Abstract
Hemopexin is a plasma protein that plays a well-established biological role in sequestering heme that is released into the plasma from hemoglobin and myoglobin as the result of intravascular or extravascular hemolysis as well as from skeletal muscle trauma or neuromuscular disease. In recent years, a variety of additional biological activities have been attributed to hemopexin, for example, hyaluronidase activity, serine protease activity, pro-inflammatory and anti-inflammatory activity as well as suppression of lymphocyte necrosis, inhibition of cellular adhesion, and binding of divalent metal ions. This review examines the challenges involved in the purification of hemopexin from plasma and in the recombinant expression of hemopexin and evaluates the questions that these challenges and the characteristics of hemopexin raise concerning the validity of many of the new activities proposed for this protein. As well, an homology model of the three-dimensional structure of human hemopexin is used to reveal that the protein lacks the catalytic triad that is characteristic of many serine proteases but that hemopexin possesses two highly exposed Arg-Gly-Glu sequences that may promote interaction with cell surfaces.
Keywords: hemopexin, serine protease, anti-inflammatory, pro-inflammatory, lymphocyte necrosis factor, metal ion binding, cell adhesion, heme binding
Introduction
Hemopexin1† is a plasma β1-glycoprotein and a positive acute phase reactant protein2,3 that binds heme that is released into the blood as the result of intra- and extra-vascular hemolysis or rhabodomyolysis and transports it principally to the liver. By sequestering heme that is released into blood and transporting it safely to the liver for catabolism, plasma hemopexin plays the dual role of promoting the metabolic processing of heme and its iron as well as inhibiting the toxicity resulting from the oxidative catalytic activity of heme. These biological consequences of heme binding by circulating hemopexin are well documented and have been reviewed extensively.4–7 Physical–chemical understanding of hemopexin and the complex it forms with heme were advanced significantly by crystallographic determination of the three-dimensional structure of the rabbit hemopexin-heme complex by Baker and coworkers.1,8 This structure established that hemopexin is comprised of homologous N- and C-terminal domains having similar 4-bladed β-propeller folding motifs that are joined by a flexible hinge sequence. In this structure, heme is shown to bind to the protein by coordination of the heme iron to one His residue in the flexible linker region and another in the C-terminal domain.
In addition to heme binding, hemopexin has also been reported to (i) possess serine protease activity, (ii) exhibit anti- and pro-inflammatory activities, (iii) suppress the necrosis of polymorphonuclear lymphocytes, (iv) inhibit cellular adhesion, and (v) bind certain divalent metal ions. In view of the significant biological implications of these proposed diverse activities, the evidence on which they are based merits serious consideration. Before undertaking such an analysis, we consider the various means by which hemopexin has been obtained for experimental investigation, how it responds to a variety of treatments, and the limitations to the conditions that native hemopexin will tolerate in vitro. As discussed below, studies using hemopexin prepared by methods that expose the protein to conditions known to damage it have led to the proposal of new activities for hemopexin that cannot be unambiguously attributed to the native protein.
Purification of Hemopexin from Blood
A major factor in considering the data reported in the published hemopexin literature is the quality of the protein used in experimental work. Most studies of hemopexin involve protein isolated from blood plasma, and a variety of purification strategies have been used. Published evidence establishes that historically, hemopexin has been challenging to purify.9,10 Transferrin, α2-macroglobulin, histidine-rich glycoprotein (also known as histidine-proline-rich glycoprotein), and immunoglobulins are common contaminants.10–15 Over 20 years ago, Müller-Eberhard noted in a review on hemopexin that methods for its purification relied on one of three characteristics of the protein, for example, its high affinity for heme, its extensive glycosylation, or its resistance to acid denaturation.10 At present, the methods for purification of hemopexin in quantities sufficient for physical studies can be grouped into those that rely primarily on an initial precipitation step and those that rely primarily on an affinity-based procedure. Some purification schemes use both. Differential precipitation-based methods typically involve initial treatment of fresh plasma, serum, or Cohn fraction IV with perchloric acid, ammonium sulfate, or Rivanol (2-ethoxy-6,9-diamino acridine monolactate monohydrate) to precipitate many contaminating proteins. The hemopexin-enriched fraction is then resolved further by various types of ion exchange and gel filtration chromatography to achieve purification. The most commonly used strategy based on affinity involves an initial fractionation of anti-coagulated plasma with heme-agarose affinity chromatography followed by a variety of ion exchange and gel filtration fractionations. Heme affinity resins differ in the manner by which the heme is coupled to the resin owing to different synthetic strategies,14–20 but Müller-Eberhard identified one18 that exhibits minimal binding of other plasma proteins. Recently, commercial heme-affinity resins have been used almost exclusively in such purifications of hemopexin, but the nature of the heme coupling to these resins is rarely if ever defined by the vendor. A variety of other chromatographic purification protocols have been used for hemopexin on the basis of its affinity for lectins, Cibacron Blue, metal ions, or antibodies.10,21 The quality and purity of the hemopexin recovered in these affinity protocols differs and is governed not only by elution conditions but also by the length and configuration of the coupling spacer, the coordination geometry and density as well as the chemical and physical properties of the support matrix. Procedures used subsequent to purification of the protein (e.g., dialysis, lyophilization, storage, and mode of freezing) all have the potential to alter various properties of the purified hemopexin.
Recombinant Hemopexin
In principle, recombinant hemopexin is an alternative to the protein purified from blood. Only a few studies have reported the isolation of recombinant hemopexin or the use of commercial recombinant protein. The first report of recombinant hemopexin involved expression of the rabbit protein in insect cells to enable substitution of amino acid residues that might serve as axial ligands to the heme iron.22 The NMR spectra of the recombinant proteins included in that report provide the most extensive characterization of any recombinant hemopexin yet reported. Subsequently, some authors have reported the use of recombinant human hemopexin they expressed in Pichia pastoris23,24 or they obtained from vendors that expressed the protein in mammalian cell culture.25 Unfortunately, these recombinant forms of hemopexin remain essentially uncharacterized. No evidence is available that these recombinant forms of hemopexin are properly folded, change conformation upon heme binding, or exhibit the spectroscopic or functional properties observed for native hemopexin purified from blood.
A major concern in expressing native, recombinant hemopexin is the proper formation of its six disulfide bonds. Each of the two hemopexin β-propeller domains has three disulfide bonds, one of which forms between a Cys residue toward the N-terminal end of the domain and another toward the C-terminal end of the domain. This latter disulfide bond unites the N- and C-terminal sequences of each domain and has been proposed to be crucial to stabilizing the characteristic β-propeller fold of each domain.26–28 For this reason, expression must be performed with a recombinant system that can support proper folding of hemopexin, or a refolding protocol that restores the native conformation must be identified and used. Although it is possible that correct glycosylation of the recombinant protein is essential for proper folding, the crystal structures of the glycosylated and enzymatically deglycosylated rabbit protein are essentially identical.1 Nevertheless, glycosylation of human hemopexin isolated from Cohn fraction IV by heme-Sepharose affinity chromatography15 was insufficient to ensure correct refolding of hemopexin following disulfide bond reduction and reoxidation.29 Thus, the role of glycosylation in folding of recombinantly expressed hemopexin remains unknown.
Verification that any purified hemopexin is folded correctly is limited to spectroscopic criteria because hemopexin lacks an intrinsic catalytic activity that can be used for this purpose. At present, the most compelling criterion of native hemopexin formation is afforded by the characteristic positive Cotton effect at 231 nm in the far UV CD spectrum.30–32 This unusual CD band, which is observed in the spectrum of a relatively small number of proteins,33,34 is exhibited by native hemopexin and intensifies upon binding of heme30 only if the hinge sequence that links the N- and C-domains is intact.35 Although the origin of this Cotton effect remains speculative, it may result from one or more Trp residues in an unusual environment that changes in response to heme binding to the native protein, and thus may reflect a realignment of the two domains with respect to each other.1,34 The visible electronic absorption spectrum of the hemopexin-heme complex is characteristic of a heme protein with low-spin, bis-histidine axial coordination. Such visible electronic spectra by themselves do not differentiate between native and non-native hemopexin because human hemopexin possesses 19 histidyl residues, and the coordination of any two of these to the heme iron could result in a visible spectrum that is indistinguishable from that of the native hemopexin-heme complex. For example, ferriporphyrins can bind to the N-domain fragment of hemopexin35 to produce a UV–vis absorption spectrum with characteristics similar to that of the native protein, thus providing evidence for bis-His coordination.8,35,36 However, CD and NMR spectroscopy of the complexes formed with the N-terminal domain and the native protein are distinctly different.35,37
Recombinant human hemopexin produced in Escherichia coli has been reported to be either unstable and rapidly degraded22 or to be stable and able to bind heme but lacking the positive Cotton effect at 231 nm.21 Human hemopexin produced in baculovirus-infected insect cells exhibited a 413.5 nm/280 nm absorbance ratio (for an equimolar mixture of hemopexin and heme) less than that of hemopexin isolated from plasma as well as a lowered affinity for heme.22 The far UV CD spectrum of this protein was not reported. Although recombinant human hemopexin expressed in P. pastoris has been used in at least one report,23 no evidence has been provided to establish that this protein is properly folded. Commercially available recombinant human hemopexin (e.g., R&D Systems, Minneapolis, MN; Creative Biomart, Shirley, NY; Sino Biological, Beijing, China) has the potential complication of a His-tag, which in the absence of information to the contrary may bind heme or may promote oligomerization of the protein.38,39 Little or no functional evidence is provided by the vendors to validate the structural authenticity of their recombinant protein. R&D Systems cites the ability of its product to bind protoporphyrin IX as detected by fluorescence emission spectroscopy. However, this criterion is nonspecific and provides no assurance that the protein is folded properly. As protoporphyrin IX lacks a central iron atom, it can bind to the heme binding site without requiring the axial ligands that normally stabilize heme binding, but it does so far less discriminately and with far lower affinity than does heme.40,41
Modification of Hemopexin during Purification
Conformational changes that may not be reversible
The low pH generally used to elute hemopexin from heme or antibody affinity resins (typically pH 2.0–2.514,25,42–44) is sufficient to unfold the protein.30 Although the ability of human hemopexin to bind heme following exposure to low pH largely recovers within several hours after return to neutrality,45 more than 48 h can be required for the protein to elute similarly to native hemopexin during hydrophobic chromatography.15 The positive Cotton effect at 231 nm in the CD spectrum of human hemopexin decreases in intensity as the pH is reduced below 3.1, it is abolished at pH 2.1, and only ∼75% of the original positive ellipticity is regained upon incubation at pH 7.4 for several hours (S.-I. Takayama, unpublished data). Human hemopexin purified by fractionation with ammonium sulfate or low pH is also susceptible to aggregation10,46,47 and to loss of sialic acid residues.48
The structure of hemopexin greatly affects its susceptibility to proteolysis. The native rabbit and human hemopexin-heme complexes are resistant to proteolysis,35,49 although the human hemopexin-heme complex can be hydrolyzed by trypsin at Lys101.21,49 The corresponding heme-free proteins are also relatively resistant to proteolysis except for the linker region that connects the N- and C- domains, which is susceptible to cleavage upon limited hydrolysis with trypsin or plasmin.1,35,50 However, human hemopexin that is prepared by the acidified acetone procedure51 is rapidly cleaved into many fragments by trypsin.49
Use of correctly folded hemopexin is as important to the assessment of its interaction(s) with cells and other biological activities as it is in all other functional studies of hemopexin. Conformational modification of or loss of carbohydrate from hemopexin could increase the hydrophobic character of the protein surface that could in turn enable abnormal or detrimental interactions with cell surfaces. This possibility is suggested by a report that apo-hemoglobin and its subunits, which are known to have large hydrophobic surface areas, mediate damage to the plasma membrane of endothelial cells.52
Potential chemical and structural modification of hemopexin
Mammalian hemopexins exhibit considerable similarity in amino acid and carbohydrate composition.10 Human hemopexin has a molecular weight of ∼59 kDa, of which 80% (49,292 Da) is attributable to its 439 amino acid residues and the remaining 20% is attributable to glycosylation in the form of bi- or tri-antennary carbohydrate structures including a currently unique N-terminal glycan.49,50,53,54 Human hemopexin is nearly monomorphic insofar as it exhibits polymorphisms only among black populations where very low levels of two other alleles have been reported.5,55,56 Confusion over the molecular weight of native human hemopexin may arise from apparent molecular weights reported on the basis of SDS-PAGE analyses because hemopexin exhibits an apparent molecular weight that is ∼25% greater under reducing conditions than that observed under nonreducing conditions.57 Many reports fail to indicate which of these conditions was used. The multiple hemopexin bands observed by isoelectric focusing of plasma samples are attributed to carbohydrate variability, especially the extent of sialylation.55 This variation in carbohydrate content results in a group of bands for human hemopexin observable on SDS-PAGE that can be attributed to at least 5 species with pI values of 5.46–6.36 that span ∼7 kDa range in mass (typically 58–65 kDa and 73–80 kDa under nonreducing and reducing conditions, respectively).57 The two most abundant forms observed in these reports differ by <2 kDa.55,57,58 Similar behavior is exhibited by other mammalian hemopexins. For example, reducing SDS-PAGE of rat hemopexin indicate an apparent molecular weight of 71 ± 3 kDa.59 The glycosylation of hemopexin as well as the extent of its sialylation varies from species to species60–62 and is highly complex and heterogeneous for the human protein. The human hemopexin glycan profile is representative of the variable, liver-specific glycoproteome.62 At present, however, the dependence of the functional properties of hemopexin on carbohydrate structure and its modification remains largely uncharacterized.
