes referred to as a cnaB-type fold. These IgG variants are seen across all of the Gram-positive surface adhesins, and the following sections discuss the structural details of these surface proteins and pilins and the biological implications of their structural arrangements.
Figure 2.

Topology and structure of the IgG constant domain and its variant DEv-IgG, IgG-rev, seen in the MSCRAMMs and pilins. The core β-strands are labeled A–G in a rainbow style color (red to violet). (a) IgG constant domain contains a four-stranded β-sheet I (A, B, E, D) on one side of the barrel and a three-stranded β-sheet II (C, F, G) on the other side. (b) The DEv-IgG fold is similar to IgG-C fold but the additional secondary structural elements (gray) are present between the strands D and E. A possible intra-domain isopeptide bond between A and F strands is shown by a block line. The DEv-IgG fold is first observed in the domains of CNA A-region, hence called as cnaA-type (c) The IgG-rev fold contains a four- (DAGF) and three- (EBC) or three- (DAG) and four- (FEBC) stranded β-sheets similar to IgG-C fold but their arrangements are different. A possible intra-domain isopeptide bond between A and G strands is shown by a block line. The IgG-rev fold is first observed in domains of CNA B repeats, and sometimes described as cnaB-type. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Gram-Positive Pilins
Pili in Gram-positive bacteria were first identified in Corynebacterium renale,23 followed by others such as Acitnomyces, Ruminococcus, Enterococcus, Clostridia, and species of Streptococcus (Refs. 6 7 24 25). In most of the Gram-positive pathogens, the genes for the pilus components are arranged in pilus islands (PIs) that encode one major (backbone) pilin, one or two minor (ancillary) pilins, and one or more pilus-specific and one house-keeping sortases. The pilus-specific sortases catalyze the covalent polymerization of the pilins by forming an inter-molecular amide bond between the Thr residue at the C-terminus sorting (LPXTG) motif of one pilin and a Lys residue side chain present in the pilin motif (YPKN) of another, and the house-keeping sortase is responsible for anchoring the assembled pili onto the cell wall peptidoglycan, as first demonstrated for C. diphtheriae. 7 8
Intra-domain isopeptide bonds in Gram-positive pilins
Interestingly, Gram-positive pilins harbor intra-molecular isopeptide bonds, which are covalent bonds between the side chains of Lys and Asn, and the bond formation is catalyzed by proximal Asp or Glu residues.26 Mutations that abrogate the formation of such isopeptide bonds have been shown to affect the thermal stability of pilins and decrease their resistance to proteases. 16 27 In contrast, the pilin folding occurs in the oxidizing environment of the periplasm of the Gram-negative bacteria, where the pilins exhibit a jelly-roll fold with two β-sheets and carry disulfide bonds that connect the β-strands of two sheets or the β-strands of the same β-sheet. Interestingly, the isopeptide bond formation pattern is structurally and spatially conserved across the pilins and for a few MSCRAMMs, although they have poor sequence homology (<18%). Two types of isopeptide bonds are seen: the D-type, which is frequently observed in DEv-IgG folds, and the E-type, which is observed in IgG-rev folds [Figs. 2(b, c)]. The D-type isopeptide bond connects two β-sheets, where the A strand having Lys (sheet I) and the F strand hosting Asn (sheet II) are antiparallel, with a conserved Asp residue present on the C strand (sheet II). The E-type isopeptide bond connects two adjacent parallel strands of the same sheet (I), the Lys on the first A strand and the Asn on the last G strand, with a conserved Glu residue present on the E strand of sheet II. In addition, the isopeptide bonds identified in the Gram-positive pilins and MSCRAMMs have two isomeric forms: cis and trans configurations. Interestingly, the predicted inter-molecular isopeptide bonds catalyzed by sortases have yet to be observed, as no crystal structure of isopeptide-linked multiple pilins is available. However, similar inter-molecular isopeptide bonds are known to exist, such as those observed during ubiquitination 28 and transglutamination 29 that dictate the stability of the respective participants.
Minor/ancillary pilins
We can classify the minor pilins into two groups, base pilins and tip pilins, based on their size and, to some extent, on their position and function (adhesive nature). The base pilin, shown to be involved in cell wall anchoring30 and also decorated along the pili in some pathogens, for example, in C. diptheriae. 31 It is usually made of one or two IgG-rev domains and is of a small size compared to a tip pilin. A proline-rich C-terminal tail, which is shown to participate in cell wall anchoring, is also observed in the base pilin. 15 30 The tip pilin/major ancillary pilin exhibits an adhesive function and consist of more than two types of domains, which include a vWFA (von Willibrand factor A) fold and an IgG-like fold. Unlike the major pilins, the minor pilins do not have a regular pilin motif. The tip pilin is incorporated into the pilus tip via its C-terminal LPXTG-like motif, whereas the base pilin requires a Lys residue for its incorporation at the pilus base because its C-terminal LPXTG-like motif is thought to be involved in the cell wall anchoring. Recent structural studies on minor pilin GBS52 15 and minor pilin FctB of GAS 30 and homology model building studies (unpublished) suggest that they could be incorporated via a Lys residue, which is found at the C-terminal half in the case of single-domain minor pilins (SpaB of C. diphtheriae and FctB of Streptococcus pyogenes) or near the domain linker in the case of minor pilins having two or more domains, such as GBS52 of Streptococcus agalactiae and, likely, RrgC of Streptococcus pneumoniae. The three-component (one major and two minor pilins) pilus assembly model, first proposed for C. diphtheriae, is consistent with the three-component pili of other Gram-positive pathogens such as S. agalactiae, S. pneumoniae, S. pyogenes, and Enterococcus faecalis.
Structure of the tip pilins
In S. pneumoniae, the pilus-forming LPXTG-containing proteins (RrgA, RrgB, and RrgC) are encoded by an rlrA islet. The major pilin RrgB is necessary for the pilus formation, while the major ancillary pilin RrgA is located at the tip of pilus and is shown to be essential for pilus adhesion. The crystal structure of RrgA17 shows three IgG-like domains (N1, N2, and N4) and a vWFA domain (N3), similar to an integrin I domain [Fig. 3(a)]. The topologies of N1 and N4 are similar and are variants of the IgG-rev fold, having two additional strands at the C-terminal end that align with the edge of each sheet that resemble four- and five-stranded β-sheets. The N1 is made of two parts of the same chain, where the last strand G is the link between the N2 and N4 domains. The N4 domain has an E-type isopeptide bond in a trans configuration, and the N2 domain is a variant of the DEv-IgG fold, with its typical D-type isopeptide bond also in a trans configuration. The N3 domain is inserted between the B and C strands of N2 and also in the N1 domain between its A and G strands. Thus, with additional strands between its D and E (D′, D″) and two more between A and G, the N2 domain belongs to an 11-stranded DEv-IgG fold.
Figure 3.
