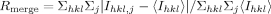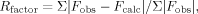Abstract
UDP-hexose 4-epimerases play a pivotal role in lipopolysaccharide (LPS) biosynthesis and Leloir pathway. These epimerases are classified into three groups based on whether they recognize nonacetylated UDP-hexoses (Group 1), both N-acetylated and nonacetylated UDP-hexoses (Group 2) or only N-acetylated UDP-hexoses (Group 3). Although the catalysis has been investigated extensively, yet a definitive model rationalizing the substrate specificity of all the three groups on a common platform is largely lacking. In this work, we present the crystal structure of WbgU, a novel UDP-hexose 4-epimerase that belongs to the Group 3. WbgU is involved in biosynthetic pathway of the unusual glycan 2-deoxy-L-altruronic acid that is found in the LPS of the pathogen Pleisomonas shigelloides. A model that defines its substrate specificity is proposed on the basis of the active site architecture. Representatives from all the three groups are then compared to rationalize their substrate specificity. This investigation reveals that the Group 3 active site architecture is markedly different from the “conserved scaffold” of the Group 1 and the Group 2 epimerases and highlights the interactions potentially responsible for the origin of specificity of the Group 3 epimerases toward N-acetylated hexoses. This study provides a platform for further engineering of the UDP-hexose 4-epimerases, leads to a deeper understanding of the LPS biosynthesis and carbohydrate recognition by proteins. It may also have implications in development of novel antibiotics and more economic synthesis of UDP-GalNAc and downstream products such as carbohydrate based vaccines.
Keywords: lipopolysaccharide, N-acetylglucosamine, Rossmann fold, 4-epimerase, galactose metabolism
Introduction
UDP-hexose 4-epimerases belong to the short chain dehydrogenase/reductase enzyme superfamily.1 These enzymes typically have distinct N- and C-terminal domains. The N-terminal domain is a modified Rossmann fold that binds one molecule of NAD(H) with the relatively small C-terminal domain being mainly responsible for substrate binding.2 These enzymes are important in myriad biological pathways such as lipopolysaccharide (LPS) biosynthesis3,4 and galactose metabolism.5 LPS is implicated in several facets of host-pathogen interactions such as resistance to serum-mediated killing, phagocytosis and killing by cationic peptides6–10 whereas a malfunctioning UDP-hexose 4-epimerase can lead to epimerase-deficiency galactosemia in humans.11,12 General details of the epimerization reaction have been thoroughly investigated by Wilson and Hogness,13 Bauer et al.,2 Thoden et al.,14 Liu et al.,15 Wohlers et al.11 among others. Briefly, the reaction proceeds via abstraction of a proton from the 4'-hydroxyl group by a tyrosine acting as an active site base, its pKa being lowered by a neighboring lysine. A serine residue is thought to facilitate this abstraction by formation of a low barrier hydrogen bond, hence the term SYK catalytic triad.14 This proton abstraction and subsequent stereospecific hydride transfer to NAD+ leads to formation of a keto intermediate. A net rotation of 180° in this keto intermediate allows a nonstereospecific return of hydride at the 4' position resulting in formation of either of the epimers. These details are illustrated in Scheme 1 of the Supporting Information.
Despite the conservation of catalytic residues and the mechanism of catalysis, UDP-hexose 4-epimerases (henceforth referred to as “the epimerases”) can recognize different UDP-hexose substrates in a specific manner. Ishiyama et al. classified the epimerases into three different groups based on their substrate specificity: Group 1 epimerases recognize nonacetylated UDP-hexoses; Group 2 epimerases are promiscuous toward N-acetylated and nonacetylated UDP-hexoses and Group 3 epimerases strongly prefer N-acetylated UDP-hexoses.4 In the context of current work, WbgU belongs to the Group 3 epimerases and is critical for synthesis of 2-deoxy-L-altruronic acid (2Ac-AltUA) in Plesiomonas shigelloides.3 2Ac-AltUA, in turn, is one of the two glycans that form the O-unit of the LPS of this Gram-negative bacterium. Previous work on the substrate specificity of the epimerases has been mainly focused on the Group 1 and the Group 2 epimerases, WbpP being the only representative of the group 3 epimerases that has been investigated for its substrate specificity.4,16 In this work, we first propose a structure based model to define substrate specificity of the Group 3 epimerases. We then put this model of substrate recognition in the perspective of substrate recognition by the Group 1 and the Group 2 epimerases and finally, we attempt to consolidate certain previously reported investigations regarding UDP-hexose 4-epimerases.
Results
Overall structure
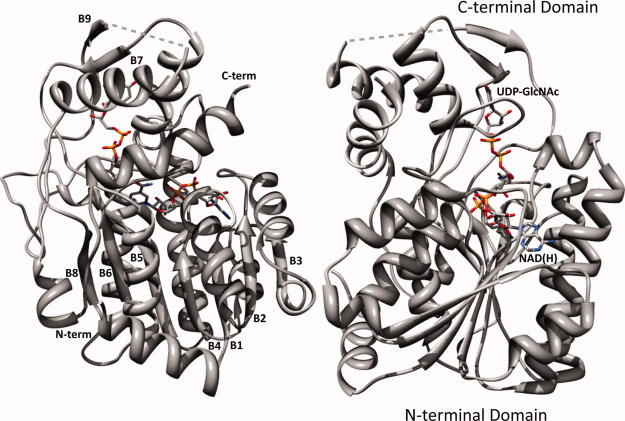
WbgU crystallizes in the space group P32 with generally well defined electron density, exception being a short region between His287 and Ile293. The electron density for the cofactor NAD(H) as well as the substrate UDP-GlcNAc is also fairly well defined (Fig. 1). The data processing and refinement statistics are listed in Table I. The asymmetric unit of the WbgU crystal structure contains a tetramer. Two of the four subunits of this tetramer are related by a 2-fold symmetry axis that associates them via a 4-helix bundle (Fig. 1, Supporting Information). Since there is a unique axis of symmetry in the asymmetric unit and since WbgU is known already known to exist as a dimer in solution, 3 this is the most likely dimeric interface observed in solution state. The 4-helix bundle formed at the dimeric interface is the commonly observed structural motif in several other epimerases,2,4 thus further corroborating this position as the physiological dimeric interface. The overall tertiary structure of WbgU is comprised of two domains, N-terminal domain and C-terminal domain (Fig. 2). The N-terminal domain is a Rossmann type fold with strand order 3214567 and contains two “topological switch points,” where the helices connecting the consecutive β strands switch planes. The first topological switch point is formed near carboxy edge of Strand 2 and Strand 3 and results in formation of a crevice containing the adenine moiety of NAD(H). The second topological switch point in the β sheet is at the carboxy edge of Strand 6. This switch results formation of another crevice between carboxy edge of Strand 5 and Strand 6. GlcNAc moiety of the substrate UDP-GlcNAc sits in this crevice. The C-terminal domain contains substrate binding region of the active site and is contributed by Phe194-Tyr238, Ala264-Ala308, Ile323-Gly342. NAD(H) sits on and parallel to the carboxy edge of the β sheet with UDP-GlcNAc approximately perpendicular to it.