Exposure of hemopexin to acidic pH has the potential to result in a number of chemical modifications to the structure of human hemopexin, so the acidic conditions involved in some precipitation methods or in elution from affinity resins are a concern. For example, human hemopexin is susceptible to hydrolysis in dilute acid (pH 2)50 with the N-domain having most of the susceptible peptide bonds63; however, hydrolysis in dilute acid is relatively slow, so long reaction times are generally required for the reaction to proceed to a significant extent. On the other hand, the acidic conditions involved in isolation of hemopexin with some heme-agarose or antibody affinity matrices can lead to degradation of the carbohydrate portion of the glycoprotein,15,64 deamidation,65,66 or even dimerization/polymerization of the protein.46,47,67 Repeated exposure to pH 4 followed by return to neutral pH has been reported to result in progressive desialylation of hemopexin.48 Other chemical modifications have been reported for human hemopexin, for example, exposure of hemopexin to Ni2+ can result in hydrolysis of a susceptible peptide bond in the linker sequence.68 Carbonylation of hemopexin has been detected in patients with sporadic Alzheimer's disease69 and with idiopathic pulmonary fibrosis.70 Although the cause of this oxidation remains unknown, it raises the possibility that hemopexin could also be susceptible to oxidation under the conditions of some purification protocols.
Alternative Activities Proposed for Hemopexin
Hemopexin as a hyaluronidase
Of the catalytic or binding activities attributed to hemopexin that are unrelated to the ability of the protein to bind heme, a particularly notable proposal was that hemopexin is a hyaluronidase71 that can hydrolyze the high molecular weight glycosaminoglycan hyaluronic acid that is one of the most abundant constituents of the vertebrate extracellular matrix.72 The apparent identity of porcine hemopexin and porcine liver hyaluronidase led to the conclusion that hemopexin is a hyaluronidase.71 This finding was based on cloned complementary DNA corresponding to an abundant protein in a purified porcine liver fraction that exhibited hyaluronidase activity. Hrkal et al. determined later73 that human hemopexin does not possess hyaluronidase activity but that hyaluronidase, which is electrophoretically distinct from hemopexin, contaminated nearly half of their hemopexin preparations. Nevertheless, these authors found that a portion (10–60%) of their purified hemopexin could bind hyaluronic acid. In an agarose matrix,73 the pattern of hemopexin-hyaluronic acid interaction exhibited microheterogeneity that varied from one serum sample to another as well as from one hemopexin preparation to another. Subsequently, however, purified hemopexin was found to be unable to bind hyaluronic acid in solution.4 Therefore, although the hemopexin C-domain sequence contains a consensus motif (residues 348–356 in human hemopexin) that has been proposed to be sufficient for hyaluronic acid binding,74 solution requirements for promoting interaction of hemopexin with hyaluronic acid are unresolved. Examination of a homology model for human hemopexin with heme bound indicates that the motif suggested for hyaluronic acid binding is not highly exposed in the C-domain and that only the argininyl residues of that sequence are exposed on the surface of the protein (Fig. 1). As well, hemopexin appears to lack the binding groove motif that has been reported to occur in protein receptors for hyaluronic acid.76 Nevertheless, it is clear that hemopexin does not hydrolyze hyaluronic acid and so does not possess intrinsic hyaluronidase activity.
Figure 1.
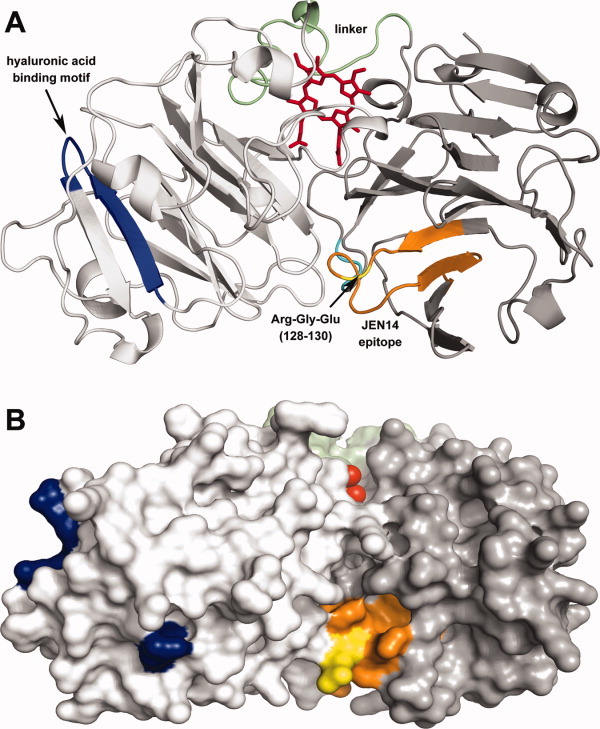
Homology model of the human hemopexin-heme complex21 in identical orientations rendered as a (A) ribbon diagram and (B) surface diagram. The C-domain (white), N-domain (gray), linker sequence (green), and heme (red) are colored as indicated: The hyaluronic acid binding motif [348–358 (blue)], the Arg-Gly-Glu sequence [128–130 (yellow)] at the interface of the N- and C-domains, and residues 123–143 (orange) of the JEN14 epitope75 are also indicated. This figure was prepared with PyMol (Schrödinger, San Diego).
Hemopexin as a serine protease
In 1988, Bakker et al. observed that samples of plasma from patients with minimal change nephritic syndrome (MCNS) in relapse increase vascular permeability following intra-dermal administration to rats.77 These patients were known to exhibit a loss of polyanionic glomerular sialoglyco-proteins (GPA).78 As part of this study, these authors isolated a fraction from normal human plasma denoted as 100KF (containing several proteins of apparent molecular weight 80–100 kDa) that increased vascular permeability and also resulted in a loss of GPA staining following perfusion into rat kidneys ex vivo, thereby mimicking the findings from MCNS patients in relapse. However, the plasma of MCNS patients in relapse contained less of this 100KF fraction than did plasma from healthy control subjects. In view of the issues related to electrophoresis of hemopexin discussed above, SDS-PAGE analysis of the 100KF fraction (Ref. 77, Fig. 2) clearly shows that the majority of protein in this preparation cannot be native human hemopexin. Similar fractionation of rat serum had previously produced a protein fraction of apparent molecular weight 120 kDa that also increased vascular permeability following intra-dermal administration to rats and when incubated with kidney sections in vitro also decreased GPA staining.79 Subsequently, this group reported that 100KF/hemopexin preparations from human serum also reduced staining of ecto-ATPases in glomeruli of kidney sections44 as well as induced proteinuria and fusion of epithelial foot processes in glomerular capillary segments following intra-renal infusion in vivo in rats.80 This latter observation was expanded in a recent study24 that used cultured epithelial cells (podocytes) to provide evidence that led to the conclusion that their hemopexin preparations cause reorganization of actin fibers in podocytes through a nephrin-dependent signaling cascade.
Figure 2.