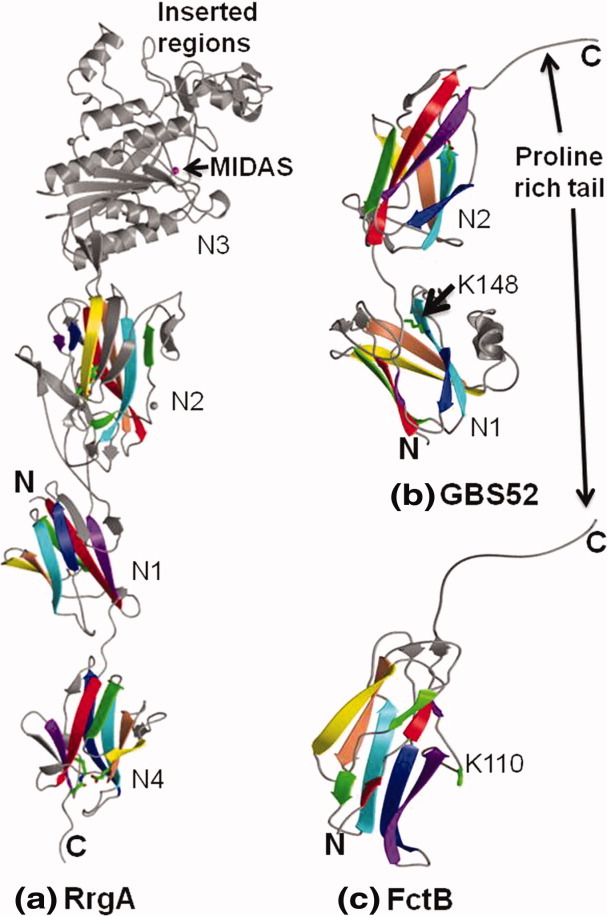
Ribbon representation of minor pilin crystal structures. The IgG-like domains are shown in rainbow style as in Figure 2. The intra-domain isopeptide bonds are shown in sticks. (a) Tip pilin RrgA of S. pneumoniae display two domains of IgG-rev fold (N1 and N4), a DEv-IgG fold domain (N2) and a domain of vWFA fold (N3) with a metal-ion-dependent adhesion site (MIDAS) and two inserted regions. The N2 and N4 domains contain a D- and E-type isopeptide bonds, respectively. (b) Minor pilin GBS52 of S. agalactiae contains two domains (N1 and N2) of IgG-rev fold. The N2 domain has an E-type isopeptide bond and proline-rich C-terminal tail. A lysine (K148) from a pilin-like motif (IYPK) in the domain linker was suggested for its incorporation into the pilus shaft. (c) Minor FctB of S. pyogenes is made of single domain of IgG-rev fold. A lysine (K110) in the Ω-loop was suggested for it incorporation into the pilus shaft. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
The N3 domain consists of a central six-stranded β-sheet surrounded by α helices, as observed in the I-domains of integrins32 and the vWFA-like domains of complement convertases. 33 34 The presence of N- and C-termini on one side of the molecule and a MIDAS (metal-ion-dependent adhesion site) on the opposite side are characteristic features of vWFA domains, which are implicated in metal-mediated adhesion and signaling. In addition, the RrgA N3 domain displays two extra regions inserted on either side of the MIDAS, one with two β hairpins and two long loops and another having one short hairpin region and two short helices. A similar vWFA variant fold is also predicted for the tip pilins of S. agalactiae and C. diphteriae. 35 Based on their structural homology with many adhesive prokaryotic protein vWFA domains, we can suggest a possible metal-mediated adhesive role for these tip pilins. However, we cannot yet speculate on the role of the above-mentioned insertions around the MIDAS or the nature of these tip pilin targets.
Structures of the other ancillary pilins
Genomic analysis showed the presence of two similar PIs in S. agalactiae or group B Streptococcus (GBS) that encode a number of LPXTG motif-bearing proteins and associated sortase enzymes.36–38 The major pilin GBS80 forms the pilus shaft, whereas the minor (associated) pilin GBS52 was recently shown to be an adhesin, 15 37 the deletion of which significantly reduced the GBS adherence to pulmonary epithelial cells.
The structure of GBS5215 revealed the presence of two IgG-rev domains [Fig. 3(b)]. Significant additional features include two hook-shaped loops in the N1 domain, an extended hydrophobic proline-rich C-terminus tail of the N2 domain and a predominantly hydrophobic interface between the N1 and N2 domains that are linked head-to-tail. The N2 domain has an E-type isopeptide bond in a trans configuration. Biochemical studies revealed that the N2 domain of GBS52 is necessary and sufficient for GBS adhesion to lung epithelial cells. 15 Interestingly, the GBS52 structure also shows a pilin-like motif (IYPK) at the N1–N2 domain interface like the major pilins, suggesting that the GBS52 could be incorporated via the Lys residue similar to a major pilin polymerization.
S. pyogenes or group A streptococcus (GAS) encodes a minor pilin FctB (strain 90/360S), whose structure revealed a single domain of an IgG-rev fold with an extended proline-rich C-terminal end,30 as seen in GBS52. Similar to GBS52, the strands DAG form a three-stranded β-sheet, whereas strands CBEF form a four-stranded β-sheet, but the G strand is interrupted by an Ω loop shared between two sheets. Thus, the N-terminus of the G strand aligns with the four-stranded β-sheet, resembling a five-stranded β-sheet [Fig. 3(c)]. The Lys110 in the Ω loop was suggested for the incorporation of FctB into the pilus shaft. 30 Though FctB does not contain an intramolecular isopeptide bond, an internal hydrogen bond in an identical position of the conserved hydrophobic core has been identified.
Major/backbone pilins
The crystal structures of some major pilins (S. agalactiae GBS80, Bacillus cereus BcpA, and S. pneumonia RrgB) are available only after limited proteolysis that removes the N-terminal N1 domain at the critical pilin motif (YPKN), often found on a linker that connects two N-terminal domains (NTDs), by clipping between the Lys and Asn residues.18 39 40 The pilin motif Lys residue is involved in the inter-molecular isopeptide bond, and the neighboring Asn residue participates in the N1 domain intramolecular/domain isopeptide bond. In a significant discovery, Budzik et al. (2009) revealed that during the pilus polymerization, the intramolecular isopeptide bond in the N1 domain forms only after the major pilin precursor participates in Lys-Thr inter-molecular amide bond formation, 41 leading to a polymerized and stable pilus shaft. Hence, in the absence of the intra-domain isopeptide bond, even with all the essential ingredients in place, the respective pilins and their N1 domains are susceptible for proteolysis. This could possibly explain the available crystal structures for full-length C. diphtheriae SpaA, which does not have an Asn at the pilin motif (YPKHQ), and major pilin GAS Spy0128, which also exhibits a similarly deficient pilin motif (GSKVP). 16 26
S. agalactiae GBS80
The major pilin GBS80 from the strain 2603V/R (PI-1) of S. agalactiae or GBS is predicted to have three IgG-like domains, N1, N2, and N3, and in the available crystal structure of GBS80N2N3 (PDB ID: 3PF2), obtained after limited proteolysis of recombinant full-length GBS80, the N2 domain exhibits an DEv-IgG fold, whereas the N3 domain is an IgG-rev fold [Fig. 4(a)]. The N2 domain has a D-type isopeptide bond in a cis configuration, whereas the N3 domain contains an E-type isopeptide bond in a trans configuration. The flexibility between the two domains is restricted by the presence of a short, three-stranded β-sheet, which is formed by the N2–N3 domain linker and associated neighboring loops. There are two hook-shaped loops, the BC loop of the N3 domain and the D″E loop of the N2 domain, on its opposite sides, giving a symmetrical shape to the molecule. Two high-affinity calcium-binding sites in the N2 domain, one close to its isopeptide bond and another in the FG loop, are observed. Interestingly, the Lys210, which forms the isopeptide bond, also participates in the metal coordination via its main chain carbonyl along with neighboring acidic residue side chains (Asp209, Asp211, and Asp218) and two water molecules. No water molecules are seen coordinating with the FG loop calcium ion that has eight coordinating oxygen atoms from the side chains of Glu244, Asp358, Thr360, Asp362, Asn366, and the main chain carbonyls of Thr360 and Ala364. The crystal packing of the GBS80N2N3 structure suggests that the pilus shaft could be formed by the N2 and N3 domains, which have a strong interface, arranged in a head-to-tail fashion, and homology model building suggests N1 domain association in a supporting role by its lateral (∼20° bend) tilt.
