Abstract
FOXP1 belongs to the P-subfamily of forkhead transcription factors and contains a conserved forkhead DNA-binding domain. According to size exclusion chromatography analysis, the forkhead domain of FOXP1 existed as a mixture of monomer and dimer. The dissociation constants of the forkhead domain of wild-type, C61S, and C61Y mutants of FOXP1 were 27.3, 28.8, and 332.0 μM, respectively. In contrast, FOXP1 A39P mutant formed only a monomer. NMR analysis also showed that FOXP1 C61S and C61Y mutants existed as a mixture. The solution structure of FOXP1 A39P/C61Y mutant was similar to the X-ray structure of the FOXP2 monomer. Comparison of backbone dynamics of FOXP1 A39P/C61Y and C61Y mutants showed that the residues preceding helix 3, the hinge region, exhibited the largest conformational exchange in FOXP1 monomer. The A39 residue of FOXP1 dimer has a lower order parameter with internal motion on the ps-ns timescale, suggesting that the dynamics of the hinge region of FOXP1 are important in the formation of the swapped dimer. The analysis also showed that the residues exhibiting the motions on the ps-ns and μs-ms timescales were located at the DNA-binding surface of FOXP1, suggesting the interactions between FOXP1 and DNA may be highly dynamic.
Keywords: dynamics, forkhead DNA-binding protein, NMR, structure, swapped dimer
Introduction
Forkhead box (FOX)-containing transcription factors are defined from a conserved winged-helix/forkhead DNA-binding domain containing ∼100 amino acids.1–4 They are expressed in many eukaryotic organisms and act as regulatory keys in embryogenesis, tumorigenesis, or the maintenance of differentiated cell states.5–8 FOXP proteins are newly defined P-subfamily of forkhead transcription factors. To date, four FOXP1 proteins have been found and they are as follows: FOXP1, FOXP2, FOXP3, and FOXP4 contain 677, 740, 431, and 680 amino acids, respectively.9–12 They all contain multiple domains—including a glutamine-rich region, a zinc finger, a leucine zipper, and a forkhead DNA-binding domain [Fig. 1(A)]. FOXP proteins are distinct from other members of the forkhead family in that they have a C-terminal forkhead DNA-binding domain and a C2H2 zinc finger domain.13 14 The aberrant expression and gene mutations of FOXP proteins are found in many human diseases.10 15 16 Two examples are the loss of heterozygosity of FOXP1 gene that is found in breast cancer; and the loss of FOXP1 expression, which is associated with a shorter survival, indicating that FOXP1 functions as a tumor suppressor.9 17–19 In contrast, the chromosomal translocation of FOXP1 gene is identified in several types of lymphomas; and deregulated expression of FOXP1 is relevant to poor prognosis, suggesting that FOXP1 may be an oncogene.20–24 The forkhead domain of FOXP1 shares 88, 76, and 89% identity with that of FOXP2, FOXP3, and FOXP4, respectively. In particular, several disease-causing mutations on the forkhead domain of FOXP2 and FOXP3 are identified.10 11 25 26
Figure 1.
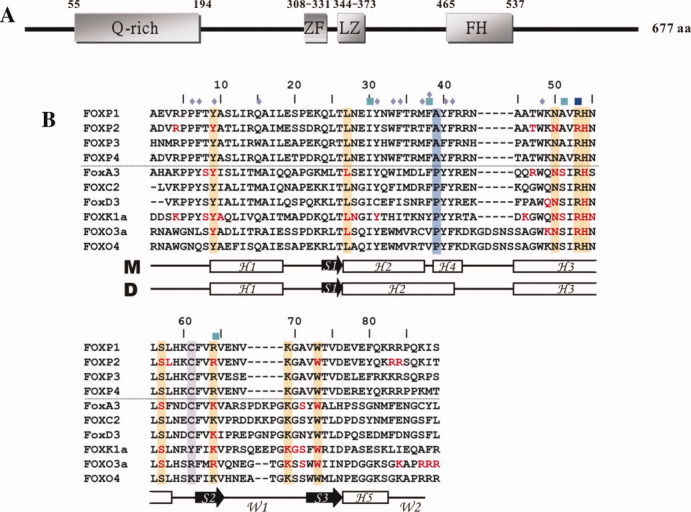
Schematic representation of FOXP1 and sequence alignment of its forkhead domain with other FOX proteins. (A) Schematic domain structure of FOXP1. The glutamine-rich region (Q-rich), the zinc finger domain (ZF), leucine zipper domain (LZ), and forkhead DNA-binding domain (FH) are shown. (B) Sequence alignment of the forkhead domains. Sequence alignment of winged-helix/forkhead proteins: FOXP1, FOXP2, FOXP3, FOXP4, FoxA3, FOXC3, FoxD3, FOXK1a, FOXO3a, and FOXO4. The DNA-binding residues are colored in red. The conserved residues of the forkhead DNA-binding domain, the A39, and C61 residues are shaded in orange, blue, and purple. The residues involved in intermolecular interactions in swapped dimer are denoted by a blue rhombus. Missense mutations of FOXP2 and FOXP3 are shown in dark blue and cyan boxes, respectively. Schematic diagrams of the secondary structures of the monomer (M) and swapped dimer (D) are also shown below the sequence.