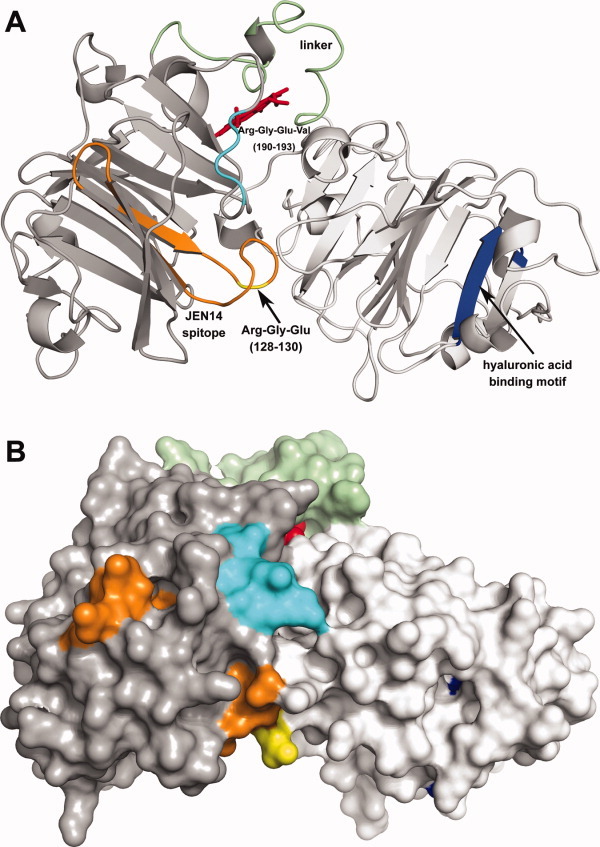
Homology model of the human hemopexin-heme complex21 (rotated ∼ 180° about the Y-axis with respect to Fig. 1) rendered as a (A) ribbon diagram and (B) surface diagram. The C-domain (white), N-domain (gray), linker sequence (green), and heme (red) are colored as indicated: The Arg-Gly-Glu-Val sequence [190–193 (cyan)] is located on the surface toward the center of the N-domain, whereas the Arg-Gly-Glu sequence [128–130 (yellow)] is at the interface of the N- and C-domains. The JEN14 epitope [123–143 (orange)]75 and hyaluronic acid binding motif [348–356 (blue)] are shown. This figure was prepared with PyMol (Schrödinger, San Diego).
Various types of hemopexin preparations have been used in publications from the Bakker group, including recombinant hemopexin, and each merits consideration. Initial studies concerned the 100KF material that contained hemopexin and impurities as described above, and subsequent studies involved purified hemopexin preparations that SDS-PAGE analysis showed to contain various amounts of protein contaminants when stained with Ponceau S,44 a protein stain that is less sensitive than Coomassie Blue.81 A protocol for further fractionation of the 100KF fraction44 used Blue-Sepharose chromatography,11 a procedure shown to produce hemopexin contaminated with hyaluronidase,73 to obtain samples containing hemopexin as well as multiple protein bands of >85 kDa. An alternative approach was also taken to fractionate a human 100KF preparation further by chromatography on a rabbit anti-hemopexin antibody affinity column, but the resulting product exhibited substantial cleavage following this treatment [Ref. 44, Fig. 3(B)]. In at least one report,23 recombinant hemopexin was expressed in P. pastoris, but the only evidence for purity of the isolated protein was a Western blot with a band consistent with a molecular weight of 85 kDa, and no evidence for the presence of native hemopexin was provided. In various publications from this group, the reported molecular weight of “active hemopexin” varies over the range 45–127 kDa44 but is usually in the range of 70–100 kDa.80 However, more recent experiments from this group82 failed to report either the source or purification method used to obtain their “active hemopexin.”
The mechanism by which the four biological effects described [i.e., proteinuria, diminished staining of GPA, diminished activity of ecto-apyrase (i.e., ecto-ATPase) in kidney glomeruli (assays described in Ref. 44), and fusion of epithelial cell foot processes] occur has been attributed to a serine protease activity that Bakker et al. propose for hemopexin.23 This activity was first proposed following the observation that soybean trypsin inhibitor, anti-thrombin III, and ɛ-aminocaproic acid all inhibited the ability of the 120 kDa fraction from rat serum (vide supra) to diminish staining of GPA and resulted in a 30% inhibition of the vascular response.79 Subsequently, Bakker et al. proposed that hemopexin in normal human plasma occurs in an inactive form23,24 that is activated during isolation to a form of hemopexin with a serine protease activity similar to that detected in the plasma of MCNS patients in relapse83 or with other on-going pathology.84 Recently, this group also reported that the plasma of pregnant (but not pre-eclamptic) women produces reduced staining of glomerular ecto-apyrase82,85 and that the plasma of some renal transplant recipients has increased protease activity.84 However, attributing these plasma activities to hemopexin or, moreover, solely to a modification of hemopexin without more direct evidence remains presumptive.
Additional support for the conclusion that hemopexin is a serine protease was fostered by further reports from the Bakker group that a variety of protease inhibitors can block the biological effects of hemopexin observed in a selection of assays. These results have not always been consistent and even led to the proposal of multiple enzyme activities occurring as a complex in the 100KF preparations.86 For example, the diminished staining of ecto-apyrase caused by hemopexin preparations was initially reported to be unaffected by PMSF,86 a conclusion that was later reversed by the group.23 The effects of hemopexin preparations on GPA and ecto-apyrase activities in vitro have been reported to be inhibited by α2-macroglobulin80 and antithrombin III,87 but more recently, the protease activity of hemopexin samples was reported to not be inhibited by α2-macroglobulin.84 Antithrombin III is a 432 residue glycoprotein (pI ∼5, Ref. 88) with four bi-antennary complex carbohydrate chains89 and with a molecular weight of 58 kDa, so it is conceivable that this protein might compete with hemopexin for binding sites on glomerular cells and the glomerular basement membrane. Whether hemopexin exposed to 80°C for 20–60 min82,86 is an appropriate denatured protein control in these assays23,24,80,86 is questionable because exposure to high temperature produces polymeric hemopexin.90 This polymerization has been attributed to disulfide interchange as well as to crosslinks that once formed are not reversed by DTT.91 These polymeric products are likely to present surface characteristics that are considerably different from those of native hemopexin thus altering binding interactions with cells.90 Attempts to demonstrate that the actin reorganization initiated by hemopexin preparations is the result of hemopexin serine protease activity also led to ambiguous results. Although the serine protease inhibitor 4-(2-aminoethyl)benzenesulfonyl fluoride hydrochloride (AEBSF) (0.5 mM) decreased actin reorganization in podocytes treated with hemopexin (0.8 μM), this reagent alone albeit at higher concentration (≥1.0 mM) exhibited significant independent effects on actin reorganization.24 Furthermore, exposure of the podocytes to just 10% plasma completely blocked the effect of the protein isolate on actin fibers.
The most direct strategy designed to establish the serine protease activity of hemopexin preparations involved the use of synthetic substrates.23 However, very high concentrations of recombinant hemopexin (∼8 μM), and very long reaction times (up to 25 h) were required to produce significant absorbance changes at 405 nm23 relative to the typical conditions used for these substrates with known proteases (e.g., reaction times of several minutes, 37°C, sub-micromolar enzyme concentration92–94). No evidence was provided in these assays that the very weak activity exhibited by recombinant hemopexin could be inhibited by nonhydrolyzable synthetic analogue substrates. The conditions used in these kinetic experiments raise the possibility that the absorbance changes used to validate existence of this activity might result from an increase in light scattering caused by the aggregation of recombinant hemopexin during exposure to 37°C for a day. Without additional kinetic evidence, it seems unlikely that this exceedingly low, putative hemopexin protease activity is sufficient to produce the biological effects attributed to it, for example, increased proteinuria observed within 5 min after renal perfusion of rats with recombinant hemopexin23 or within less than 18 min after perfusion with the 100KF material.44 The proteinuria following perfusion with recombinant hemopexin decreased after 10 min.23 The relatively prompt response and rapid reversibility for these biological effects of hemopexin isolates could be more consistent with a binding process rather than modification of cellular proteins.
Two additional observations concerning the proposed serine protease activity of hemopexin merit comment in view of the well-documented physical and chemical properties of hemopexin. First, the serine protease activity of hemopexin is reported to be inhibited by an 80-fold molar excess of ADP.87 In this work, however, it was not established whether ADP interacts with the “enzyme” (hemopexin) or the “substrate” (cellular components, including ecto-ATPase). It would not be surprising that ADP could interact with hemopexin because hemopexin binds to Cibacron Blue affinity resins,11 which are used routinely to purify proteins that bind NADH,95,96 the structure of which is ADP coupled to ribosylated nicotinamide. Although hemopexin does not possess the classic ADP-binding βαβ-fold,97 the high salt concentrations required to elute hemopexin from Cibacron blue resins indicates that electrostatic interactions play a major role in the binding of hemopexin to the immobilized dye.98 Notably, other groups have found that the ecto-ATPase of rat glomerular mesangial cells is remarkably resistant to trypsin, pronase, and papain.99 Furthermore, the activity of the ecto-ATPase is diminished when renal cells are incubated in serum-free medium.99 At the high concentrations of ADP (2 mM) used in these studies,87 ADP could bind to hemopexin or to cellular components, a situation that would significantly alter their electrostatic surface properties so that any binding interaction between them could be inhibited without requiring “inhibition of protease activity” by ADP.
The second observation was that in serum-free medium, both cultured podocytes and glomerular endothelial cells exhibited diminished binding of the lectin wheat germ agglutinin (presumably to extracellular glycoproteins) after treatment with hemopexin.24 However, the reduced fluorescence from wheat germ agglutinin antibodies is insufficient to distinguish loss of extracellular glycoproteins (glycocalyx) as the result of proteolysis by hemopexin (as might be shown by identification of peptides released by this activity), from metabolic events, or from competitive binding effects. This last possibility deserves particular consideration because human and rat hemopexins bind readily to wheat germ lectin-affinity resin.100,101 In view of these considerations, it seems likely that the effects of hemopexin on renal tissue are not the result of proteolytic activity but that under the experimental conditions reported, hemopexin binds to cell surface components and may induce a subsequent cascade of metabolic effects. This interaction with cell surfaces could be blocked by the presence of specific electrolytes or presence of other proteins or lectins. We conclude, therefore, that there is no compelling evidence that hemopexin possesses proteolytic activity.
Hemopexin as a pro-inflammatory or anti-inflammatory mediator
The protein isolates of Bakker et al. demonstrate toxic effects on renal tissue [i.e., diminished reactivity of the anti-inflammatory glomerular enzyme ecto-apyrase and increased endothelial permeability (summarized in Ref. 102)] that are considered to be pro-inflammatory. This result seems to conflict with the recent findings of Liang et al.25 that their hemopexin preparations diminish the upregulation of the pro-inflammatory cytokines TNF and IL-6 induced by LPS-stimulation of macrophages. This mechanism was affected by heme-free hemopexin and does not appear to involve induction of heme oxygenase-1, a protein which also has anti-inflammatory effects.