Figure 1.
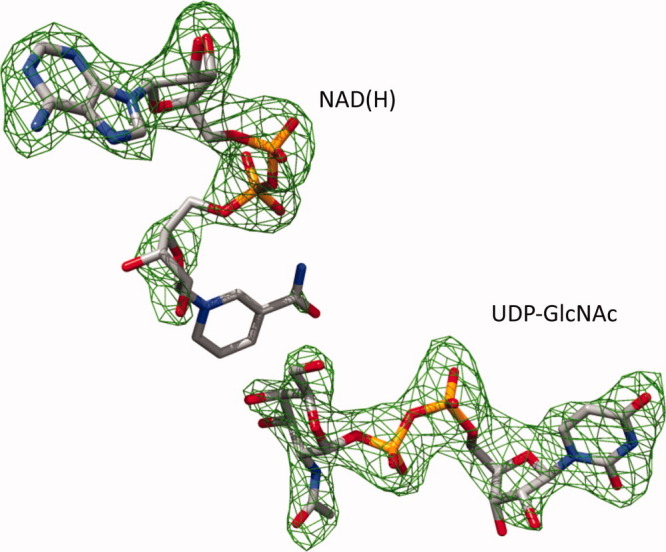
Difference electron density map at the active site of WbgU. Fo-Fc electron density map in the region of UDP-GlcNAc and NAD(H) depicted at an absolute electron density of 0.15 e/Å3 corresponding to 3.0σ cutoff. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table I.
Data Collection and Refinement Statistics
| Data collection statistics | |
| Source | Beamline 9-2 SSRL |
| Space group | P32 |
| Unit cell parameters | |
| a, b, c (Å) | 78.1, 78.1, 231.9 |
| α, β, γ (deg) | 90, 90, 120 |
| Temperature (K) | 103 |
| Wavelength (Å) | 0.97 |
| Resolution (Å) | 44.03–2.50 (2.56–2.50)a |
| Rmergeb | 0.13 (0.9) |
| Completeness (%) | 94.7 (88.8) |
| 〈I/σ(I)〉 | 13.5 (1.4) |
| Number of unique reflections | 51939 (3563) |
| Average redundancy | 4.3 (4.1) |
| B factor from Wilson plot (Å2) | 44.9 |
| PDB entry | 3LU1 |
| Refinement statistics | |
| Resolution (Å) | 44.03-2.50 |
| Number of reflections | 51878 |
| Rfactorc | 20.7 |
| Rfreec | 25.8 |
| Number of protein atoms | 10684 |
| Number of water atoms | 182 |
| Number of ligand atoms | 332 |
| RMS deviation from ideal values for bond distances (Å)d | 0.01 |
| RMS deviation from ideal values for bond angles (deg)d | 1.15 |
| Average B-factors | |
| Main chain (Å2) | 36.6 |
| Side chain and waters (Å2) | 37.0 |
| All atoms (Å2) | 36.8 |
| Ramachandran plote | |
| Favored (%) | 95.8 |
| Disallowed (%) | 0.3 |
Figure 2.
Overall tertiary structure of WbgU/NAD(H)/UDP-GlcNAc complex. N-terminal domain is a modified Rossmann type fold with the cofactor NAD(H) sitting on and parallel to the carboxy edge of the β sheet. The C-terminal domain binds the substrate UDP-GlcNAc. β strands are numbered from the N- to the C- terminal. Dashed grey lines represent the six-residue loop from His287-Ile293 that was not modeled due to poorly defined electron density. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Overall sequence and structure comparison
As expected, the polypeptide sequence of WbgU bears the closest resemblance to WbpP (67%) (Fig. 3). The sequence identity with CGne and HGal is 23% and with GalE it is 25%. The most prominent differences in WbgU polypeptide sequence, when compared to the epimerases from group 1 and group 2 are: (i) insertion of an N-terminal α-helix in WbgU at Tyr1-Pro16,† this region being completely absent in GalE, CGne and HGal; (ii) insertion of a six residue loop in GalE, CGne and HGal at Arg200-Gln201 position of WbgU; (iii) insertion of an eight residue loop GalE, CGne and HGal at the Asp229-Gly230 position of WbgU and (iv) deletion of four residues in GalE, CGne, and HGal near His292-Glu297 region of WbgU. Although the sequence identity is relatively low across the three groups, the overall similarity in tertiary structures of GalE, HGal, WbpP and WbgU is very high with an overall r.m.s.d. of the multiple structure alignment being 1.08 Å, as computed by MultiProt.19 Despite this high degree of homology in tertiary structure, local changes in the polypeptide fold are evident throughout and are most pronounced in the C-terminal domain (Fig. 2, Supporting Information).
Figure 3.