Figure 4.
Structures of major pilins. The isopeptide bond between the Lys and Asn residues and the conserved proximal Glu residue are shown by sticks. (a) N2 and N3 domains of GBS80 from S. agalactiae. N2 is DEv-IgG fold with D-type isopeptide bond, and two high-affinity metal-binding sites, whereas N3 is an IgG-rev fold with E-type isopeptide bond. (b) Spy0128 from S. pyogenes is made of two IgG-rev fold domains, each with an E-type isopeptide bond. A lysine (K161) in the Ω-loop was suggested for pilus polymerization. (c) SpaA from C. diphtheriae shows three IgG-like domains (N1, N2, and N3). N1 and N3 exhibits IgG-rev folds, whereas N2 exhibits DEv-IgG fold, and hosts a metal-binding site and D-type isopeptide bond. The N3 contains an E-type isopeptide bond while N1 does not. The pilin motif lysine (K190) is seen at N1-N2 domain linker. (d) N2, N3, and N4 domains of BcpA from B. cereus. The N2 and N4 exhibit IgG-rev fold while N3 is of DEv-IgG fold. (e) N2, N3, and N4 domains of RrgB from S. pneumonia. The N2 domain display a DEv-IgG fold, whereas N3 and N4 domains show IgG-rev fold with respective isopeptide bonds. The N2 and N3 domains are packed parallel, and N3 domain is inserted into N2 domain. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
S. pyogenes Spy0128
The full-length S. pyogenes major pilin Spy0128 (strain SF370) is made of two IgG-like domains [Fig. 4(b)], each with an E-type isopeptide bond in the cis form.26 Similar to the minor pilin FctB, the N1 domain has a Ω loop on G strand, and the N-terminal part of this strand associates with a four-stranded β-sheet (II), so it resembles an elongated five-stranded sheet. A Lys161 residue from the Ω loop has been suggested to be involved in the inter-molecular isopeptide bond formation. 26 In the N2 domain, the three-stranded β-sheet (I) becomes five-stranded due to the insertion of two strands between the E and F strands present at one edge, and two additional strands between B and C at the other edge.
C. diphtheriae SpaA
C. diphtheriae, the causative agent of pharyngeal diphtheria, was one of the earliest organisms in which the presence of pili was described,23 and their assembly is the best detailed. 8 42 43 C. diphtheriae display three types of pili that are named according to their respective major pilins and are products of sortase-mediated pilus assembly (Spa): SpaA-, SpaD-, and SpaH-type. In the SpaA-type pilus, the major pilin SpaA builds the pilus shaft, SpaB decorates the shaft, and SpaC sits at the tip of the pilus. The crystal structure of SpaA revealed the presence of three (N1, N2, and N3) domains [Fig. 4(c)]. The N1 and N3 domains have the IgG-rev fold, and the N2 domain is a DEv-IgG fold. 16 The pilin motif is located on the last strand of the N1-domain and the critical Lys190, whose mutation abrogates the polymerization of SpaA molecules during pilus shaft formation, is located close to the N1 and N2 domain interface. The N1 domain has no isopeptide bond, but the N2 domain has a D-type isopeptide bond in a cis configuration, and N3 has an E-type isopeptide bond in a trans configuration. A high-affinity Ca2+-binding site is also observed in the AB loop of the N2 domain with eight coordinating atoms, including two water molecules.
B. cereus BcpA
B. cereus is commonly associated with toxin-induced emetic and diarrheagenic syndromes and is also responsible for skin or soft tissue infections. The pilus genes of B. cereus encode a major pilin BcpA and a minor pilin BcpB, which is positioned at the tip of the fiber. BcpA contains four domains (N1, N2, N3, and N4). The crystal structure of the BcpA fragment (N2, N3, and N4), obtained by limited proteolysis, displays an IgG-rev fold for the N2 and N4 domains and a DEv-IgG fold for the N3 domain [Fig. 4(d)].18 Each domain contains an isopeptide bond, including the N1 domain, in which the isopeptide bond is shown to be formed during the pilus polymerization. The N2 and N4 domains have E-type isopeptide bonds, in trans and cis configurations, respectively, and the N3 domain has a D-type isopeptide bond in a cis configuration.
S. pneumonia RrgB
The major pilin RrgB from the TIGR4 strain of S. Pneumonia is composed of four domains (N1, N2, N3, and N4). The crystal structure of the RrgB fragment containing N2, N3, and N4 domains was obtained, again by limited proteolysis. The N2 domain displays a DEv-IgG fold, and the N3 and N4 domains show IgG-rev folds [Fig. 4(e)].40 Interestingly, the N2 and N3 domains are packed in parallel, which is different from the domain (tandem) arrangement of other pilin structures, and the N3 domain is inserted in a loop between F strand and G of the N2 domain.
In addition to the above, there are a few more pilin-like structural homologs from commensal bacteria, which are members of the oral biofilm, for example, Actinomyces strain MG-1 assembles two types of pili. Type1 consist of major pilin FimP and a tip pilin FimQ, and Type 2 pili are made of major pilin FimA and tip pilin FimB.44 The structure of FimA (to be published), obtained by limited proteolysis, also shows IgG-rev- and DEv-IgG-like folds with respective E/D type intramolecular isopeptide bonds.
Surface proteins as adhesions
The collagen-binding MSCRAMMs
Collagens are the most abundant structural proteins in vertebrates, and both eukaryotes and prokaryotes express collagen-binding adhesins. Collagen fibers are thin, long, and rope-like right-handed triple helical structures composed of three parallel left-handed polypeptide strands coiled about each other. Each polypeptide strand is composed of a repeating Gly-X-Y sequence, where X and Y can be of any amino acid; however, proline (Pro) and hydroxyproline (HPO) residues are most often found in these positions.45 The triple helical structure of the collagen presents significant constraints for structural analysis, by itself and/or when complexed with its receptors. The first and only crystal structure of a collagen-bound eukaryotic protein complex is the I-domain of integrin α2β1 in complex with a collagen-derived peptide ligand. 32 A metal ion (Mg2+) present in the I-domain mediates the protein–ligand interactions, and a Glu residue of the ligand collagen completes the I-domain metal coordination and stabilizes the complex.
There are many collagen-binding adhesins (MSCRAMMs) in Gram-positive bacteria,10 but they do not share sequence and structural homology with the collagen-binding I-domain and also do not require a metal ion for collagen binding. CNA, the collagen-binding MSCRAMM of S. aureus with an N-terminal 55-kDa non-repetitive collagen-binding A-region, followed by a C-terminal 23-kDa repetitive B-region, binds collagen with moderate affinity 46 [Fig. 2(b)].