Notwithstanding the possibility that this apparent disagreement concerning the function of hemopexin as a pro- versus anti-inflammatory mediator may be a matter of semantics (see commentary in Refs. 103 and104), comparison of the protocols used by these two groups to isolate the protein samples suggests a common mechanism for the results of both (vide infra). The active agent isolated from mouse serum described in the study of Liang et al.25 exhibited unusual properties for mouse hemopexin (see Ref. 25, Fig. 1). As determined by chromatofocusing, the apparent pI of 4.3 for their active material is low compared with the value of 7.59 (ProtParam: http://www.expasy.ch/tools/protparam.html105) predicted from the amino acid sequence of mouse hemopexin (http://www.ncbi.nlm.nih.gov/protein/AAH19901.1 aligned with the sequence of mature, human hemopexin). Although this estimate does not account for any contribution from the carbohydrate component of the protein, the experimentally determined pI values of human and rabbit hemopexin (both 5.8, Ref. 9) are less than one pI unit lower than the values predicted on the basis of amino acid sequence alone (6.4 and 6.7, respectively105). This result suggests that the mouse hemopexin isolated by Liang et al.25 has been modified. In addition, during gel filtration chromatography the active material eluted slowly and in low yield, relative to the known high content of hemopexin in serum. No data were provided concerning the molecular weight of this sample. The active material was identified as mouse hemopexin by MS analysis of a tryptic digest. However, such analyses typically assess just 25–30% of the protein sequence, so truncation of the protein cannot be detected in this manner nor can chemical modification of residues in tryptic peptides that are not analyzed. Following this mass spectroscopic identification, these authors used commercial recombinant human hemopexin or mouse hemopexin [isolated by Rivanol precipitation, acidic elution (pH 2) from a heme-agarose affinity column, and two reverse phase chromatography fractionations under unspecified elution conditions] in subsequent studies. Although the mouse hemopexin purified by this reverse-phase protocol migrated as a single band on SDS-PAGE (apparent molecular weight not indicated), no evidence was provided to establish that the product retains the structure or properties of native hemopexin. Considering as well that reverse phase chromatography (i) has not been used in previous procedures reported for purification of native hemopexin (e.g., Refs. 10 and21), (ii) typically uses solvents that are expected to denature proteins (e.g., acetonitrile106), and (iii) has been shown to cause bovine hemopexin to lose ability to bind heme,107 several reasons can be identified to conclude that native hemopexin is not responsible for the effects observed by these authors.
A notable difference between the report of Liang et al.25 and those of Bakker and coworkers24,44,80,86,87 is that the initial observations pertinent to the study of Liang et al. were made with high concentrations of plasma,108 whereas Bakker and coworkers observed their responses only with protein isolates and not with normal plasma. Although these may seem to be contradictory characteristics of the two lines of investigation, the fact that the active agent isolated by Liang et al. represented a small fraction of the hemopexin expected to occur in plasma whereas activities observed by Bakker and coworkers arose during fractionation suggests that in neither case is native hemopexin the active agent. In other words, the results of both groups could be attributed to a fraction of hemopexin that was modified, degraded or denatured. Such altered species could be capable of interaction with a variety of receptors or cellular binding sites and thereby elicit the experimentally observed effects either through competitive binding interactions or triggering of alternate molecular signaling pathways.
Hemoglobin-induced oxidative modification of LDL, considered a causative factor in atherosclerosis (e.g., Ref. 109), has been shown to be inhibited by hemopexin.110 Hemopexin can also suppress hemoglobin-mediated oxidation of lipids of atheromatous lesions,111 thereby attenuating subsequent endothelial cytotoxicity. An anti-inflammatory role for hemopexin in protecting against atherosclerosis has been proposed recently on the basis of experiments interpreted in terms of a hypothetical ternary complex of hemoglobin (Hb), haptoglobin (Hp), and hemopexin.112 The proposal for occurrence of such a complex was based on (i) the identification of a macrophage receptor (CD163) for the Hb-Hp complex, (ii) identification of a receptor [low-density lipoprotein receptor-related protein (LRP/CD91) that occurs on several cell types] for the hemopexin-heme complex,48 (iii) the presence of Hb in pro-inflammatory HDL of atherogenic/hyperlipidemic mice, and (iv) the reported association of Hp with apoA-1,113,114 the major protein component of HDL. This hypothetical ternary complex was proposed to allow removal of pro-inflammatory Hb-containing HDL mediated by the hemopexin-heme (LRP/CD91) receptor. Subsequently, Watanabe and coworkers115 reported that Hb is associated with apoA-1 containing HDL as a component of the Hb-Hp complex and that hemopexin is associated with HDL particles containing apoA-1. However, these authors also found that hemopexin attached to HDL does not appear to have heme bound (Ref. 115, center panel, Hp−/− Fig. 5), and there is currently no direct evidence that the Hb-Hp complex can release heme to hemopexin. As a result, the pro-inflammatory Hb-containing HDL particles should not be recognized by the receptor for the hemopexin-heme complex. So the proposal by Watanabe et al.115 that hemopexin could be anti-inflammatory if it prevents the interaction of Hb and Hp in HDL is not directly supported by experimental evidence.
Using hemopexin-heme-affinity resin, Hvidberg et al. showed that human hemopexin is capable of interacting with proteins in cellular membranes one of which was identified as a carboxylesterase.48 It is unclear whether those interactions or the interactions of hemopexin with apoA-1 containing HDL are highly specific or if they represent low affinity binding interactions that have been amplified by the experimental conditions in vitro. For example, Hb exhibits low affinity binding in vitro to the Hb-Hp receptor CD163 on macrophages.116 Although any number of mechanisms could be proposed as to how hemopexin associates with the apoA-1 containing HDL (e.g., through recognition of the carbohydrate portion of hemopexin or through specific or nonspecific electrostatic interactions), establishment of an anti-inflammatory role for hemopexin in this aspect of atherosclerosis requires additional evidence.
Hemopexin as a necrosis-suppressing factor
Suzuki et al. found that serum prevents the rapid death of polymorphonuclear leukocytes (PMNs) that otherwise occurs following exposure to the protein kinase C activator phorbol 12-myristate 13-acetate (PMA).42 Three species of purified hemopexins (bovine, porcine, and human) substantially suppressed the necrosis of PMA-activated PMNs but only if other macromolecules, presumably required for regulation of osmotic pressure under the conditions required for the culture, were present.42 The human hemopexin used in these studies exhibited the expected apparent molecular weight and banding pattern (see discussion above) on analysis by SDS-PAGE and contained only a trace of lower molecular weight components that cross-reacted with polyclonal anti-hemopexin antibodies. Both hemopexin and the hemopexin-heme complex were found to suppress necrosis, leading the authors to discount their original premise,42 which was that hemopexin protects cells by sequestering free heme released into the extracellular milieu by oxidative breakdown, thereby preventing self-damage of PMA-exposed PMNs caused by heme-aggravated oxidative cytotoxicity. As a result, the mechanism by which hemopexin suppresses necrosis in this system remained undefined.
However, a receptor for the hemopexin-heme complex on the cell surface of PMNs had previously been described by Okazaki et al.117 Their findings suggest that following receptor mediated uptake of hemopexin-heme by PMNs, hemopexin is recycled to the medium. Thus, the original mechanism proposed by Suzuki et al.42 would accommodate this result because hemopexin may have become available from the hemopexin-heme complex to which the cells were initially exposed. As a result, the necrosis-suppressing activity of hemopexin can be most simply explained as another consequence of the heme-sequestering antioxidant capacity of hemopexin.
Hemopexin as an inhibitor of cellular adhesion
Further studies by Suzuki et al. to define better the role of hemopexin in regulating the cytotoxic functions of PMA-activated PMNs (vide supra) led them to examine the cellular adhesion properties of activated PMNs.43 In serum, Ca2+ is required for cell adhesion while under serum-free conditions, Mg2+ is required. Hemopexin and the hemopexin-heme complex were found to inhibit the adhesion of PMNs activated by PMA or Mn2+ to fibrinogen-coated surfaces under serum-free conditions. However, activated PMNs suspended in serum showed marked adhesion to such surfaces in the presence of hemopexin. This adhesion was proposed to result from competition between hemopexin and unidentified adhesion promoting factors (i.e., Ca2+ plus one or more components of <30 kDa). The apparent requirement for metal ions led Suzuki et al. to suggest43 that hemopexin may inhibit cell adhesion by interaction of the hemopexin β-propeller domains (a structural fold common to many proteins that mediate protein–protein interactions28,36) with β2-integrin or with β2-integrin regulatory proteins.
However, another more specific mode of interaction whereby hemopexin may inhibit cellular adhesion should be considered. Stanley has compared118,119 hemopexin with another member of a pexin gene family, S-protein (also known as vitronectin, serum spreading factor, or protein-X). He noted that both have cell binding sites but that only S-protein has the Arg-Gly-Asp tripeptide sequence common to fibrinogen, fibronectin, and von Willebrand factor that interacts specifically with a cell surface receptor.120,121 However, it should be noted that human hemopexin has two related sequences, for example, Arg-Gly-Glu sequences at residues 128–130 and 190–192. One lies within an epitope (residues 123–143) of rabbit hemopexin recognized by the monoclonal antibody JEN14. This region (Fig. 1) was proposed to be a receptor recognition site because the antibody blocks the binding of the hemopexin-heme complex to cells.75,122 As well, a sequence similar to the longer Arg-Gly-Asp-Val of S-protein also occurs in human hemopexin (Arg-Gly-Glu-Val, residues 190–193) that merits consideration as a potential site for interaction of human hemopexin with cellular surfaces. Precedence for involvement of an Arg-Gly-Glu motif in binding to cell surfaces is provided by viral protein recognition by cell surfaces.123
The observation that hemopexin fails to inhibit adhesion of activated PMNs to fibrinogen coated surfaces in the presence of serum does not necessitate the existence of as yet unidentified cellular adhesion promoting factors. A much simpler explanation is that the abundance of electrolytes and proteins present in serum can compete for cellular sites that also interact with hemopexin (see discussions above).