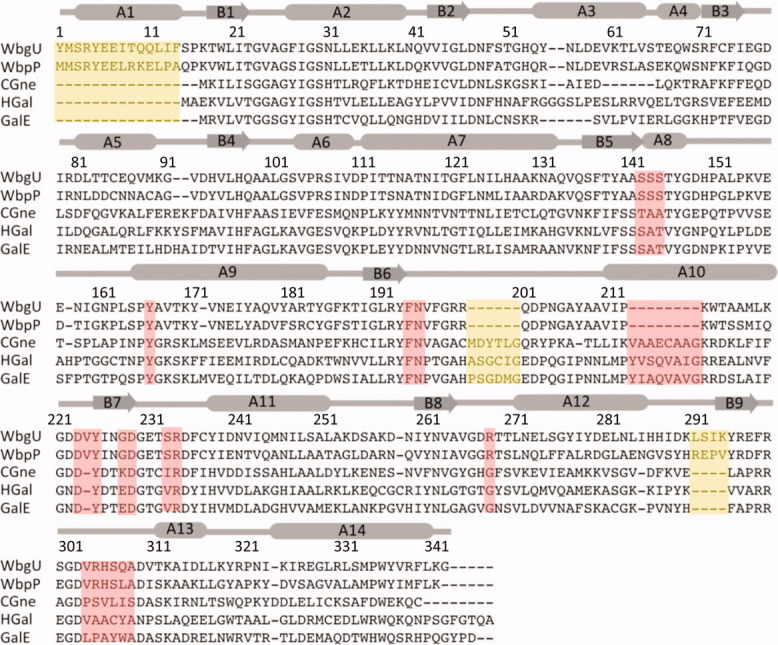
Multiple sequence alignment between 5 different UDP-hexose 4-epimerases. Regions deemed important for determination of substrate specificity are highlighted in pink. Regions highlighted in yellow are distinct structural variations that do not have a direct bearing on substrate binding or catalysis. WbgU is UDP-GalNAc 4-epimerase from P. shigelloides; WbpP is UDP-GlcNAc 4-epimerase from P. aeruginosa; CGne is UDP-Glc/GlcNAc 4-epimerase (Gne) from C. jejuni; HGal is UDP-Glc/GlcNAc 4-epimerase from Homo sapiens; GalE is UDP-Glc 4-epimerase from E.coli. The numbering and the secondary structure assignment corresponds the structure of WbgU. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Active site and its comparison across the three groups
The cofactor NAD(H) is bound at the carboxy edge of the β sheet of the N-terminal domain. This region is fairly conserved across all the three groups. The catalytically important Tyr166 and Ser142 are also highly conserved. The substrate binding region is contributed by the residues Ser142-Ser144, Phe194-Asn195, Ala209-Trp214, Tyr225-Arg234, and Glu297-Ala308 [Figs. 4(a, b)]. This region contains potentially important variations (Fig. 5): Ser143 in WbgU is substituted Ala in CGne, HGal and GalE; Ser233 in WbgU is substituted by Ile in CGne and by Val in GalE and HGal; Arg268 in WbgU is substituted by Gly in CGne, HGal and GalE; Arg304 is substituted by Ser in CGne, Ala in HGal and Pro in GalE; His305 in WbgU is substituted by Val in CGne and by Ala in GalE and HGal and Ser306 of WbgU is substituted by a Cys in the group 2 and by a Tyr in the group 1 epimerases. The values estimated for active site volume and surface area of all the 4 experimentally obtained structures, that is, GalE, HGal, WbgU, and WbpP reveal that the Group 2 epimerases have the largest active site volumes and surface areas with the Group 3 having the smallest (Table II).
Figure 4.
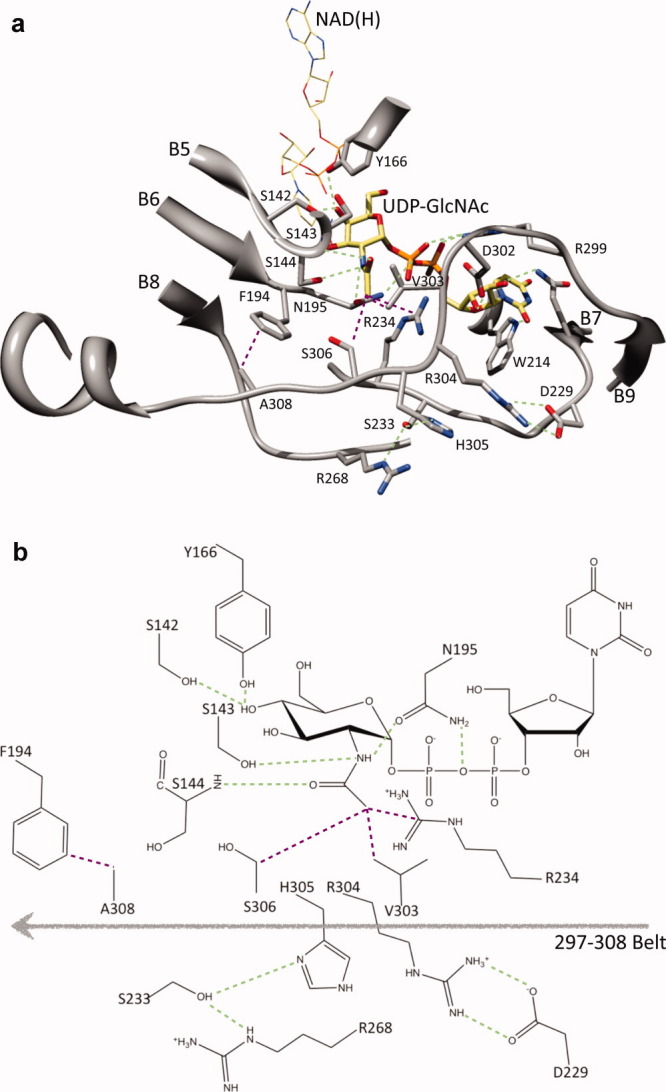
Architecture of the substrate binding region of WbgU. A salt bridge between Arg304 and Asp229 on one side and a hydrogen bonding network formed between His305, Ser233, and Arg268 launch the substrate binding region in a conformation that results in the formation of a hydrophobic cluster contributed by Val303, Arg234, and Ser303. This hydrophobic cluster directly interact with the CH3- group of the GlcNAc moiety. In addition, this conformation results in an interaction between Ala308 and Phe194. This interaction places Asn195 in a hydrogen bond with NH- group of the GlcNAc moiety and the oxy bridge of the diphospho moiety. In addition, a hydrogen bond is formed between Ser143 and NH- group of the GlcNAc moiety and between main chain NH- of Ser144 and CO- group of the GlcNAc moiety. (a) Important interactions with the GlcNAc moiety of the substrate UDP-GlcNAc are highlighted as green (polar interactions) or purple (nonpolar interactions). Tyr166 and Ser142 are hydrogen bonded to 4' hydroxyl group. This region defines the catalytic motif and is highly conserved as is Asn195. The regions defining the substrate binding site are mainly contributed by residues 209-214, 225-234 and 297-308. (b) A schematic representation of the interactions deemed most important in the substrate recognition and catalysis. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Figure 5.