The ligand-binding A-region
The A-region of CNA is composed of N1, N2, and N3 domains and the N2 domain; the minimal collagen-binding region21 47 displays a single domain having a DEv-IgG fold with one D-type isopeptide bond. Three strands and a helix are observed as the additional elements at the edge of the sheets; thus, each sheet consists of five anti-parallel β-strands: β-sheet I (A,B,E,D,D*) and β-sheet II (D′,D″,C,F,G) [Fig. 2(b)]. The molecular surface of the N2 domain revealed an apparent groove that is 1–5 Å deep and 10–15 Å wide on β-sheet I, complementary to the triple helical collagen motif. 13 21 A similar model was proposed for ACE, 48 the collagen-binding MSCRAMM of E. faecalis, where the ACE19 subdomain exhibits 95% sequence identity with the CNA N2 domain. However, ACE is made of only two domains, N1 and N2, and has the same affinity toward collagen as CNA. 49
The crystal structures of CNAN2N3 determined in the apo form and in complex with a synthetic, collagen-like triple-helical peptide50 revealed N1 and N2 domains exhibiting a DEv-IgG fold. 51 In the apo form, the N1 and N2 domains are connected by a long flexible linker, which creates a distinct hole between the two domains [Figs. 5(a,b)]. Surprisingly, the C-terminus of the N2 domain extends toward and into the N1 domain and forms a β-strand that complements one of its β-sheets [Figs. 5(a,b)].
Figure 5.
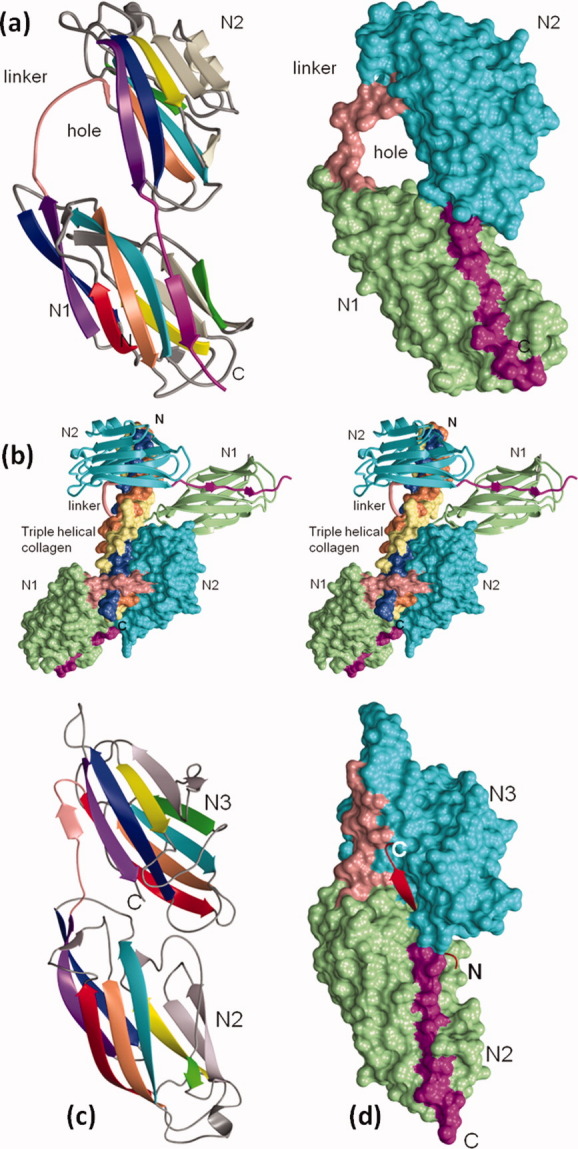
Crystal structures of MSCRAMMs as apo- and in complex with ligands. (a) Ribbon and surface representation collagen-binding domains (N1 and N2) of CNA. In the ribbon diagram, the N1 and N2 domains are shown in rainbow colors, whereas in the surface diagram they are shown in light green and cyan, respectively. A long flexible linker (in pink) connects the N1 and N2 domains. The C-terminus of the N2 domain extends toward and into the N1 domain and forms a β-strand (in magenta) that complements one of the β-sheets of the N1 domain. The linker is partially disordered in the apo structure and it is modeled to show the hole between the N1 and N2 domains. (b) Stereo representation of CNAN1N2 in complex with collagen-like triple helical peptide. The three chains of the collagen peptide (in the surface representation) are leading (L) (coral), middle (M) (light yellow), and trailing (T) (light blue), as viewed from their N-termini. Two CNA molecules [one in the ribbon and another in surface representation and colored as in Fig. 5(a)] interact with the collagen peptide in an identical manner. The leading and trailing chains of the peptide interact with the N2 domain and the middle chain with N1. The N1-N2 linker covers the leading and trailing chains and holds the rope-like ligand in place. (c) Ribbon representation of N2 and N3 domains of SdrG. N2 and N3 domains are shown in rainbow colors. The N2-N3 linker is represented in pink and the C-terminal is disordered in the apo structure. (d) Surface representation of SdrGN2N3 in complex with the fibrinogen peptide. The N2 domain is in light green, N3 is in cyan and the inter-domain linker is in pink, whereas the Fg peptide is shown as a ribbon in red. The C-terminal latch region is shown in magenta. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Unlike the prokaryotic I-domain-bound collagen complex,52 hydrophobic interactions dominate the CNAN2N3-collagen peptide complex. In the complex structure, 50 the collagen triple helix is seen penetrating the hole between the N1 and N2 domains of two CNA molecules [Fig. 5(b)], where one molecule is seen translated along the helical axis from the other by adopting the twist and pitch of the triple helix. The two CNAN2N3 molecules exhibit identical interactions with collagen, which are primarily hydrophobic, in addition to several hydrogen bonds and van der Waals contacts. The inter-domain “hole” is about 12–5 Å in diameter [Fig. 5(a,b)], into which the GPO repeating units of the collagen peptide fit snugly, leaving no extra space to accommodate any other larger side chain for X and Y amino acids, suggesting high specificity.