Hemopexin as a metal ion binding protein
The first indication that human hemopexin can bind divalent metal cations was the report of Cu2+ contamination in purified monomeric hemopexin preparations.47 The observations of Porath et al.124–126 and of Andersson127 that hemopexin is one of a small number of plasma proteins that binds to an immobilized metal ion affinity resin provided more direct evidence that hemopexin can interact with certain metal ions. The ability of hemopexin to bind a variety of metal ions has been used in development of an efficient protocol for purification of human hemopexin from blood plasma cryosupernate by means of immobilized metal ion affinity chromatography.57 The binding of Zn2+, Cu2+, Ni2+, Co2+, and Mn2+ to both human hemopexin and the corresponding hemopexin-heme complex has been characterized.57,58 All except Mn2+ were found to lower the thermal stability (Tm) of both hemopexin and the hemopexin-heme complex.58,91,128 Other effects of metal ions on the structure of hemopexin and the hemopexin-heme complex have been investigated by thermally induced difference absorption spectroscopy91 and NMR spectroscopy,57 respectively. Acidic patches at the narrow opening of the central tunnels in the β-propellers have been proposed as potential binding sites for metal ions, particularly Ca2+.1,8 An homology model for the structure of the human hemopexin-heme complex based on the structure of the rabbit protein was used to identify several additional, putative binding sites for metal ions on human hemopexin21 and has led to speculation that hemopexin and the hemopexin-heme complex might play a role in transport of metal ions and limitation of metal ion toxicity in disease states.7,21,57,129 More recent experimental studies have suggested additional functional interactions of metal ions with hemopexin. For example, following receptor mediated uptake of hemopexin-heme by hepatoma cells, there is a rapid decrease in the levels of the copper chaperone CCS1 that is consistent with an increase in the intracellular concentration of Cu2+.130 Cu2+, but not Ca2+, also inhibits rebinding of deuteroheme to rabbit or human hemopexin following exposure to acidic pH (4.2) and chloride levels proposed to simulate endosomal conditions, leading to the proposition that Cu2+ facilitates heme export from the endosome.130 On the other hand, at neutral pH Ca2+ promotes the binding of heme68 and increases the thermal stability (Tm) of the hemopexin-heme complex.21 All of these observations, as well as specific ion effects on the dissociation of heme from the hemopexin-heme complex,57,68,131 suggest a means by which the interaction of the protein with metal ions may promote transfer and binding of heme to hemopexin in blood plasma, release of heme from the hemopexin-heme complex in the endosome, and, as a result, promotion of the known anti-oxidant capacity of hemopexin. Nevertheless, there is no experimental evidence at present that metal ion binding to hemopexin comprises a new functional property of the protein in the transport of metal ions in vivo.
Summary
Although hemopexin has been studied continuously for over 40 years, it continues to present an experimental challenge because purification of the native protein from blood plasma requires considerable care and because no well-defined, established recombinant expression protocol that produces protein shown to be native has been reported. Nevertheless, viable purification strategies have been reported for preparation of pure, native hemopexin from plasma. Although the purity of a hemopexin sample can be assessed in part by SDS-PAGE analysis, additional information is required to demonstrate that the sample retains the structure and properties of the native protein. As hemopexin lacks an established catalytic activity that can be used to validate the quality of the purified protein, only spectroscopic criteria are available for this purpose. As discussed above, the most compelling spectroscopic evidence for retention of the native structure of hemopexin is observation of the positive Cotton effect in the far UV CD spectrum of the protein that is enhanced upon heme binding. Attribution of new biological activities to hemopexin can be accepted only if it is demonstrated that the protein responsible for the activity has the native structure of hemopexin. In the absence of such evidence, it remains possible that any newly observed biological activity is a property of a chemically modified or improperly folded form of hemopexin.
The information provided in the three-dimensional structure of hemopexin1 is a tremendous resource that only a few subsequent studies of hemopexin (e.g., Refs. 21,58,68, and91) have attempted to use to aid in the interpretation of experimental results. Notably, examination of the homology model of the human hemopexin-heme complex reveals that hemopexin lacks the classic serine protease catalytic triad (His, Ser, and Asp)132 though the presence of other structural types of serine protease active sites133 are not so readily ruled out. This observation provides a structural basis for the conclusion drawn above that hemopexin is not a serine protease. In the hemopexin-heme complex, the consensus motif suggested to be sufficient for binding to hyaluronic acid is arranged such that only the arginyl residues are accessible on the surface of the C-domain (Figs. 1 and 2). This three-dimensional structure reveals that the Arg-Gly-Glu sequence (residues 190–192) proposed above to act as a site for interaction with cell surfaces is highly exposed on the surface of the protein and near the JEN14 epitope [residues 123–143, which include the Arg-Gly-Glu sequence (128–130)] that was predicted to be one site recognized by the rabbit hemopexin-heme receptor.
As summarized here, identification of new activities for hemopexin can be a difficult undertaking. Some of the cytoprotective effects reported for this protein may not represent new activities but more likely reflect the well established biological role of hemopexin in sequestering oxidatively active heme. Early reports of a hyaluronidase activity for hemopexin were subsequently corrected. The analysis provided here indicates that current evidence is insufficient to support the conclusion that hemopexin possesses proteolytic activity or to unequivocally substantiate several of the pro- or anti-inflammatory activities reported for the protein. Some of these observations for alternative activities of hemopexin may have arisen from the presence of hemopexin at a high local concentration under experimental conditions in vitro that do not reflect the ionic conditions that normally occur in vivo so that weak binding interactions dominate. Although some of the experimental results discussed here demonstrate a response to bio-active samples that may be derived from hemopexin and that affect cellular surfaces and signaling pathways, insufficient evidence is available to conclude that these activities are attributable to native hemopexin or are related to physiological processes involving native hemopexin or its heme complex. Nevertheless, re-evaluation of some of these proposed activities (e.g., anti-inflammatory activity) with alternative experimental strategies that avoid the difficulties outlined here may ultimately vindicate some of the conclusions that are challenged in this review.
Acknowledgments
We thank Professor Thomas Poulos for helpful discussions.
Footnotes
The term hemopexin refers in this report to the hemopexin glycoprotein without ferriprotoporphyrin IX bound, and the term hemopexin-heme complex refers to hemopexin with ferriprotoporphyrin IX bound at a site equivalent to that identified in the three-dimensional structure of rabbit hemopexin.
References
- 1.Paoli M, Anderson BF, Baker HM, Morgan WT, Smith A, Baker EN. Crystal structure of hemopexin reveals a novel high-affinity heme site formed between two β-propeller domains. Nat Struct Biol. 1999;6:926–931. doi: 10.1038/13294. [DOI] [PubMed] [Google Scholar]
- 2.Grabar P, de Vaux St-Cyr C, Cleve H. Presence of β1-B-globulin in the perchloric acid extracts of human normal serums. Bull Soc Chim Biol. 1960;42:853–856. [PubMed] [Google Scholar]
- 3.Immenschuh S, Song DX, Satoh H, Müller-Eberhard U. The type II hemopexin interleukin-6 response element predominates the transcriptional regulation of the hemopexin acute phase responsiveness. Biochem Biophys Res Commun. 1995;207:202–208. doi: 10.1006/bbrc.1995.1173. [DOI] [PubMed] [Google Scholar]
- 4.Morgan WT, Smith A. Binding and transport of iron-porphyrins by hemopexin. Adv Inorg Chem. 2001;51:205–241. [Google Scholar]
- 5.Delanghe JR, Langlois MR. Hemopexa review of biological aspects and the role in laboratory medicine. Clin Chim Acta. 2001;312:13–23. doi: 10.1016/s0009-8981(01)00586-1. [DOI] [PubMed] [Google Scholar]
- 6.Ascenzi P, Fasano M. Heme-hemopexa ‘chronosteric’ heme-protein. IUBMB Life. 2007;59:700–708. doi: 10.1080/15216540701689666. [DOI] [PubMed] [Google Scholar]
- 7.Tolosano E, Fagoonee S, Morello N, Vinchi F, Fiorito V. Heme scavenging and the other facets of hemopexin. Antioxid Redox Signal. 2010;12:305–320. doi: 10.1089/ars.2009.2787. [DOI] [PubMed] [Google Scholar]
- 8.Faber HR, Groom CR, Baker HM, Morgan WT, Smith A, Baker EN. 1.8 Å crystal structure of the C-terminal domain of rabbit serum haemopexin. Structure. 1995;3:551–559. doi: 10.1016/s0969-2126(01)00189-7. [DOI] [PubMed] [Google Scholar]
- 9.Morgan WT. Porphyrin-binding proteins in serum. Ann NY Acad Sci. 1975;244:624–650. doi: 10.1111/j.1749-6632.1975.tb41558.x. [DOI] [PubMed] [Google Scholar]