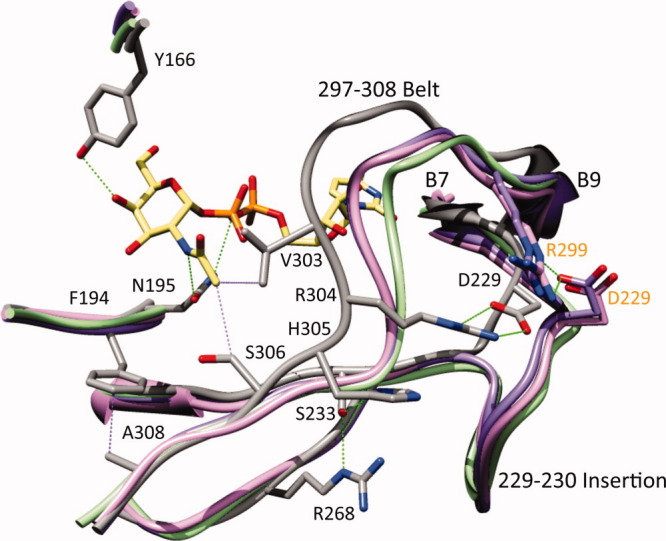
Substrate recognition by the group 1 and the group 2 UDP-hexose 4-epimerases from the perspective of the group 3. His305 in WbgU is substituted by Val in CGne and by Ala in GalE and HGal; Ser233 in WbgU is substituted by Ile in CGne and by Val in GalE and HGal; Arg268 in WbgU is substituted by Gly in CGne, HGal, and GalE (side chains are not shown for clarity). Similarly Arg304 is conserved in WbgU and WbpP whereas being substituted by Ser in CGne, Ala in HGal and Pro in GalE. The loss of the hydrogen bonding network at His305 position and the salt bridge at Arg304 position result in an altered conformation of substrate binding loop in the Group 1 and the Group 2 epimerases. In addition, the insertion of an 8 residue loop at the Asp229 causes formation of a salt bridge with Arg299. The absence of the polar interactions in the Group 1 and the Group 2 epimerases at the 304 and 305 positions in combination with the presence of a salt bridge at the 8 residue insertion between Asp229-Arg299, thus dictates the conformation of the substrate binding region in the group 1 and the group 2 epimerases. The common architecture of the substrate binding region in the group 1 and the group 2 epimerases has one important variation: Ser306 of WbgU (and WbpP) is substituted by a Cys in the Group 2 and by a Tyr in the group 1 hence restricting the access of bulky N-acetyl group to the active site of the group 1 epimerases, which in turn makes them specific towards the nonacetylated substrates. The labels and numbering in black color correspond to WbgU; the labels and numbering in orange color correspond to HGal. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Table II.
Active Site Dimensions of UDP-hexose 4-epimerases
| Protein | Classification | PDB id | Vol (Å3) | SA (Å2) |
|---|---|---|---|---|
| GalE15 | Group 1 | 1NAH | 3700 | 2600 |
| HGal14 | Group 2 | 1HZJ | 4000 | 2800 |
| WbpP4 | Group 3 | 1SB8 | 2900 | 2100 |
| WbgU | Group 3 | 3LU1 | 2600 | 1900 |
Discussion
The catalysis carried out by UDP-hexose 4-epimerases has been extensively investigated using biochemical and structural studies. However, a well-defined rational model ceases to exist for the substrate specificity. This is especially true for the Group 3 epimerases that are highly specific toward N-acetylated hexoses. In this work, we investigated the structure of WbgU, a novel UDP-GalNAc 4-epimerase that belongs to the Group 3. This investigation led us to propose a structure based model that rationalizes the substrate specificity of the Group 3 epimerases. According to this model, the region between Glu297 and Ala308 provides the scaffold that defines the substrate specificity of WbgU. We call this region “297-308 belt” (Fig. 4). Two sets of interactions stabilize the conformation of the 297-308 belt: first, a salt bridge formed between Arg304 and Asp229 and second, a hydrogen bond network between His305, Ser233, and Arg268.
The 297-308 belt, in turn, resorts to a conformation that leads to several interactions potentially critical in determination of the substrate specificity (Fig. 4). First, it leads to formation of a hydrophobic cluster contributed by Arg234, Val303, and Ser306. This hydrophobic cluster interacts with the CH3- group of the N-acetyl moiety of the UDP-GlcNAc. Second, it causes the relatively conserved Ala308 to interact with Phe194. This interaction places the loop containing Phe194 and Asn195 in a position that causes the Asn195 to hydrogen bond with NH- group of the GlcNAc moiety on one side and the oxy-bridge of the diphospho moiety of UDP-GlcNAc on the other side. Thirdly, the conformation of the 294-308 belt orients the substrate in such a manner that the NH- group of the GlcNAc moiety hydrogen bonds with Ser143 and the CO- group of the GlcNAc moiety also forms a weak hydrogen bond (3.5 Å) with the main chain NH- of Ser144. Thus, the change in conformation of the 297-308 belt in the Group 3 epimerases (relative to the Group 1 and the Group 2 epimerases) is transmitted to other regions of the substrate binding pocket, directly by the interaction between Ala308 and Phe194 and indirectly via the substrate to render the observed specificity. It is also noted that Ser143 in the Group 3 epimerases is substituted by an Ala in the Group 1 and the Group 2 epimerases and could be a critical variation.