The inter-domain linker region, which is essential for the confinement of collagen,21 covers only one chain of the bound peptide and has no specific contacts with the other two chains of the triple helical motif. Hydrophobic interactions between the residues of the N2 domain trench region and the Pro residues of the collagen peptide and polar interactions involving the HPO dominate this collagen-receptor complex. A single 310 helix, observed in the apo-CBD35 structure adjacent to the N1 and N2 domain linkers, was transformed into a β-strand upon ligand binding, which results in an apparent shrinkage of the inter-domain hole to fit snugly around the bound ligand. 50
The B-region of CNA
The B-region of CNA in S. aureus (strain FDA574), which has no role in collagen-binding,53 contains three 23-kDA repeating units (B1, B2, and B3) [Fig. 2(c)]. The B1 structure has two similar domains (D1 and D2) placed side by side, and each domain displays a four- (DAGF) and three-strand (CBE) arrangement, similar to the IgG-rev fold observed in pilin domains. 22 Each D domain of the IgG-rev fold has an E-type isopeptide bond. In the B1B2 structure, the B1 (D1 and D2) and B2 (D3 and D4), with almost identical primary sequences (99.5%), display similar domain structures. A Gly residue present in the D1–D2 and D3–D4 linkers provides the needed flexibility between the D subdomains, whereas the linker joining B1 and B2 repeats (D2–D3), without such a Gly residue, have restricted flexibility. The restricted movements between the B repeats and the apparent flexibility between the D subdomains of individual repeats may facilitate the B-region to act as a “stalk” that projects the A-region away from the bacterial cell surface and positions it for binding collagen. 53
The “collagen hug” model
The closed conformation of E. faecalis ACE40, stabilized by a engineered disulfide link between the latch sequence (C-terminal extension of N2 domain) and the bottom of the N1 domain, did not bind type I collagen, proving that only an open conformation of ACE (and CNA) is capable of binding collagen.49 A multistep “Collagen Hug” model was proposed based on the fact that collagens are long (∼200 Å) rope-like structures that exhibit side chains of different sizes, shapes, and polarity for amino acids at the X and Y positions in G-X-Y repeating units and, thus, could not be “threaded” through the “hole” seen between the N1 and N2 domains of the apo-CNA35 structure. Upon ligand binding, initiated by low-affinity interactions between the residues present in the shallow “trench” of N2 domain and the ligand, the inter-domain linker wraps around the triple helical ligand and repositions the N1 domain to cover it. The inter-domain “hole” then shrinks like a clamp, triggered by some specific conformational changes, and CNA snugly tightens around the bound collagen. The N2 domain's C-terminal extension (connected to N3 domain in CNA) occupies the “latch” space available in the N1 domain β-sheet I to further secure the ligand. 49 The Collagen Hug model also suggests that CNA can only bind to monomeric forms of collagen, which are naturally occurring or generated through tissue injury. 54 55
The Fg-binding MSCRAMMs
Fg is a glycoprotein found in blood plasma that plays key roles in hemostatis and coagulation56 and is composed of six polypeptide chains (two α-, two β- and two γ-chains) arranged in a symmetrical dimeric complex structure. The C-terminus of the Fg γ-chain is targeted by the pathogen S. aureus, resulting in Fg-dependent cell clumping and tissue adherence, 11 and the same region also interacts with the platelet integrin αIIβ3 during Fg-dependent platelet adherence and aggregation. 57
Clumping factor A (ClfA) of S. aureus was the first MSCRAMM identified targeting the γ-chain of Fg, and ClfB was identified later to bind specifically to the Fg α-chain.10 Both ClfA and ClfB contain a Ser-Asp repeat region (R region) in the C-terminal part of the protein in addition to all other common features observed in a typical MSCRAMM. The Fg-binding activity of ClfA is affected by the divalent cations Ca2+ and Mn2+, analogous to an αIIβ3 integrin. 58 59 Higher concentrations of both cations inhibit Fg-dependent bacterial clumping and binding of the recombinant ClfA to isolated Fg. 51
The ligand-binding region of ClfA
The crystal structure of the stable and minimum Fg-binding fragment of ClfA51 revealed two compact domains (N2 and N3) with a DEv-IgG fold. The recombinant ClfA N1 and N2 domains fail to bind Fg when expressed independently, suggesting a cooperative ligand-binding role for both domains.
The “dock, lock, and latch” model
The crystal structures of the Fg-binding A region N2N3 segment of Staphylococcus epidermidis MSCRAMM SdrG,60 determined as apo- and in complex with a synthetic peptide similar to the N-terminal 6–20 of human Fg β-chain 60 [Fig. 5(c,d)], revealed two domains (N2 and N3) having the topology of a DEv-IgG fold. A similar arrangement is also seen in the recently determined peptide-bound ClfAN2N3 complex structure. 61 The Fg-derived peptides in both ClfA and SdrG complexes are in an extended conformation and fit snugly into a cleft (∼30 Å in length) present between the respective N2 and N3 domains. With the C-terminus of N3 domain directed into the solvent, the apo-SdrGN2N3 and apo-ClfaN2N3 crystal structures exhibit an unoccupied space in the respective N2 domain β-sheet I, sufficient for an extra β-stand between their respective D and E strands. However, in the complex structure, once the ligand peptide is positioned in the “cleft” region and stabilized by hydrogen bonds and hydrophobic interactions, the N3 C-terminus is redirected and forms a regular β-strand that is inserted in the unoccupied space between the D and E strands of the above-mentioned N2 domain β-sheet. Based on these observed conformational changes, a “dock, lock, and latch” (DLL) model was proposed in which the ligand binding takes place in multiple steps: the ligand peptide docks into the cleft, followed by structural rearrangement at the C-terminus, which crosses over the binding cleft and locks the bound peptide in place. The resulting hydrogen bonds between the peptide and adhesin secure the ligand in the binding pocket, and the final step is latching, which stabilizes the complex by the insertion of the N3 C-terminus as a strand between the D and E strands of N2 domain, completing the intra-molecular β-strand complementation. The TYTFTDYVD-like motif present in the back of the latching cleft is conserved across MSCRAMMs such as ClfA, ClfB, CNA, and ACE, suggesting that all Gram-positive bacterial adhesins might be using similar ligand-binding mechanism.
Significantly, a stabilized closed conformation of SdrGN2N3 (locked in closed conformation by an engineered disulfide link) does not bind to Fg, whereas a similarly engineered ClfA does bind the ligand,61 suggesting that for SdrG, an open conformation is required for the initial docking of the ligand peptide but not for ClfA. The ClfAN2N3 in a closed conformation bound the ligand with higher affinity compared to the wild-type ClfAN2N3, and this observation raises the possibility that Fg peptide binding to ClfA involves a different mechanism or a variation of DLL.
Fn-binding MSCRAMMs
Fn is a ∼440-kDa glycoprotein found in the ECM and body fluids of vertebrates, and it is involved in many cellular processes including cell adhesion, growth, migration, tissue repair, and blood clotting. Many MSCRAMMs of staphylococci and streptococci target Fn use D repeats in their C-terminal region. The MSCRAMM-binding site present in the N-terminal domain of Fn contains five sequential type I repeats (1–5F1). An NMR structure of the 1–2F1 module pair complexed with the FnBP peptide (B3) from S. dysgalactiae was determined, and a tandem β-zipper model was proposed for such binding, where the 1F1- and 2F1-binding motif in B3 forms an additional antiparallel β-strand on sequential F1 modules.62–64
The “tandem β-zipper” model
Two Fn-binding MSCRAMMs (FnBPA and FnBPB) were identified in S. aureus65 that contain multiple Fn-binding repeats (FnBRs) in the C-terminal region, six of which bind the Fn NTD with high affinity. The crystal structures of the FnBPA-1 and FnBPA-5 peptides (STAFF1 and STATT5) with F1 module pairs (2–3F1, 4–5F1) were solved as (2–3F1-STATT1, 4–5F1-STAFF1, 2–3F1-STATT5, and 4–5F1-STAFF5) complexes. 66 The FnBP peptide, with four short conserved motifs, binds Fn by adding an antiparallel strand to the three-stranded β-sheet of the four sequential F1 modules. 62 The B3 peptide-bound F1 module structures are very similar to peptide-free forms. The FnBPA-1/Fn NTD and FnBPA-5/Fn NTD interactions are dominated by hydrophobic and electrostatic interactions with a large buried surface area (∼4300 Å2). Interestingly, the FnBPA also binds to Fg via its N-terminal A-region, and this bi-functionality of FnBPA may facilitate disease progression by synergistic and/or cumulative adhesion 67 to the host ECM.