- 10.Müller-Eberhard U. Hemopexin. Methods Enzymol. 1988;163:536–565. doi: 10.1016/0076-6879(88)63049-7. [DOI] [PubMed] [Google Scholar]
- 11.Hrkal Z, Cabart P, Kalousek I. Isolation of human haemopexin in apo-form by chromatography on S-sepharose fast flow and blue sepharose CL-6B. Biomed Chromatogr. 1992;6:212–214. doi: 10.1002/bmc.1130060412. [DOI] [PubMed] [Google Scholar]
- 12.Jensen PE, Birkenmeier G, Stigbrand T. Zinc chelates bind human hemopexin. Acta Chem Scand. 1991;45:537–538. doi: 10.3891/acta.chem.scand.45-0537. [DOI] [PubMed] [Google Scholar]
- 13.Morgan WT. The histidine-rich glycoprotein of serum has a domain rich in histidine, proline, and glycine that binds heme and metals. Biochemistry. 1985;24:1496–1501. doi: 10.1021/bi00327a031. [DOI] [PubMed] [Google Scholar]
- 14.Suttnar J, Hrkal Z, Vodrazka Z. Affinity chromatography of serum haemopexin. J Chromatogr. 1977;131:453–457. doi: 10.1016/s0021-9673(00)80969-3. [DOI] [PubMed] [Google Scholar]
- 15.Strop P, Borvak J, Kasicka V, Prusik Z, Moravek L. Isolation of human haemopexin by bioaffinity chromatography on haeme-sepharose. J Chromatogr. 1981;214:317–325. doi: 10.1016/s0021-9673(00)80560-9. [DOI] [PubMed] [Google Scholar]
- 16.Suttnar J, Hrkal Z, Vodrazka Z, Rejnkova J. Haeme-Sepharose 4B as a chromatographic matrix for the isolation of haemopexin from human serum. J Chromatogr. 1979;169:500–504. doi: 10.1016/0021-9673(75)85091-6. [DOI] [PubMed] [Google Scholar]
- 17.Olsen KW. A new method for affinity chromatography of heme-binding protein synthesis and characterization of hematin- and hematoporphyrin-agarose. Anal Biochem. 1980;109:250–254. doi: 10.1016/0003-2697(80)90644-2. [DOI] [PubMed] [Google Scholar]
- 18.Tsutsui K, Mueller GC. Affinity chromatography of heme-binding proteins: An improved method for the synthesis of hemin-agarose. Anal Biochem. 1982;121:244–250. doi: 10.1016/0003-2697(82)90475-4. [DOI] [PubMed] [Google Scholar]
- 19.Tsutsui K. Affinity chromatography of heme-binding proteins: synthesis of hemin-agarose. Methods Enzymol. 1986;123:331–338. doi: 10.1016/s0076-6879(86)23039-6. [DOI] [PubMed] [Google Scholar]
- 20.Olsen KW. Affinity chromatography of heme-binding proteins: synthesis and characterization of hematin- and hematoporphyrin-agarose. Methods Enzymol. 1986;123:324–331. doi: 10.1016/s0076-6879(86)23038-4. [DOI] [PubMed] [Google Scholar]
- 21.Mauk MR, Rosell FI, Mauk AG. Structural modelling of metal ion binding to human haemopexin. Nat Prod Rep. 2007;24:523–532. doi: 10.1039/b604184c. [DOI] [PubMed] [Google Scholar]
- 22.Satoh T, Satoh H, Iwahara S, Hrkal Z, Peyton DH, Müller-Eberhard U. Roles of heme iron-coordinating histidine residues of human hemopexin expressed in baculovirus-infected insect cells. Proc Natl Acad Sci USA. 1994;91:8423–8427. doi: 10.1073/pnas.91.18.8423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bakker WW, Borghuis T, Harmsen MC, van den Berg A, Kema IP, Niezen KE, Kapojos JJ. Protease activity of plasma hemopexin. Kidney Int. 2005;68:603–610. doi: 10.1111/j.1523-1755.2005.00438.x. [DOI] [PubMed] [Google Scholar]
- 24.Lennon R, Singh A, Welsh GI, Coward RJ, Satchell S, Ni L, Mathieson PW, Bakker WW, Saleem MA. Hemopexin induces nephrin-dependent reorganization of the actin cytoskeleton in podocytes. J Am Soc Nephrol. 2008;19:2140–2149. doi: 10.1681/ASN.2007080940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liang X, Lin T, Sun G, Beasley-Topliffe L, Cavaillon JM, Warren HS. Hemopexin down-regulates LPS-induced proinflammatory cytokines from macrophages. J Leukoc Biol. 2009;86:229–235. doi: 10.1189/jlb.1208742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baker SC, Saunders NF, Willis AC, Ferguson SJ, Hajdu J, Fulop V. Cytochrome cd1 structure: unusual haem environments in a nitrite reductase and analysis of factors contributing to β-propeller folds. J Mol Biol. 1997;269:440–455. doi: 10.1006/jmbi.1997.1070. [DOI] [PubMed] [Google Scholar]
- 27.Gomis-Rüth XF. Hemopexin domains. In: Messerschmidt A, Bode W, Cygler M, editors. Handbook of metalloproteins. New York: Wiley; 2004. pp. 631–646. [Google Scholar]
- 28.Smith TF, Gaitatzes C, Saxena K, Neer EJ. The WD repeat: a common architecture for diverse functions. Trends Biochem Sci. 1999;24:181–185. doi: 10.1016/s0968-0004(99)01384-5. [DOI] [PubMed] [Google Scholar]
- 29.Suttnar J, Hrkal Z. The reactivity of the disulfide bonds of human serum haemopexin. Int J Biochem. 1986;18:283–284. doi: 10.1016/0020-711x(86)90120-5. [DOI] [PubMed] [Google Scholar]
- 30.Morgan WT, Müller-Eberhard U. Interactions of porphyrins with rabbit hemopexin. J Biol Chem. 1972;247:7181–7187. [PubMed] [Google Scholar]
- 31.Wu ML, Morgan WT. Characterization of hemopexin and its interaction with heme by differential scanning calorimetry and circular dichroism. Biochemistry. 1993;32:7216–7222. doi: 10.1021/bi00079a018. [DOI] [PubMed] [Google Scholar]
- 32.Wu ML, Morgan WT. Conformational analysis of hemopexin by Fourier-transform infrared and circular dichroism spectroscopy. Proteins. 1994;20:185–190. doi: 10.1002/prot.340200208. [DOI] [PubMed] [Google Scholar]
- 33.Hider RC, Kupryszewski G, Rekowski P, Lammek B. Origin of the positive 225-230 nm circular dichroism band in proteins. Its application to conformational analysis. Biophys Chem. 1988;31:45–51. doi: 10.1016/0301-4622(88)80007-3. [DOI] [PubMed] [Google Scholar]
- 34.Woody RW. Contributions of tryptophan side chains to the far-ultraviolet circular dichroism of proteins. Eur Biophys J. 1994;23:253–262. doi: 10.1007/BF00213575. [DOI] [PubMed] [Google Scholar]
- 35.Morgan WT, Smith A. Domain structure of rabbit hemopexin. Isolation and characterization of a heme-binding glycopeptide. J Biol Chem. 1984;259:12001–12006. [PubMed] [Google Scholar]
- 36.Baker HM, Anderson BF, Baker EN. Dealing with iron: common structural principles in proteins that transport iron and heme. Proc Natl Acad Sci USA. 2003;100:3579–3583. doi: 10.1073/pnas.0637295100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cox MC, Le Brun N, Thomson AJ, Smith A, Morgan WT, Moore GR. MCD, EPR and NMR spectroscopic studies of rabbit hemopexin and its heme binding domain. Biochim Biophys Acta. 1995;1253:215–223. doi: 10.1016/0167-4838(95)00163-4. [DOI] [PubMed] [Google Scholar]
- 38.Freydank AC, Brandt W, Drager B. Protein structure modeling indicates hexahistidine-tag interference with enzyme activity. Proteins. 2008;72:173–183. doi: 10.1002/prot.21905. [DOI] [PubMed] [Google Scholar]
- 39.Chant A, Kraemer-Pecore CM, Watkin R, Kneale GG. Attachment of a histidine tag to the minimal zinc finger protein of the Aspergillus nidulans gene regulatory protein AreA causes a conformational change at the DNA-binding site. Protein Expr Purif. 2005;39:152–159. doi: 10.1016/j.pep.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 40.Seery VL, Müller-Eberhard U. Binding of porphyrins to rabbit hemopexin and albumin. J Biol Chem. 1973;248:3796–3800. [PubMed] [Google Scholar]
- 41.Morgan WT, Smith A, Koskelo P. The interaction of human serum albumin and hemopexin with porphyrins. Biochim Biophys Acta. 1980;624:271–285. doi: 10.1016/0005-2795(80)90246-9. [DOI] [PubMed] [Google Scholar]
- 42.Suzuki K, Kato H, Sakuma Y, Namiki H. Hemopexins suppress phorbol ester-induced necrosis of polymorphonuclear leucocytes. Cell Struct Funct. 2001;26:235–241. doi: 10.1247/csf.26.235. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki K, Kobayashi N, Doi T, Hijikata T, Machida I, Namiki H. Inhibition of Mg2+-dependent adhesion of polymorphonuclear leukocytes by serum hemopexdifferences in divalent-cation dependency of cell adhesion in the presence and absence of serum. Cell Struct Funct. 2003;28:243–523. doi: 10.1247/csf.28.243. [DOI] [PubMed] [Google Scholar]
- 44.Cheung PK, Stulp B, Immenschuh S, Borghuis T, Baller JF, Bakker WW. Is 100KF an isoform of hemopexin? Immunochemical characterization of the vasoactive plasma factor 100KF. J Am Soc Nephrol. 1999;10:1700–1708. doi: 10.1681/ASN.V1081700. [DOI] [PubMed] [Google Scholar]
- 45.Hrkal Z, Kodicek MB, Vodrazka Z. Kinetics of the conformational changes of hemopexin in acid media. Ann Clin Res. 1976;8(Suppl 17):239–243. [PubMed] [Google Scholar]
- 46.Müller-Eberhard U, English EC. Purification and partial characterization of human hemopexin. J Lab Clin Med. 1967;70:619–626. [PubMed] [Google Scholar]
- 47.Aisen P, Leibman A, Harris DC, Moss T. Human hemopexin. Preparation and magnetic properties. J Biol Chem. 1974;249:6824–6827. [PubMed] [Google Scholar]
- 48.Hvidberg V, Maniecki MB, Jacobsen C, Hojrup P, Moller HJ, Moestrup SK. Identification of the receptor scavenging hemopexin-heme complexes. Blood. 2005;106:2572–2579. doi: 10.1182/blood-2005-03-1185. [DOI] [PubMed] [Google Scholar]
- 49.Takahashi N, Takahashi Y, Putnam FW. Structure of human hemopexO-glycosyl and N-glycosyl sites and unusual clustering of tryptophan residues. Proc Natl Acad Sci USA. 1984;81:2021–2025. doi: 10.1073/pnas.81.7.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Takahashi N, Takahashi Y, Putnam FW. Complete amino acid sequence of human hemopexin, the heme-binding protein of serum. Proc Natl Acad Sci USA. 1985;82:73–77. doi: 10.1073/pnas.82.1.73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Theorell H, Åkeson A. Reversible splitting of a homogeneous horse myoglobin. Ann Acad Sci Fennicae Ser A. 1955;II:303–312. [Google Scholar]