A comparison of WbgU with the Group 1 (GalE), the Group 2 (CGne and HGal) and the Group 3 (WbpP) epimerases further reinforced our model. It was found that all the aforesaid residues proposed to be important in determination of substrate specificity of the Group 3 epimerases are conserved in the Group 3 (WbgU and WbpP) epimerases whereas being variant among the Group 1 (GalE) and the Group 2 (CGne, HGal) epimerases (Figs. 3 and 5). The hydrogen bond network corresponding to the His305 position of WbgU is absent in the group 1 and the group 2 epimerases and so is the salt bridge between Arg304 and Asp229. These variations combined with the insertion of an 8 residue loop in the group 1 and the group 2 epimerases at the position corresponding to Asp229-Val230 of WbgU, result in formation of a salt bridge between the Asp229 and Arg299 (numbering follows HGal) in the Group 1 and the Group 2 epimerases. This salt bridge and several van der Waals interactions render a conformation to this region that is very similar in the Group 1 and the Group 2 epimerases while being markedly different from the Group 3. This is also indicated by the fact that the 297-308 belt in the Group 3 is rotated by ∼10° from the corresponding region of the Group 1 and the Group 2 epimerases, the measurement being done from the C-terminal α-helix (Fig. 2, Supporting Information). The surface topology of the active site is very similar in both the Group 1 and the Group 2 epimerases while being significantly different from the Group 3 epimerases (Fig. 3, Supporting Information). The volume of active site is also much lesser in the Group 3 epimerases when compared with the Group 1 and the Group 2 epimerases (Table II). Thus, it follows that the substrate binding region of the Group 1 and the Group 2 epimerases are built on a very similar main chain scaffold, which is markedly different from the corresponding region in the Group 3 epimerases.
Finally, we attempted to consolidate some results previously reported in the literature, using our model of substrate recognition. First, if the substrate binding scaffold of the Group 1 epimerases and the Group 2 epimerases is so similar then what causes their substrate specificities to be different, that is, the Group 1 epimerases being specific toward nonacetylated UDP-hexoses, whereas the Group 2 epimerases having similar specificity toward nonacetylated and N-acetylated UDP-hexoses? The key seems to be the variation at Ser306 position. Ser306 in WbgU is substituted by a Cys in HGal and by a Tyr in GalE. Since the main chain scaffold of the Group 1 and the Group 2 epimerases is very similar, the bulkier side chain of this Tyr in the substrate binding region of the Group 1 epimerases can easily result in a greater specificity toward less bulkier substrates, that is, nonacetylated hexoses. The importance of this variation has been reported previously by a Tyr299Cys mutation engineered in the Group 1 epimerase GalE that switched its specificity from the Group 1 to Group 2,20 by a Ser306Tyr mutation engineered into the Group 2 epimerase from E. coli O86:B7 that switched its specificity from the Group 2 to the Group 121 and the corresponding Cys307Tyr mutation in HGal that switched its activity from the Group 2 to the Group 1.20,22 Second, can a single point mutation of Ser306Tyr also switch the substrate specificity from the Group 3 to the Group 1? Based on our model of substrate recognition, a single point mutation of Ser306Tyr can not be engineered in the Group 3 epimerases since it would place the Tyr residue in steric clashes in all of the allowed rotamer conformations. This immediately explains the loss in activity of this mutant reported for the Group 3 epimerase WbpP.4
To conclude, a general basis of the substrate specificity is apparent among the Group 1 and the Group 2 epimerases while the altered architecture of active site of the Group 3 epimerases causes the corresponding substrate specificity to be defined in a significantly different manner. This investigation provides a platform for structure-guided engineering of UDP-hexose 4-epimerases, which, in turn can lead to more economic production of UDP-GalNAc and downstream products such as carbohydrate based vaccines. It also enriches our comprehension of the LPS biosynthesis. A UDP-hexose 4-epimerase from Trypanosoma brucei, a protozoan causing a variety of tropical diseases, has been reported to be essential for survival of its host.23,24 Thus, a deeper understanding of substrate recognition by UDP-hexose 4-epimerases has potential implications in structure-based development of novel antibiotics. In addition, these results may also provide useful insights regarding other 4-epimerases. For example, a GlcNAc-P-P-Und/GalNAc-P-P-Und epimerase was recently reported to be critical for LPS biosynthesis in E.coli O157.25 Based on our studies and previous literature available regarding UDP-hexose 4-epimerases, it is very likely that the C-terminal domain of the GlcNAc-P-P-Und/GalNAc-P-P-Und epimerase sits close to the inner side of the inner membrane allowing GlcNAc-P-P-/GalNAc-P-P-moiety of the GlcNAc-P-P-Und/GalNAc-P-P-Und to wedge itself in the substrate binding region, whereas the N-terminal domain containing the cofactor sits further away from inner membrane.