Conclusions
The structural biology of Gram-positive adhesins is an emerging field, and considerable progress has been made in this field in a relatively short time. The structural understanding of the bacterial tools used for adhesion will facilitate the design of new classes of antimicrobials, whose necessity is obvious in light of prevailing antibiotic resistance. Starting from the crystal structure of the surface protein CNA ligand-binding domain,21 a steady stream of adhesins and their host ligand complex crystal structures have been determined and they have made significant contributions to our understanding. Recent rapid progress in determining the structures of individual pilins, starting with the minor pilin GBS52 of S. agalactiae 15 and studies to decipher their spatial position in respective pili and role in host adhesion, are broadening our understanding of microbial adhesion in general, and specifically providing insight into Gram-positive pili assembly. By employing variants of topologically simple human immunoglobulin IgG constant domain as the building block for their adhesive proteins, microbes have gained a tactical advantage and seem to be a few steps ahead of the human immune arsenal, forcing us to invent or design new therapies. The microbes have perfected the utilization of IgG domains for adhesion and have further improved their adaptability by employing a “donor strand complementation” technique, inter-molecular in the case of Gram-negatives and intra-molecular by Gram-positives, for pilus assembly and ligand adhesion, respectively. Furthermore, the Gram-positives utilize the IgG domains, hosting inter- and intra-molecular isopeptide bonds for resistance against proteolytic degradation and for pili polylemarization, respectively, and the later reactions are catalyzed by the unique and conserved cysteine transpeptidase sortase, which sets them apart from Gram-negative pathogens.
References
- 1.Kline KA, Dodson KW, Caparon MG, Hultgren SJ. A tale of two pili: assembly and function of pili in bacteria. Trends Microbiol. 2010;18:224–232. doi: 10.1016/j.tim.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sauer FG, Mulvey MA, Schilling JD, Martinez JJ, Hultgren SJ. Bacterial pili: molecular mechanisms of pathogenesis. Curr Opin Microbiol. 2000;3:65–72. doi: 10.1016/s1369-5274(99)00053-3. [DOI] [PubMed] [Google Scholar]
- 3.Waksman G, Hultgren SJ. Structural biology of the chaperone-usher pathway of pilus biogenesis. Nat Rev Microbiol. 2009;7:765–774. doi: 10.1038/nrmicro2220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Choudhury D, Thompson A, Stojanoff V, Langermann S, Pinkner J, Hultgren SJ, Knight SD. X-ray structure of the FimC-FimH chaperone-adhesin complex from uropathogenic Escherichia coli. Science. 1999;285:1061–1066. doi: 10.1126/science.285.5430.1061. [DOI] [PubMed] [Google Scholar]
- 5.Scott JR, Zahner D. Pili with strong attachments: Gram-positive bacteria do it differently. Mol Microbiol. 2006;62:320–330. doi: 10.1111/j.1365-2958.2006.05279.x. [DOI] [PubMed] [Google Scholar]
- 6.Telford JL, Barocchi MA, Margarit I, Rappuoli R, Grandi G. Pili in Gram-positive pathogens. Nat Rev Microbiol. 2006;4:509–519. doi: 10.1038/nrmicro1443. [DOI] [PubMed] [Google Scholar]
- 7.Ton-That H, Schneewind O. Assembly of pili in Gram-positive bacteria. Trends Microbiol. 2004;12:228–234. doi: 10.1016/j.tim.2004.03.004. [DOI] [PubMed] [Google Scholar]
- 8.Ton-That H, Schneewind O. Assembly of pili on the surface of Corynebacterium diphtheriae. Mol Microbiol. 2003;50:1429–1438. doi: 10.1046/j.1365-2958.2003.03782.x. [DOI] [PubMed] [Google Scholar]
- 9.Cossart P, Jonquieres R. Sortase, a universal target for therapeutic agents against Gram-positive bacteria? Proc Natl Acad Sci USA. 2000;97:5013–5015. doi: 10.1073/pnas.97.10.5013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Patti JM, Allen BL, McGavin MJ, Höök M. MSCRAMM-mediated adherence of microorganisms to host tissues. Annu Rev Microbiol. 1994;48:585–617. doi: 10.1146/annurev.mi.48.100194.003101. [DOI] [PubMed] [Google Scholar]
- 11.Hawiger J, Timmons S, Strong DD, Cottrell BA, Riley M, Doolittle RF. Identification of a region of human fibrinogen interacting with staphylococcal clumping factor. Biochemistry. 1982;21:1407–1413. doi: 10.1021/bi00535a047. [DOI] [PubMed] [Google Scholar]
- 12.Kuusela P. Fibronectin binds to Staphylococcus aureus. Nature. 1978;276:718–720. doi: 10.1038/276718a0. [DOI] [PubMed] [Google Scholar]
- 13.Speziale P, Raucci G, Visai L, Switalski LM, Timpl R, Höök M. Binding of collagen to Staphylococcus aureus Cowan 1. J Bacteriol. 1986;167:77–81. doi: 10.1128/jb.167.1.77-81.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schneewind O, Model P, Fischetti VA. Sorting of protein A to the staphylococcal cell wall. Cell. 1992;70:267–281. doi: 10.1016/0092-8674(92)90101-h. [DOI] [PubMed] [Google Scholar]
- 15.Krishnan V, Gaspar AH, Ye N, Mandlik A, Ton-That H, Narayana SV. An IgG-like domain in the minor pilin GBS52 of Streptococcus agalactiae mediates lung epithelial cell adhesion. Structure. 2007;15:893–903. doi: 10.1016/j.str.2007.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kang HJ, Paterson NG, Gaspar AH, Ton-That H, Baker EN. The Corynebacterium diphtheriae shaft pilin SpaA is built of tandem Ig-like modules with stabilizing isopeptide and disulfide bonds. Proc Natl Acad Sci USA. 2009;106:16967–16971. doi: 10.1073/pnas.0906826106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Izoré T, Contreras-Martel C, El Mortaji L, Manzano C, Terrasse R, Vernet T, Di Guilmi AM, Dessen A. Structural basis of host cell recognition by the pilus adhesin from Streptococcus pneumoniae. Structure. 2010;18:106–115. doi: 10.1016/j.str.2009.10.019. [DOI] [PubMed] [Google Scholar]
- 18.Budzik JM, Poor CB, Faull KF, Whitelegge JP, He C, Schneewind O. Intramolecular amide bonds stabilize pili on the surface of bacilli. Proc Natl Acad Sci USA. 2009;106:19992–19997. doi: 10.1073/pnas.0910887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bork P, Holm L, Sander C. The immunoglobulin fold. Structural classification, sequence patterns and common core. J Mol Biol. 1994;242:309–320. doi: 10.1006/jmbi.1994.1582. [DOI] [PubMed] [Google Scholar]
- 20.Harpaz Y, Chothia C. Many of the immunoglobulin superfamily domains in cell adhesion molecules and surface receptors belong to a new structural set which is close to that containing variable domains. J Mol Biol. 1994;238:528–539. doi: 10.1006/jmbi.1994.1312. [DOI] [PubMed] [Google Scholar]