- 52.Tsemakhovich VA, Bamm VV, Shaklai M, Shaklai N. Vascular damage by unstable hemoglobins: the role of heme-depleted globin. Arch Biochem Biophys. 2005;436:307–315. doi: 10.1016/j.abb.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 53.Altruda F, Poli V, Restagno G, Argos P, Cortese R, Silengo L. The primary structure of human hemopexin deduced from cDNA sequence: evidence for internal, repeating homology. Nucleic Acids Res. 1985;13:3841–3859. doi: 10.1093/nar/13.11.3841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hrkal Z, Müller-Eberhard U. Partial characterization of the heme-binding serum glycoproteins rabbit and human hemopexin. Biochemistry. 1971;10:1746–1750. doi: 10.1021/bi00786a002. [DOI] [PubMed] [Google Scholar]
- 55.Kamboh MI, Ferrell RE. Genetic studies of low-abundance human plasma proteins. VI. Polymorphism of hemopexin. Am J Hum Genet. 1987;41:645–653. [PMC free article] [PubMed] [Google Scholar]
- 56.Kamboh MI, Bunker CH, Nwankwo MU, Ferrell RE. Hemopexa unique genetic polymorphism in populations of African ancestry. Hum Biol. 1993;65:655–660. [PubMed] [Google Scholar]
- 57.Mauk MR, Rosell FI, Lelj-Garolla B, Moore GR, Mauk AG. Metal ion binding to human hemopexin. Biochemistry. 2005;44:1864–1871. doi: 10.1021/bi0481747. [DOI] [PubMed] [Google Scholar]
- 58.Mauk MR, Rosell FI, Mauk AG. Chromatographically distinguishable heme insertion isoforms of human hemopexin. Biochemistry. 2007;46:15033–15041. doi: 10.1021/bi701821a. [DOI] [PubMed] [Google Scholar]
- 59.Goldfarb V, Trimble RB, De Falco M, Liem HH, Metcalfe SA, Wellner D, Müller-Eberhard U. An avian serum α1-glycoprotein, hemopexin, differing significantly in both amino acid and carbohydrate composition from mammalian (β-glycoprotein) counterparts. Biochemistry. 1986;25:6555–6562. doi: 10.1021/bi00369a033. [DOI] [PubMed] [Google Scholar]
- 60.Coddeville B, Stratil A, Wieruszeski JM, Oliver RW, Green BN, Spik G. Characterization of sheep hemopexin glycovariants. Glycoconj J. 1995;12:645–650. doi: 10.1007/BF00731260. [DOI] [PubMed] [Google Scholar]
- 61.van Gelder W, Huijskes-Heins MI, Hukshorn CJ, de Jeu-Jaspars CM, van Noort WL, van Eijk HG. Isolation, purification and characterization of porcine serum transferrin and hemopexin. Comp Biochem Physiol B Biochem Mol Biol. 1995;111:171–179. doi: 10.1016/0305-0491(94)00255-s. [DOI] [PubMed] [Google Scholar]
- 62.Debruyne EN, Vanderschaeghe D, Van Vlierberghe H, Vanhecke A, Callewaert N, Delanghe JR. Diagnostic value of the hemopexin N-glycan profile in hepatocellular carcinoma patients. Clin Chem. 2010;56:823–831. doi: 10.1373/clinchem.2009.139295. [DOI] [PubMed] [Google Scholar]
- 63.Inglis AS. Cleavage at aspartic acid. Methods Enzymol. 1983;91:324–332. doi: 10.1016/s0076-6879(83)91030-3. [DOI] [PubMed] [Google Scholar]
- 64.BeMiller JN. Acid-catalyzed hydrolysis of glycosides. Adv Carbohydr Chem Biochem. 1967;22:85–91. doi: 10.1016/s0096-5332(08)60151-4. [DOI] [PubMed] [Google Scholar]
- 65.Flatmark T. On the heterogeneity of beef heart cytochrome c. 3. A kinetic study of the non-enzymic deamidation of the main subfractions (Cy I-Cy 3) Acta Chem Scand. 1966;20:1487–1496. doi: 10.3891/acta.chem.scand.20-1487. [DOI] [PubMed] [Google Scholar]
- 66.Robinson NE, Robinson AB. Prediction of protein deamidation rates from primary and three-dimensional structure. Proc Natl Acad Sci USA. 2001;98:4367–4372. doi: 10.1073/pnas.071066498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Wang W, Antonsen K, Wang YJ, Wang DQ. pH dependent effect of glycosylation on protein stability. Eur J Pharm Sci. 2008;33:120–127. doi: 10.1016/j.ejps.2007.10.008. [DOI] [PubMed] [Google Scholar]
- 68.Mauk MR, Mauk AG. Metal ions and electrolytes regulate the dissociation of heme from human hemopexin at physiological pH. J Biol Chem. 2010;285:20499–20506. doi: 10.1074/jbc.M110.123406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yu HL, Chertkow HM, Bergman H, Schipper HM. Aberrant profiles of native and oxidized glycoproteins in Alzheimer plasma. Proteomics. 2003;3:2240–2248. doi: 10.1002/pmic.200300475. [DOI] [PubMed] [Google Scholar]
- 70.Rottoli P, Magi B, Cianti R, Bargagli E, Vagaggini C, Nikiforakis N, Pallini V, Bini L. Carbonylated proteins in bronchoalveolar lavage of patients with sarcoidosis, pulmonary fibrosis associated with systemic sclerosis and idiopathic pulmonary fibrosis. Proteomics. 2005;5:2612–2618. doi: 10.1002/pmic.200401206. [DOI] [PubMed] [Google Scholar]
- 71.Zhu L, Hope TJ, Hall J, Davies A, Stern M, Müller-Eberhard U, Stern R, Parslow TG. Molecular cloning of a mammalian hyaluronidase reveals identity with hemopexin, a serum heme-binding protein. J Biol Chem. 1994;269:32092–32097. [PubMed] [Google Scholar]
- 72.Laurent TC, Fraser JR. Hyaluronan. FASEB J. 1992;6:2397–2404. [PubMed] [Google Scholar]
- 73.Hrkal Z, Kuzelova K, Müller-Eberhard U, Stern R. Hyaluronan-binding properties of human serum hemopexin. FEBS Lett. 1996;383:72–74. doi: 10.1016/0014-5793(96)00225-6. [DOI] [PubMed] [Google Scholar]
- 74.Yang B, Yang BL, Savani RC, Turley EA. Identification of a common hyaluronan binding motif in the hyaluronan binding proteins RHAMM, CD44 and link protein. EMBO J. 1994;13:286–296. doi: 10.1002/j.1460-2075.1994.tb06261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Morgan WT, Muster P, Tatum F, Kao SM, Alam J, Smith A. Identification of the histidine residues of hemopexin that coordinate with heme-iron and of a receptor-binding region. J Biol Chem. 1993;268:6256–6262. [PubMed] [Google Scholar]
- 76.Banerji S, Hide BR, James JR, Noble ME, Jackson DG. Distinctive properties of the hyaluronan-binding domain in the lymphatic endothelial receptor Lyve-1 and their implications for receptor function. J Biol Chem. 2010;285:10724–10735. doi: 10.1074/jbc.M109.047647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Bakker WW, Baller JF, van Luijk WH. A kallikrein-like molecule and plasma vasoactivity in minimal change disease. Increased turnover in relapse versus remission. Contrib Nephrol. 1988;67:31–36. [PubMed] [Google Scholar]
- 78.Blau EB, Haas JE. Glomerular sialic acid and proteinuria in human renal disease. Lab Invest. 1973;28:477–481. [PubMed] [Google Scholar]
- 79.Bakker WW, Roskam G, Hardonk MJ, Vos JT, Bleumink E. The glomerular polyanion (GPA) of the rat kidney. III. Further characterization of a vaso-active serum factor which reduces GPA. Br J Exp Pathol. 1985;66:47–55. [PMC free article] [PubMed] [Google Scholar]
- 80.Cheung PK, Klok PA, Baller JF, Bakker WW. Induction of experimental proteinuria in vivo following infusion of human plasma hemopexin. Kidney Int. 2000;57:1512–1520. doi: 10.1046/j.1523-1755.2000.00996.x. [DOI] [PubMed] [Google Scholar]
- 81.Yonan CR, Duong PT, Chang FN. High-efficiency staining of proteins on different blot membranes. Anal Biochem. 2005;338:159–161. doi: 10.1016/j.ab.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 82.Bakker WW, Henning RH, van Son WJ, van Pampus MG, Aarnoudse JG, Niezen-Koning KE, Borghuis T, Jongman RM, van Goor H, Poelstra K, Navis G, Faas MM. Vascular contraction and preeclampsia: downregulation of the angiotensin receptor 1 by hemopexin in vitro. Hypertension. 2009;53:959–964. doi: 10.1161/HYPERTENSIONAHA.108.127951. [DOI] [PubMed] [Google Scholar]
- 83.Bakker WW, van Dael CM, Pierik LJ, van Wijk JA, Nauta J, Borghuis T, Kapojos JJ. Altered activity of plasma hemopexin in patients with minimal change disease in relapse. Pediatr Nephrol. 2005;20:1410–1415. doi: 10.1007/s00467-005-1936-3. [DOI] [PubMed] [Google Scholar]
- 84.Krikken JA, van Ree RM, Klooster A, Seelen MA, Borghuis T, Lems SP, Schouten JP, Bakker WW, Gans RO, Navis G, Bakker SJ. High plasma hemopexin activity is an independent risk factor for late graft failure in renal transplant recipients. Transpl Int. 2010;23:805–812. doi: 10.1111/j.1432-2277.2010.01055.x. [DOI] [PubMed] [Google Scholar]
- 85.Bakker WW, Donker RB, Timmer A, van Pampus MG, van Son WJ, Aarnoudse JG, van Goor H, Niezen-Koning KE, Navis G, Borghuis T, Jongman RM, Faas MM. Plasma hemopexin activity in pregnancy and preeclampsia. Hypertens Pregnancy. 2007;26:227–239. doi: 10.1080/10641950701274896. [DOI] [PubMed] [Google Scholar]
- 86.Cheung PK, Baller JF, Bakker WW. Oxygen-dependent injury by a human plasma factor associated with minimal change disease. Pediatr Nephrol. 1998;12:452–458. doi: 10.1007/s004670050486. [DOI] [PubMed] [Google Scholar]
- 87.Kapojos JJ, Poelstra K, Borghuis T, Banas B, Bakker WW. Regulation of plasma hemopexin activity by stimulated endothelial or mesangial cells. Nephron Physiol. 2004;96:P1–P10. doi: 10.1159/000075574. [DOI] [PubMed] [Google Scholar]
- 88.Dönges R, Römisch J, Stauss H, Brazel D. Separation of antithrombin III variants by micellar electrokinetic chromatography. J Chromatogr A. 2001;924:307–313. doi: 10.1016/s0021-9673(01)00827-5. [DOI] [PubMed] [Google Scholar]
- 89.Franzén LE, Svensson S, Larm O. Structural studies on the carbohydrate portion of human antithrombin III. J Biol Chem. 1980;255:5090–5093. [PubMed] [Google Scholar]
- 90.Jensen LB, Dam J, Teisner B. Identification and removal of polymer- and aggregate-forming proteins in human plasma albumin preparations. Vox Sang. 1994;67:125–131. doi: 10.1111/j.1423-0410.1994.tb01646.x. [DOI] [PubMed] [Google Scholar]
- 91.Rosell FI, Mauk MR, Mauk AG. Effects of metal ion binding on structural dynamics of human hemopexin. Biochemistry. 2007;46:9301–9309. doi: 10.1021/bi7008079. [DOI] [PubMed] [Google Scholar]
- 92.Gallimore MJ, Friberger P. Simple chromogenic peptide substrate assays for determining prekallikrein, kallikrein inhibition and kallikrein “like” activity in human plasma. Thromb Res. 1982;25:293–298. [PubMed] [Google Scholar]