Materials and Methods
Cloning, expression, and purification
WbgU was cloned, expressed, and purified with some modifications to the protocol described previously.3 Briefly, wbgU was inserted between XhoI and BamHI restriction sites of pET-15b plasmid (Novagen) with an N-terminal (His)6 fusion tag. The construct pET/wbgU was transformed into BL21(DE3) (Novagen) for protein expression. A small scale culture of these cells grown overnight in LB medium was transferred to large-scale LB medium in a ratio of 1:100. The large scale culture was allowed to grow to an OD595 of 0.8 at 30°C with constant shaking. Following this, the culture was induced with 0.15 mM IPTG and allowed to grow for 5 more hours. The cells were then harvested and stored at −0°C till further use. The pellet thus obtained, was washed and dissolved in buffer A (50 mM Tris-HCl pH 8.9, 500 mM NaCl and 5 mM imidazole), and sonicated over ice after addition of PMSF to a final concentration of 2 mM. The lysate was centrifuged for 40 min at 31,000g and the supernatant was loaded onto a 5 mL nickel-sepharose high performance affinity column (GE healthcare) previously equilibrated with buffer A. The column was washed with 20 column volumes of buffer B (50 mM Tris-HCl pH 8.9, 500 mM NaCl, and 50 mM imidazole) and eluted in buffer C (50 mM Tris-HCl pH 8.9, 500 mM NaCl and 500 mM imidazole). The buffer was exchanged to buffer D (200 mM glycine pH 10.0, 40 mM NaCl) and the protein concentrated to 25 mg/mL using Amicon Ultra centrifugal filter unit (Millipore). UDP-GlcNAc was added to a final concentration of 15 mM and the protein sample was allowed to equilibrate overnight at 4°C.
Crystallization, data collection and data processing
The WbgU/UDP-GlcNAc sample prepared above was used to search for condition(s) that could lead to crystallization using home made screens. The first crystals were obtained in 35% PEG 400, 200 mM ammonium sulfate using a hanging drop vapor diffusion set up. The drop was prepared by mixing 2 μL protein sample with 2 μL crystallization reagent and was allowed to equilibrate with the well solution containing 1000 μL crystallization reagent. The protein failed to crystallize in absence of UDP-GlcNAc. The best diffracting crystals could be obtained after optimization using crystallization screens (Hampton Research, CA) in a microbatch under oil set up,26,27 where 1 μL of WbgU/UDP-GlcNAc sample was set up with 1 μL of crystallization reagent (250 mM ammonium sulfate, 100 mM Bis-Tris pH 6.5, 25% w/v PEG 3,350) overlaid with 200 μL of mineral oil. These crystals grew to maximum dimensions of 300 μm × 300 μm × 100 μm within 3 weeks. After equilibration in a harvest buffer (250 mM ammonium sulfate, 0.1M BIS-TRIS pH 6.5, 30% w/v PEG 3,350) for 40 min, the crystals were transferred to cryoprotection buffer (250 mM ammonium sulfate, 0.1M BIS-TRIS pH 6.5, 30% w/v PEG 3,350, 5% glycerol), flash frozen in nylon cryoloops (Hampton Research) and stored in liquid nitrogen. A 2.5 Å resolution data set was collected at the Stanford Synchrotron Radiation Lightsource (SSRL). The data were processed with MOSFLM28 in the space group P3 (a = 78.1 Å, b = 78.1 Å, c = 231.9 Å). The data processing statistics are listed in Table I.
Structure determination
A sequence alignment was generated between WbgU and WbpP using CLUSTALW.29 A search model was prepared from crystal structure of WbpP4 by mutating the mismatched residues to alanines. Molecular replacement using Phaser30 was used to find a solution to the phase problem. A unique solution could be obtained in the enatiomorphic space group P32 with four monomers in the asymmetric unit that corresponds to a Matthews coefficient (VM) of 2.6231 and solvent content of 53%. This solution had a final log-likelihood gain of 6014.3 and an Rfactor of 41.2%. 7 cycles of maximum-likelihood based refinement reduced Rfactor/Rfree to 29.3%/33.5%, where Rfree was calculated from 5% of the reflections. All the refinement was conducted using REFMAC.32 In the first round of manual model building using COOT,33 variations were made in this refined model according to the amino acid sequence of WbgU. At the same time, modifications in main chain and side chains were made according to the 2mFo-DFc electron density map. Iterative cycles of model building followed by real and reciprocal space refinement reduced Rfactor/Rfree to 24.9%/29.8%. At this point well-defined electron density was observed in the 2mFo-DFc map for NAD(H) and UDP-GlcNAc in all the subunits, which were then modeled. Subsequent alternating cycles of model building along with real and reciprocal space refinement eventually improved electron density for some regions of the polypeptide and revealed the electron density for some glycine molecules, Na+ and SO42− ions. Lastly, solvent molecules were added and a final round of refinement was done in a similar fashion as above. The final model consists of four molecules each of WbgU, UDP-GlcNAc, NAD(H) and glycine, three SO42− ions, two Na+ ions and a total of 182 solvent molecules. The electron density was found to be well defined for most part, notable exception being a loop region extending from His287-Ile293. The average B-factor of the C-terminal domain is higher (47.5 Å2) than the average B-factor N-terminal domain (32.5 Å2). MolProbity18 was used to validate the quality of the final model. Details of the data collection and final refinement statistics are listed in Table I.
Sequence and structure comparisons
Five epimerases were used to draw out the sequence comparison with at least one representative from each of the groups: (i) UDP-Gal 4-epimerase from Escherichia coli (GalE‡) (Group 1)2; (ii) UDP-Glc/GlcNAc 4-epimerase from Campylobacter jejuni (CGne) (Group 2)34; (iii) UDP-Glc/GlcNAc 4-epimerase from humans (HGal) (Group 2)11; (iv) UDP-GlcNAc 4-epimerase from Pseudomonas aeruginosa (WbpP) (Group 3)35 and (v) UDP-GlcNAc 4-epimerase from Plesiomonas shigelloides O17 (WbgU) (Group 3)3. The multiple sequence alignment was performed using T-Coffee.36 Among these five protein sequences, crystal structures have been reported for GalE,37 HGal,38 WbpP,4 and WbgU (this work). The crystal structure of CGne is not available yet. Its polypeptide sequence was submitted to I-TASSER server39 for structure prediction without providing any reference structural template. I-TASSER server could predict a model with a high C-score of 1.62. Together these five structures were then used to prepare a structure alignment using UCSF Chimera.40 The axial rotation of C-termial domains of the Group 3 epimerases relative to the Group 1 and the Group 2 epimerases was determined by computing axes passing through the geometric centroid of atoms corresponding to the main chain N atoms of Arg338, Val210, and Val303 of WbgU using UCSF Chimera.40 Finally, active site volumes were calculated for the 4 experimentally determined structures (GalE, HGal, WbpP, and WbpP) using CASTp41 with precision approximated as per the discussion by Novotny et al.42 All the molecular graphics were prepared using UCSF Chimera40,43 and POV-Ray.44
Acknowledgments
Diffraction data were collected at the 9-2 beamline at SSRL, which is operated by the Department of Energy, Office of Basic Energy Sciences.