Many 3D structures of the forkhead domain of FOX proteins—including FoxA3, FOXC2, FoxD3, FOXK1a, FOXO3a, FOXO4, and FOXP2—have been determined by X-ray crystallography and NMR spectroscopy.27–34 The sequence alignment of these proteins showed that the hinge region between H2 and H3 and the wing 1 and C-terminal regions were most diverse [Fig. 1(B)]. In general, they exhibit a compact α/β structure consisting of three α-helices (H1, H2, and H3), three β strands (S1, S2, and S3), and two wings (W1 and W2).27 35 Unlike 3D structures of the FOX protein/DNA complexes, the X-ray structure of the FOXP2/DNA complex exhibits a monomer and swapped dimer on binding to DNA.36 The monomeric form of FOXP2 has a canonical winged-helix fold with secondary structures arranged in the order H1-S1-H2-H4-H3-S2-S3-H5. In contrast to other forkhead proteins, the hinge region between H2 and H3 and the wing 2 region of FOXP2 form two short α helices. The difference between two FOXP2 conformers is that two short 11-residue H2 and 4-residue H4 of FOXP2 monomer are replaced with a long 15-residue H2 of the swapped dimer. Because both the conformational interchange between monomer and dimer and the interaction between protein and DNA are involved in a dynamic process, we propose to study dynamics and structure–function relationships of FOXP1 using NMR spectroscopy.
To study dynamics and structure–function relationships of FOXP1 monomer and dimer, we expressed the DNA-binding domain of FOXP1 and its mutant proteins in E. coli. We also determined the monomer–dimer dissociation constants, 3D structure, and backbone dynamics of FOXP1 and its mutant proteins. Our study shows that dynamic properties and the amino acid content of the hinge region between H2 and H3 of forkhead DNA-binding domain of FOXP1 are important in the formation of the swapped dimer and its DNA binding.
Results
The forkhead domain of FOXP1 exists in a mixture of monomer and dimer
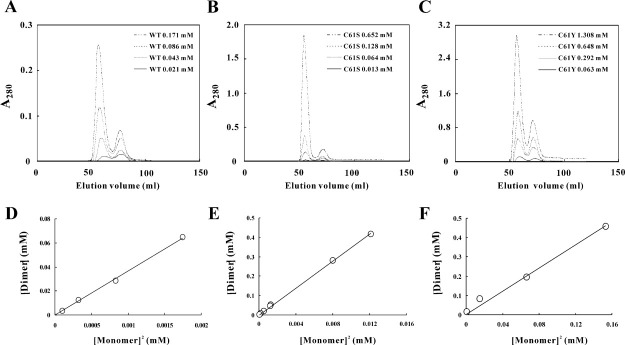
The Bio-Gel P-30 size-exclusion chromatography was used to analyze the forkhead domain of FOXP1 and its mutant proteins, which were purified to homogeneity by SP Sepharose cation chromatography and C18 reversed-phase HPLC. Our analysis showed that the forkhead domain of FOXP1 existed as a mixture of monomer and dimer, and the ratio between monomer and dimer was concentration-dependent [Fig. 2(A)]. Similar to wild-type FOXP1, the C61S and C61Y mutants also existed as a mixture of monomer and dimer [Fig. 2(B,C)]. In contrast, the A39P/C61S and A39P/C61Y mutants formed a monomer only up to the concentration of 2 mM. The dissociation constants of FOXP1 and its mutants were calculated using a simple dissociation model: D 2M, where M and D represent monomer and dimer (Table I). The dissociation constants of the forkhead domain of wild-type, C61S, and C61Y mutants of FOXP1 were 27.3, 28.8, and 332.0 μM, respectively. The mutation of C61 to S did not affect its dimer formation; however, the mutation of C61 to Y caused a 12-fold increase of dissociation constant, suggesting that hydrophobic interaction may be important in stabilizing the formation of monomer.