- 21.Symersky J, Patti JM, Carson M, House-Pompeo K, Teale M, Moore D, Jin L, Schneider A, DeLucas LJ, Höök M, Narayana SV. Structure of the collagen-binding domain from a Staphylococcus aureus adhesin. Nat Struct Biol. 1997;4:833–838. doi: 10.1038/nsb1097-833. [DOI] [PubMed] [Google Scholar]
- 22.Deivanayagam CC, Rich RL, Carson M, Owens RT, Danthuluri S, Bice T, Höök M, Narayana SV. Novel fold and assembly of the repetitive B region of the Staphylococcus aureus collagen-binding surface protein. Structure. 2000;8:67–78. doi: 10.1016/s0969-2126(00)00081-2. [DOI] [PubMed] [Google Scholar]
- 23.Yanagawa R, Otsuki K, Tokui T. Electron microscopy of fine structure of Corynebacterium renale with special reference to pili. Jpn J Vet Res. 1968;16:31–37. [PubMed] [Google Scholar]
- 24.Mandlik A, Swierczynski A, Das A, Ton-That H. Pili in Gram-positive bacteria: assembly, involvement in colonization and biofilm development. Trends Microbiol. 2008;16:33–40. doi: 10.1016/j.tim.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Proft T, Baker EN. Pili in Gram-negative and Gram-positive bacteria—structure, assembly and their role in disease. Cell Mol Life Sci. 2009;66:613–635. doi: 10.1007/s00018-008-8477-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kang HJ, Coulibaly F, Clow F, Proft T, Baker EN. Stabilizing isopeptide bonds revealed in Gram-positive bacterial pilus structure. Science. 2007;318:1625–1628. doi: 10.1126/science.1145806. [DOI] [PubMed] [Google Scholar]
- 27.Kang HJ, Baker EN. Intramolecular isopeptide bonds give thermodynamic and proteolytic stability to the major pilin protein of Streptococcus pyogenes. J Biol Chem. 2009;284:20729–20737. doi: 10.1074/jbc.M109.014514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pickart CM. Mechanisms underlying ubiquitination. Annu Rev Biochem. 2001;70:503–533. doi: 10.1146/annurev.biochem.70.1.503. [DOI] [PubMed] [Google Scholar]
- 29.Greenberg CS, Birckbichler PJ, Rice RH. Transglutaminases: multifunctional cross-linking enzymes that stabilize tissues. FASEB J. 1991;5:3071–3077. doi: 10.1096/fasebj.5.15.1683845. [DOI] [PubMed] [Google Scholar]
- 30.Linke C, Young PG, Kang HJ, Bunker RD, Middleditch MJ, Caradoc-Davies TT, Proft T, Baker EN. Crystal structure of the minor pilin FctB reveals determinants of Group A streptococcal pilus anchoring. J Biol Chem. 2010;285:20381–20389. doi: 10.1074/jbc.M109.089680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mandlik A, Das A, Ton-That H. The molecular switch that activates the cell wall anchoring step of pilus assembly in Gram-positive bacteria. Proc Natl Acad Sci USA. 2008;105:14147–14152. doi: 10.1073/pnas.0806350105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;101:47–56. doi: 10.1016/S0092-8674(00)80622-4. [DOI] [PubMed] [Google Scholar]
- 33.Krishnan V, Xu Y, Macon K, Volanakis JE, Narayana SV. The crystal structure of C2a, the catalytic fragment of classical pathway C3 and C5 convertase of human complement. J Mol Biol. 2007;367:224–233. doi: 10.1016/j.jmb.2006.12.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ponnuraj K, Xu Y, Macon K, Moore D, Volanakis JE, Narayana SV. Structural analysis of engineered Bb fragment of complement factor B: insights into the activation mechanism of the alternative pathway C3-convertase. Mol Cell. 2004;14:17–28. doi: 10.1016/s1097-2765(04)00160-1. [DOI] [PubMed] [Google Scholar]
- 35.Konto-Ghiorghi Y, Mairey E, Mallet A, Dumenil G, Caliot E, Trieu-Cuot P, Dramsi S. Dual role for pilus in adherence to epithelial cells and biofilm formation in Streptococcus agalactiae. PLoS Pathog. 2009;5:e1000422. doi: 10.1371/journal.ppat.1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lauer P, Rinaudo CD, Soriani M, Margarit I, Maione D, Rosini R, Taddei AR, Mora M, Rappuoli R, Grandi G, Telford JL. Genome analysis reveals pili in Group B Streptococcus. Science. 2005;309:105. doi: 10.1126/science.1111563. [DOI] [PubMed] [Google Scholar]
- 37.Dramsi S, Caliot E, Bonne I, Guadagnini S, Prevost MC, Kojadinovic M, Lalioui L, Poyart C, Trieu-Cuot P. Assembly and role of pili in group B streptococci. Mol Microbiol. 2006;60:1401–1413. doi: 10.1111/j.1365-2958.2006.05190.x. [DOI] [PubMed] [Google Scholar]
- 38.Rosini R, Rinaudo CD, Soriani M, Lauer P, Mora M, Maione D, Taddei A, Santi I, Ghezzo C, Brettoni C, Buccato S, Margarit I, Grandi G, Telford JL. Identification of novel genomic islands coding for antigenic pilus-like structures in Streptococcus agalactiae. Mol Microbiol. 2006;61:126–141. doi: 10.1111/j.1365-2958.2006.05225.x. [DOI] [PubMed] [Google Scholar]
- 39.Vengadesan K, Ma X, Dwivedi P, Ton-That H, Narayana SV. Purification, crystallization and halide phasing of a Streptococcus agalactiae backbone pilin GBS80 fragment. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010;66:1666–1669. doi: 10.1107/S1744309110043101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Spraggon G, Koesema E, Scarselli M, Malito E, Biagini M, Norais N, Emolo C, Barocchi MA, Giusti F, Hilleringmann M, Rappuoli R, Lesley S, Covacci A, Masignani V, Ferlenghi I. Supramolecular organization of the repetitive backbone unit of the Streptococcus pneumoniae pilus. PLoS One. 2010;5:e10919. doi: 10.1371/journal.pone.0010919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Budzik JM, Oh SY, Schneewind O. Sortase D forms the covalent bond that links BcpB to the tip of Bacillus cereus pili. J Biol Chem. 2009;284:12989–12997. doi: 10.1074/jbc.M900927200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Swierczynski A, Ton-That H. Type III pilus of corynebacteria: Pilus length is determined by the level of its major pilin subunit. J Bacteriol. 2006;188:6318–6325. doi: 10.1128/JB.00606-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gaspar AH, Ton-That H. Assembly of distinct pilus structures on the surface of Corynebacterium diphtheriae. J Bacteriol. 2006;188:1526–1533. doi: 10.1128/JB.188.4.1526-1533.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mishra A, Das A, Cisar JO, Ton-That H. Sortase-catalyzed assembly of distinct heteromeric fimbriae in Actinomyces naeslundii. J Bacteriol. 2007;189:3156–3165. doi: 10.1128/JB.01952-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Shoulders MD, Raines RT. Collagen structure and stability. Annu Rev Biochem. 2009;78:929–958. doi: 10.1146/annurev.biochem.77.032207.120833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.House-Pompeo K, Boles JO, Höök M. Characterization of bacterial adhesin interactions with extracellular matrix components utilizing biosensor technology. Methods: A Companion to Meth Enzymol. 1994;6:134–142. [Google Scholar]