Glossary
Abbreviations:
- GalE
UDP-Galactose 4-epimerase from Escherichia coli
- HGal
UDP-Galactose 4-epimerase from Homo sapiens
- LPS
lipopolysaccharide
- Und
undecaprenol.
Footnotes
Unless otherwise specified, all amino acid numbering follows the crystal structure of WbgU.
PDB id codes used: GalE:1NAH; HGal:1HZJ; WbpP:1SB8.
Supplementary material
References
- 1.Allard S, Giraud M, Naismith J. Epimerases: structure, function and mechanism. Cell Mol Life Sci. 2001;58:1650–1665. doi: 10.1007/PL00000803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bauer AJ, Rayment I, Frey PA, Holden HM. The molecular structure of UDP-galactose 4-epimerase from Escherichia coli determined at 2.5 A resolution. Proteins. 1992;12:372–381. doi: 10.1002/prot.340120409. [DOI] [PubMed] [Google Scholar]
- 3.Kowal P, Wang P. New UDP-GlcNAc C4 epimerase involved in the biosynthesis of 2-acetamino-2-deoxy-l-altruronic acid in the O-antigen repeating units of Plesiomonas shigelloides O17. Biochemistry. 2002;41:15410–15414. doi: 10.1021/bi026384i. [DOI] [PubMed] [Google Scholar]
- 4.Ishiyama N, Creuzenet C, Lam JS, Berghuis AM. Crystal structure of WbpP, a genuine UDP-N-acetylglucosamine 4-epimerase from Pseudomonas aeruginosa: substrate specificity in udp-hexose 4-epimerases. J Biol Chem. 2004;279:22635–22642. doi: 10.1074/jbc.M401642200. [DOI] [PubMed] [Google Scholar]
- 5.Holden HM, Rayment I, Thoden JB. Structure and function of enzymes of the Leloir pathway for galactose metabolism. J Biol Chem. 2003;278:43885–43888. doi: 10.1074/jbc.R300025200. [DOI] [PubMed] [Google Scholar]
- 6.Holst O. The structures of core regions from enterobacterial lipopolysaccharides-an update. FEMS Microbiol Lett. 2007;271:3–11. doi: 10.1111/j.1574-6968.2007.00708.x. [DOI] [PubMed] [Google Scholar]
- 7.Cuthbertson L, Mainprize IL, Naismith JH, Whitfield C. Pivotal roles of the outer membrane polysaccharide export and polysaccharide copolymerase protein families in export of extracellular polysaccharides in gram-negative bacteria. Microbiol Mol Biol Rev. 2009;73:155–177. doi: 10.1128/MMBR.00024-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Van Den Bosch L, Manning PA, Morona R. Regulation of O-antigen chain length is required for Shigella flexneri virulence. Mol Microbiol. 1997;23:765–775. doi: 10.1046/j.1365-2958.1997.2541625.x. [DOI] [PubMed] [Google Scholar]
- 9.Murray GL, Attridge SR, Morona R. Altering the length of the lipopolysaccharide O antigen has an impact on the interaction of Salmonella enterica serovar Typhimurium with macrophages and complement. J Bacteriol. 2006;188:2735–2739. doi: 10.1128/JB.188.7.2735-2739.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hong M, Payne SM. Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol Microbiol. 1997;24:779–791. doi: 10.1046/j.1365-2958.1997.3731744.x. [DOI] [PubMed] [Google Scholar]
- 11.Wohlers T, Christacos N, Harreman M, Fridovich-Keil J. Identification and characterization of a mutation, in the human UDP-galactose-4-epimerase gene, associated with generalized epimerase-deficiency galactosemia. Am J Human Gen. 1999;64:462–470. doi: 10.1086/302263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Maceratesi P, Daude N, Dallapiccola B, Novelli G, Allen R, Okano Y, Reichardt J. Human UDP-galactose 4' epimerase (GALE) gene and identification of five missense mutations in patients with epimerase-deficiency galactosemia. Mol Genet Metab. 1998;63:26–30. doi: 10.1006/mgme.1997.2645. [DOI] [PubMed] [Google Scholar]
- 13.Wilson DB, Hogness DS. The enzymes of the galactose operon in Escherichia coli. I. Purification and characterization of uridine diphosphogalactose 4-epimerase. J Biol Chem. 1964;239:2469–2481. [PubMed] [Google Scholar]
- 14.Thoden J, Wohlers T, Fridovich-Keil J, Holden H. Crystallographic evidence for Tyr 157 functioning as the active site base in human UDP-galactose 4-epimerase. Biochemistry. 2000;39:5691–5701. doi: 10.1021/bi000215l. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Thoden JB, Kim J, Berger E, Gulick AM, Ruzicka FJ, Holden HM, Frey PA. Mechanistic roles of tyrosine 149 and serine 124 in UDP-galactose 4-epimerase from Escherichia coli. Biochemistry. 1997;36:10675–10684. doi: 10.1021/bi970430a. [DOI] [PubMed] [Google Scholar]
- 16.Demendi M, Ishiyama N, Lam JS, Berghuis AM, Creuzenet C. Towards a better understanding of the substrate specificity of the UDP-N-acetylglucosamine C4 epimerase WbpP. Biochem J. 2005;389:173–180. doi: 10.1042/BJ20050263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engh RA, Huber R. Accurate bond and angle parameters for X-ray protein structure refinement. Acta Cryst. 1991;A47:392–400. [Google Scholar]
- 18.Chen VB, Arendall WB, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Cryst D. 2010:12–21. doi: 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Shatsky M, Nussinov R, Wolfson HJ. A method for simultaneous alignment of multiple protein structures. Proteins. 2004;56:143–156. doi: 10.1002/prot.10628. [DOI] [PubMed] [Google Scholar]