- 93.Andrew M, Manno M, Karpatkin M. Demonstration of kallikrein-like protease activity in nonactivated plasma of patients with Cooley's anemia. Blood. 1983;61:232–237. [PubMed] [Google Scholar]
- 94.Ewald GA, Eisenberg PR. Plasmin-mediated activation of contact system in response to pharmacological thrombolysis. Circulation. 1995;91:28–36. doi: 10.1161/01.cir.91.1.28. [DOI] [PubMed] [Google Scholar]
- 95.Stellwagen E. Use of blue dextran as a probe for the nicotinamide adenine dinucleotide domain in proteins. Acc Chem Res. 1977;10:92–98. [Google Scholar]
- 96.Stellwagen E. Chromatography on immobilized reactive dyes. Methods Enzymol. 1990;182:343–357. doi: 10.1016/0076-6879(90)82030-6. [DOI] [PubMed] [Google Scholar]
- 97.Wierenga RK, Terpstra P, Hol WG. Prediction of the occurrence of the ADP-binding βαβ-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986;187:101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- 98.Chambers BB, Dunlap RB. Interaction of dihydrofolate reductase from amethopterin-resistant Lactobacillus casei with cibacron blue, blue dextran, and affi-gel blue. J Biol Chem. 1979;254:6515–6521. [PubMed] [Google Scholar]
- 99.Stefanovic V, Vlahovic P. Divalent cation-activated ecto-ATPase activity of rat glomerular mesangial cells. Arch Physiol Biochem. 1995;103:15–20. doi: 10.3109/13813459509007557. [DOI] [PubMed] [Google Scholar]
- 100.Vretblad P, Hjorth R. The use of wheat-germ lectin-Sepharose for the purification of human haemopexin. Biochem J. 1977;167:759–764. doi: 10.1042/bj1670759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Smith A, Morgan WT. Haem transport to the liver by haemopexin. Receptor-mediated uptake with recycling of the protein. Biochem J. 1979;182:47–54. doi: 10.1042/bj1820047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bakker WW, Melgert BN, Faas MM. Hemopexanti-inflammatory, pro-inflammatory, or both? J Leukoc Biol. 2010;87:1–2. doi: 10.1189/jlb.0809560. [DOI] [PubMed] [Google Scholar]
- 103.Fink MP. Editorial: hemopexnewest member of the anti-inflammatory mediator club. J Leukoc Biol. 2009;86:203–204. doi: 10.1189/jlb.0309137. [DOI] [PubMed] [Google Scholar]
- 104.Warren HS, Lin T. Response to the letter of Drs. Bakker, Melgert, and Faas regarding our parent article. J Leukoc Biol. 2010;87:3. [Google Scholar]
- 105.Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPASy Server. In: Walker JM, editor. The proteomics protocols handbook. Totowa: Humana Press; 2005. pp. 571–607. [Google Scholar]
- 106.Gekko K, Ohmae E, Kameyama K, Takagi T. Acetonitrile-protein interactions: amino acid solubility and preferential solvation. Biochim Biophys Acta. 1998;1387:195–205. doi: 10.1016/s0167-4838(98)00121-6. [DOI] [PubMed] [Google Scholar]
- 107.Noiva R, Pete MJ, Babin DR. Bovine serum hemopexproperties of the protein from a single animal. Comp Biochem Physiol B Biochem Mol Biol. 1987;88:341–347. doi: 10.1016/0305-0491(87)90125-8. [DOI] [PubMed] [Google Scholar]
- 108.Heumann D, Adachi Y, Le Roy D, Ohno N, Yadomae T, Glauser MP, Calandra T. Role of plasma, lipopolysaccharide-binding protein, and CD14 in response of mouse peritoneal exudate macrophages to endotoxin. Infect Immun. 2001;69:378–385. doi: 10.1128/IAI.69.1.378-385.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Esterbauer H, Gebicki J, Puhl H, Jurgens G. The role of lipid peroxidation and antioxidants in oxidative modification of LDL. Free Radic Biol Med. 1992;13:341–390. doi: 10.1016/0891-5849(92)90181-f. [DOI] [PubMed] [Google Scholar]
- 110.Miller YI, Smith A, Morgan WT, Shaklai N. Role of hemopexin in protection of low-density lipoprotein against hemoglobin-induced oxidation. Biochemistry. 1996;35:13112–13117. doi: 10.1021/bi960737u. [DOI] [PubMed] [Google Scholar]
- 111.Nagy E, Eaton JW, Jeney V, Soares MP, Varga Z, Galajda Z, Szentmiklosi J, Mehes G, Csonka T, Smith A, Vercellotti GM, Balla G, Balla J. Red cells, hemoglobin, heme, iron, and atherogenesis. Arterioscler Thromb Vasc Biol. 2010;30:1347–1353. doi: 10.1161/ATVBAHA.110.206433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Watanabe J, Chou KJ, Liao JC, Miao Y, Meng HH, Ge H, Grijalva V, Hama S, Kozak K, Buga G, Whitelegge JP, Lee TD, Farias-Eisner R, Navab M, Fogelman AM, Reddy ST. Differential association of hemoglobin with proinflammatory high density lipoproteins in atherogenic/hyperlipidemic mice. A novel biomarker of atherosclerosis. J Biol Chem. 2007;282:23698–23707. doi: 10.1074/jbc.M702163200. [DOI] [PubMed] [Google Scholar]
- 113.Kunitake ST, Carilli CT, Lau K, Protter AA, Naya-Vigne J, Kane JP. Identification of proteins associated with apolipoprotein A-I-containing lipoproteins purified by selected-affinity immunosorption. Biochemistry. 1994;33:1988–1993. doi: 10.1021/bi00174a003. [DOI] [PubMed] [Google Scholar]
- 114.Spagnuolo MS, Cigliano L, D'Andrea LD, Pedone C, Abrescia P. Assignment of the binding site for haptoglobin on apolipoprotein A-I. J Biol Chem. 2005;280:1193–1198. doi: 10.1074/jbc.M411390200. [DOI] [PubMed] [Google Scholar]
- 115.Watanabe J, Grijalva V, Hama S, Barbour K, Berger FG, Navab M, Fogelman AM, Reddy ST. Hemoglobin and its scavenger protein haptoglobin associate with apoA-1-containing particles and influence the inflammatory properties and function of high density lipoprotein. J Biol Chem. 2009;284:18292–18301. doi: 10.1074/jbc.M109.017202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Schaer DJ, Schaer CA, Buehler PW, Boykins RA, Schoedon G, Alayash AI, Schaffner A. CD163 is the macrophage scavenger receptor for native and chemically modified hemoglobins in the absence of haptoglobin. Blood. 2006;107:373–380. doi: 10.1182/blood-2005-03-1014. [DOI] [PubMed] [Google Scholar]
- 117.Okazaki H, Taketani S, Kohno H, Tokunaga R, Kobayashi Y. The hemopexin receptor on the cell surface of human polymorphonuclear leukocytes. Cell Struct Funct. 1989;14:129–140. doi: 10.1247/csf.14.129. [DOI] [PubMed] [Google Scholar]
- 118.Stanley KK. Homology with hemopexin suggests a possible scavenging function for S-protein/ vitronectin. FEBS Lett. 1986;199:249–253. doi: 10.1016/0014-5793(86)80489-6. [DOI] [PubMed] [Google Scholar]
- 119.Jenne D, Stanley KK. Nucleotide sequence and organization of the human S-protein gene: repeating peptide motifs in the “pexin” family and a model for their evolution. Biochemistry. 1987;26:6735–6742. doi: 10.1021/bi00395a024. [DOI] [PubMed] [Google Scholar]
- 120.Suzuki S, Argraves WS, Pytela R, Arai H, Krusius T, Pierschbacher MD, Ruoslahti E. cDNA and amino acid sequences of the cell adhesion protein receptor recognizing vitronectin reveal a transmembrane domain and homologies with other adhesion protein receptors. Proc Natl Acad Sci USA. 1986;83:8614–8618. doi: 10.1073/pnas.83.22.8614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Terentiev AA, Moldogazieva NT. Cell adhesion proteins and α-fetoprotein. Similar structural motifs as prerequisites for common functions. Biochemistry (Mosc) 2007;72:920–935. doi: 10.1134/s0006297907090027. [DOI] [PubMed] [Google Scholar]
- 122.Morgan WT, Muster P, Tatum FM, McConnell J, Conway TP, Hensley P, Smith A. Use of hemopexin domains and monoclonal antibodies to hemopexin to probe the molecular determinants of hemopexin-mediated heme transport. J Biol Chem. 1988;263:8220–8225. [PubMed] [Google Scholar]
- 123.Rothwangl KB, Rong L. Analysis of a conserved RGE/RGD motif in HCV E2 in mediating entry. Virol J. 2009;6:12. doi: 10.1186/1743-422X-6-12. doi: 10.1186/1743-422X6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Porath J, Olin B. Immobilized metal ion affinity adsorption and immobilized metal ion affinity chromatography of biomaterials. Serum protein affinities for gel-immobilized iron and nickel ions. Biochemistry. 1983;22:1621–1630. doi: 10.1021/bi00276a015. [DOI] [PubMed] [Google Scholar]
- 125.Porath J, Olin B, Granstrand B. Immobilized-metal affinity chromatography of serum proteins on gel-immobilized group III A metal ions. Arch Biochem Biophys. 1983;225:543–547. doi: 10.1016/0003-9861(83)90065-6. [DOI] [PubMed] [Google Scholar]
- 126.Mantovaara T, Pertoft H, Porath J. Further characterization of carboxymethylated aspartic acid agarose. Purification of human α2-macroglobulin and hemopexin. Biotechnol Appl Biochem. 1991;13:371–379. [PubMed] [Google Scholar]
- 127.Andersson L. Fractionation of human serum proteins by immobilized metal affinity chromatography. J Chromatogr. 1984;315:167–174. doi: 10.1016/s0021-9673(01)90734-4. [DOI] [PubMed] [Google Scholar]
- 128.Rosell FI, Mauk MR, Mauk AG. pH- and metal ion-linked stability of the hemopexin-heme complex. Biochemistry. 2005;44:1872–1879. doi: 10.1021/bi0480077. [DOI] [PubMed] [Google Scholar]
- 129.Vidaud C, Dedieu A, Basset C, Plantevin S, Dany I, Pible O, Quemeneur E. Screening of human serum proteins for uranium binding. Chem Res Toxicol. 2005;18:946–953. doi: 10.1021/tx050038v. [DOI] [PubMed] [Google Scholar]
- 130.Smith A, Rish KR, Lovelace R, Hackney JF, Helston RM. Role for copper in the cellular and regulatory effects of heme-hemopexin. Biometals. 2009;22:421–437. doi: 10.1007/s10534-008-9178-z. [DOI] [PubMed] [Google Scholar]
- 131.Shipulina NV, Smith A, Morgan WT. Effects of reduction and ligation of heme iron on the thermal stability of heme-hemopexin complexes. J Protein Chem. 2001;20:145–154. doi: 10.1023/a:1011033625009. [DOI] [PubMed] [Google Scholar]
- 132.Carter P, Wells JA. Dissecting the catalytic triad of a serine protease. Nature. 1988;332:564–568. doi: 10.1038/332564a0. [DOI] [PubMed] [Google Scholar]
- 133.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]