- 47.Patti JM, House-Pompeo K, Boles JO, Garza N, Gurusiddappa S, Höök M. Critical residues in the ligand-binding site of the Staphylococcus aureus collagen-binding adhesin (MSCRAMM) J Biol Chem. 1995;270:12005–12011. doi: 10.1074/jbc.270.20.12005. [DOI] [PubMed] [Google Scholar]
- 48.Ponnuraj K, Narayana SV. Crystal structure of ACE19, the collagen binding subdomain of Enterococus faecalis surface protein ACE. Proteins. 2007;69:199–203. doi: 10.1002/prot.21464. [DOI] [PubMed] [Google Scholar]
- 49.Liu Q, Ponnuraj K, Xu Y, Ganesh VK, Sillanpaa J, Murray BE, Narayana SV, Höök M. The Enterococcus faecalis MSCRAMM ACE binds its ligand by the Collagen Hug model. J Biol Chem. 2007;282:19629–19637. doi: 10.1074/jbc.M611137200. [DOI] [PubMed] [Google Scholar]
- 50.Zong Y, Xu Y, Liang X, Keene DR, Höök A, Gurusiddappa S, Höök M, Narayana SV. A 'Collagen Hug' model for Staphylococcus aureus CNA binding to collagen. EMBO J. 2005;24:4224–4236. doi: 10.1038/sj.emboj.7600888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Deivanayagam CC, Wann ER, Chen W, Carson M, Rajashankar KR, Höök M, Narayana SV. A novel variant of the immunoglobulin fold in surface adhesins of Staphylococcus aureus: crystal structure of the fibrinogen-binding MSCRAMM, clumping factor A. EMBO J. 2002;21:6660–6672. doi: 10.1093/emboj/cdf619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley J, Knight CG, Farndale RW, Barnes MJ. Structure of the integrin α2β1-binding collagen peptide. J Mol Biol. 2004;335:1019–1028. doi: 10.1016/j.jmb.2003.11.030. [DOI] [PubMed] [Google Scholar]
- 53.Rich RL, Demeler B, Ashby K, Deivanayagam CC, Petrich JW, Patti JM, Narayana SV, Höök M. Domain structure of the Staphylococcus aureus collagen adhesin. Biochemistry. 1998;37:15423–15433. doi: 10.1021/bi981773r. [DOI] [PubMed] [Google Scholar]
- 54.Liotta LA, Tryggvason K, Garbisa S, Hart I, Foltz CM, Shafie S. Metastatic potential correlates with enzymatic degradation of basement membrane collagen. Nature. 1980;284:67–68. doi: 10.1038/284067a0. [DOI] [PubMed] [Google Scholar]
- 55.Kehrel B. Platelet-collagen interactions. Semin Thromb Hemost. 1995;21:123–129. doi: 10.1055/s-2007-1000386. [DOI] [PubMed] [Google Scholar]
- 56.Herrick S, Blanc-Brude O, Gray A, Laurent G. Fibrinogen. Int J Biochem Cell Biol. 1999;31:741–746. doi: 10.1016/s1357-2725(99)00032-1. [DOI] [PubMed] [Google Scholar]
- 57.Kloczewiak M, Timmons S, Lukas TJ, Hawiger J. Platelet receptor recognition site on human fibrinogen. Synthesis and structure-function relationship of peptides corresponding to the carboxy-terminal segment of the γ chain. Biochemistry. 1984;23:1767–1774. doi: 10.1021/bi00303a028. [DOI] [PubMed] [Google Scholar]
- 58.Smith JW, Piotrowicz RS, Mathis D. A mechanism for divalent cation regulation of β3-integrins. J Biol Chem. 1994;269:960–967. [PubMed] [Google Scholar]
- 59.D'Souza SE, Haas TA, Piotrowicz RS, Byers-Ward V, McGrath DE, Soule HR, Cierniewski C, Plow EF, Smith JW. Ligand and cation binding are dual functions of a discrete segment of the integrin β3 subunit: cation displacement is involved in ligand binding. Cell. 1994;79:659–667. doi: 10.1016/0092-8674(94)90551-7. [DOI] [PubMed] [Google Scholar]
- 60.Ponnuraj K, Bowden MG, Davis S, Gurusiddappa S, Moore D, Choe D, Xu Y, Höök M, Narayana SV. A “dock, lock, and latch” structural model for a staphylococcal adhesin binding to fibrinogen. Cell. 2003;115:217–228. doi: 10.1016/s0092-8674(03)00809-2. [DOI] [PubMed] [Google Scholar]
- 61.Ganesh VK, Rivera JJ, Smeds E, Ko YP, Bowden MG, Wann ER, Gurusiddappa S, Fitzgerald JR, Höök M. A structural model of the Staphylococcus aureus ClfA-fibrinogen interaction opens new avenues for the design of anti-staphylococcal therapeutics. PLoS Pathog. 2008;4:e1000226. doi: 10.1371/journal.ppat.1000226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schwarz-Linek U, Werner JM, Pickford AR, Gurusiddappa S, Kim JH, Pilka ES, Briggs JA, Gough TS, Höök M, Campbell ID, Potts JR. Pathogenic bacteria attach to human fibronectin through a tandem β-zipper. Nature. 2003;423:177–181. doi: 10.1038/nature01589. [DOI] [PubMed] [Google Scholar]
- 63.Schwarz-Linek U, Pilka ES, Pickford AR, Kim JH, Höök M, Campbell ID, Potts JR. High-affinity streptococcal binding to human fibronectin requires specific recognition of sequential F1 modules. J Biol Chem. 2004;279:39017–39025. doi: 10.1074/jbc.M405083200. [DOI] [PubMed] [Google Scholar]
- 64.Schwarz-Linek U, Höök M, Potts JR. Fibronectin-binding proteins of Gram-positive cocci. Microbes Infect. 2006;8:2291–2298. doi: 10.1016/j.micinf.2006.03.011. [DOI] [PubMed] [Google Scholar]
- 65.Jonsson K, Signas C, Muller HP, Lindberg M. Two different genes encode fibronectin binding proteins in Staphylococcus aureus. The complete nucleotide sequence and characterization of the second gene. Eur J Biochem. 1991;202:1041–1048. doi: 10.1111/j.1432-1033.1991.tb16468.x. [DOI] [PubMed] [Google Scholar]
- 66.Bingham RJ, Rudino-Pinera E, Meenan NA, Schwarz-Linek U, Turkenburg JP, Höök M, Garman EF, Potts JR. Crystal structures of fibronectin-binding sites from Staphylococcus aureus FnBPA in complex with fibronectin domains. Proc Natl Acad Sci U S A. 2008;105:12254–12258. doi: 10.1073/pnas.0803556105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wann ER, Gurusiddappa S, Höök M. The fibronectin-binding MSCRAMM FnbpA of Staphylococcus aureus is a bifunctional protein that also binds to fibrinogen. J Biol Chem. 2000;275:13863–13871. doi: 10.1074/jbc.275.18.13863. [DOI] [PubMed] [Google Scholar]
- 68.Bowden MG, Heuck AP, Ponnuraj K, Kolosova E, Choe D, Gurusiddappa S, Narayana SV, Johnson AE, Höök M. Evidence for the “dock, lock, and latch” ligand binding mechanism of the staphylococcal microbial surface component recognizing adhesive matrix molecules (MSCRAMM) SdrG. J Biol Chem. 2008;283:638–647. doi: 10.1074/jbc.M706252200. [DOI] [PubMed] [Google Scholar]
- 69.Vengadesan K, Ma X, Dwivedi P, Ton-That H, Narayana SV. A model for Group B Streptococcus pilus type 1: The structure of a 35-kDa C-terminal fragment of the major pilin GBS80. J Mol Biol. 2011;407:731–743. doi: 10.1016/j.jmb.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]