- 20.Thoden JB, Henderson JM, Fridovich-Keil JL, Holden HM. Structural analysis of the Y299C mutant of Escherichia coli UDP-galactose 4-epimerase. Teaching an old dog new tricks. J Biol Chem. 2002;277:27528–27534. doi: 10.1074/jbc.M204413200. [DOI] [PubMed] [Google Scholar]
- 21.Guo H, Li L, Wang PG. Biochemical characterization of UDP-GlcNAc/Glc 4-epimerase from Escherichia coli O86:B7. Biochemistry. 2006;45:13760–13768. doi: 10.1021/bi0612770. [DOI] [PubMed] [Google Scholar]
- 22.Schulz JM, Watson AL, Sanders R, Ross KL, Thoden JB, Holden HM, Fridovich-Keil JL. Determinants of function and substrate specificity in human UDP-galactose 4'-epimerase. J Biol Chem. 2004;279:32796–32803. doi: 10.1074/jbc.M405005200. [DOI] [PubMed] [Google Scholar]
- 23.Shaw MP, Bond CS, Roper JR, Gourley DG, Ferguson MAJ, Hunter WN. High-resolution crystal structure of Trypanosoma brucei UDP-galactose 4'-epimerase: a potential target for structure-based development of novel trypanocides. Mol Biochem Parasitol. 2003;126:173–180. doi: 10.1016/s0166-6851(02)00243-8. [DOI] [PubMed] [Google Scholar]
- 24.Roper JR, Guther MLS, Milne KG, Ferguson MAJ. Galactose metabolism is essential for the African sleeping sickness parasite Trypanosoma brucei. Proc Natl Acad Sci USA. 2002;99:5884–5889. doi: 10.1073/pnas.092669999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rush JS, Alaimo C, Robbiani R, Wacker M, Waechter CJ. A novel epimerase that converts GlcNAc-P-P-undecaprenol to GalNAc-P-P-undecaprenol in Escherichia coli O157. J Biol Chem. 2010;285:1671–1680. doi: 10.1074/jbc.M109.061630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Chayen NE. Comparative studies of protein crystallization by vapour-diffusion and microbatch techniques. Acta Cryst. 1998;D54:8–15. doi: 10.1107/s0907444997005374. [DOI] [PubMed] [Google Scholar]
- 27.D'Arcy A, Sweeney AM, Haber A. Practical aspects of using the microbatch method in screening conditions for protein crystallization. Methods. 2004;34:323–328. doi: 10.1016/j.ymeth.2004.03.023. [DOI] [PubMed] [Google Scholar]
- 28.Leslie AGW, Powel HR. 2007. Processing diffraction data with MOSFLM. Evolving Methods for Macromolecular Crystallography: The Structural Path to the Understanding of the Mechanism of Action of CBRN Agents:41.
- 29.Thompson JD, Gibson TJ, Higgins DG, et al. 2002. Multiple sequence alignment using ClustalW and ClustalX. Current protocols in bioinformatics/editoral board, Andreas D. Baxevanis [et al.] Chapter 2:Unit 2.3.
- 30.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Matthews BW. Solvent content of protein crystals. J Mol Biol. 1968;33:491–497. doi: 10.1016/0022-2836(68)90205-2. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Cryst. 1997;D53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Cryst. 2004;D60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 34.Bernatchez S, Szymanski C, Ishiyama N, Li J, Jarrell H, Lau P, Berghuis A, Young N, Wakarchuk W. A single bifunctional UDP-GlcNAc/Glc 4-epimerase supports the synthesis of three cell surface glycoconjugates in Campylobacter jejuni. J Biol Chem. 2005;280:4792. doi: 10.1074/jbc.M407767200. [DOI] [PubMed] [Google Scholar]
- 35.Creuzenet C, Belanger M, Wakarchuk WW, Lam JS. Expression, purification, and biochemical characterization of WbpP, a new UDP-GlcNAc C4 epimerase from Pseudomonas aeruginosa serotype O6. J Biol Chem. 2000;275:19060–19067. doi: 10.1074/jbc.M001171200. [DOI] [PubMed] [Google Scholar]
- 36.Notredame C, Higgins DG, Heringa J. T-Coffee: a novel method for fast and accurate multiple sequence alignment. J Mol Biol. 2000;302:205–217. doi: 10.1006/jmbi.2000.4042. [DOI] [PubMed] [Google Scholar]
- 37.Thoden JB, Frey PA, Holden HM. Crystal structures of the oxidized and reduced forms of UDP-galactose 4-epimerase isolated from Escherichia coli. Biochemistry. 1996;35:2557–2566. doi: 10.1021/bi952715y. [DOI] [PubMed] [Google Scholar]
- 38.Thoden JB, Wohlers TM, Fridovich-Keil JL, Holden HM. Human UDP-galactose 4-epimerase. Accommodation of UDP-N-acetylglucosamine within the active site. J Biol Chem. 2001;276:15131–15136. doi: 10.1074/jbc.M100220200. [DOI] [PubMed] [Google Scholar]
- 39.Zhang Y. I-TASSER: fully automated protein structure prediction in CASP8. Proteins. 2009;77(Suppl 9):100–113. doi: 10.1002/prot.22588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera-a visualization system for exploratory research and analysis. J Comp Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 41.Dundas J, Ouyang Z, Tseng J, Binkowski A, Turpaz Y, Liang J. CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res. 2006;34:W116–W118. doi: 10.1093/nar/gkl282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Novotny M, Seibert M, Kleywegt GJ. On the precision of calculated solvent-accessible surface areas. Acta Cryst. 2007;D63:270–274. doi: 10.1107/S0907444906044118. [DOI] [PubMed] [Google Scholar]
- 43.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J Struct Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 44.Orf L. Scientific visualizations with POV-Ray. Linux J. 2004;2004:2. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.