Figure 2.
Gel filtration analysis of wild-type, C61S, and C61Y mutants of FOXP1. Recombinant FOXP1 proteins are dissolved in 50 mM NaCl, 25 mM MgCl2, 50 mM arginine, 50 mM glutamate, and 20 mM phosphate buffer at pH 5.5 and are applied to Bio-Gel P-30 size-exclusion chromatography. Elution profiles of (A) wild-type, (B) C61S and (C) C61Y mutants of FOXP1 are shown. The concentrations of 0.021 mM (___), 0.043 mM (…..) 0.086 mM (••••), and 0.171 mM (-.. -) of FOXP1; 0.013 mM (___), 0.064 mM (…..), 0.128 mM (••••), and 0.652 mM (-.. -) of C61S mutant; and 0.063 mM (___), 0.292 mM (…..), 0.648 mM (••••), and 1.308 mM (-.. -) of C61Y mutant are used for the analysis. Linear fit of the monomer-dimer dissociation constants of (D) wild-type, (E) C61S mutant and (F) C61Y mutant of FOXP1. FOXP1's monomer-dimer dissociation constants and mutant proteins were calculated using the following formula: Kd = [monomer]2/[dimer]. The data were analyzed using least-squares fit software SigmaPlot.
Table I.
The Dissociation Constants of the Forkhead Domain of FOXP1 wt and Its Mutants
| Protein | Kd (M) | Kdmut/Kdwt (Fold) |
|---|---|---|
| Wild type | 2.7301 × 10−5 | 1.00 |
| C61S | 2.8762 × 10−5 | 1.05 |
| C61Y | 3.3238 × 10−4 | 12.17 |
| A39P/C61S | ND | ND |
| A39P/C61Y | ND | ND |
ND, the mutants only formed monomer up to the concentration of 2 mM.
We reinjected the separated dimer fractions (wild-type and C61Y mutant) to the size-exclusion chromatography for examining the dimer to monomer transition. The dimeric form of wild-type FOXP1 did not dissociate into monomeric form but convert into oligomeric forms (Supporting Information Fig. S1A). Similar to wild-type FOXP1, the dimeric form of C61Y mutant did not dissociate into monomeric form. However, more precipitation and less oligomers were formed after 17 days (Supporting Information Fig. S1B). These results suggest that the energy barrier between monomeric and dimeric forms of FOXP1 is very large.
Thermostability of wild-type FOXP1 and its mutants
Differential scanning calorimetry (DSC) was used to determine the melting points of wild-type FOXP1 and its C61S, C61Y, A39P/C61S, and A39P/C61Y mutants, which were 62.09 ± 0.06°C, 57.65 ± 0.46°C, 62.60 ± 0.08°C, 61.85 ± 0.30°C, and 67.60 ± 0.25°C, respectively (Supporting Information Fig. S2). The comparisons of A39P/C61S with C61S mutants and of A39P/C61Y with C61Y mutants showed that the mutation of A39 to P responded to 4.20 and 5.00°C increases in melting temperature. The comparisons of C61S and C61Y mutants and of A39P/C61S with A39P/C61Y showed that the mutation of S61 to Y responded to 4.95 and 5.75°C increases in melting temperature. These results suggest that monomeric FOXP1 A39P/C61Y mutant is more stable than dimeric FOXP1 C61S mutant, and the Y61 residue can stabilize both FOXP1 monomer and dimer.
The binding of FOXP1 to DNA
NMR titration and gel retardation analysis were used to study the interactions between the DNA-binding domain of FOXP1 and DNA. However, the FOXP1/DNA complex was precipitated under both experimental conditions. This result was consistent with the previous report that the forkhead domain of FOXP1 bound with DNA resulted in the precipitation of the DNA complex in the well of the gel.38 In this study, we found that the interaction between FOXP1 and DNA can be analyzed by the change of tryptophan and tyrosine fluorescence emission on DNA binding. When FOXP1 A39P/C61S and A39P/C61Y were exited at 285 nm, their tryptophan fluorescence emission had the maximum values at 338.5 and 336 nm, respectively [Fig. 3(A,B)]. The intensities of emission spectra were decreased on binding to DNA. The difference spectra between FOXP1 A39P/C61S and A39P/C61Y and their DNA complexes were used to determine their dissociation constants, which were 3.59 ± 0.82 and 2.19 ± 0.14 μM, respectively [Fig. 3(C,D)]. However, wild-type, C61S, and C61Y mutants of FOXP1 were not determined because of the formation of monomer/dimer mixture.
Figure 3.
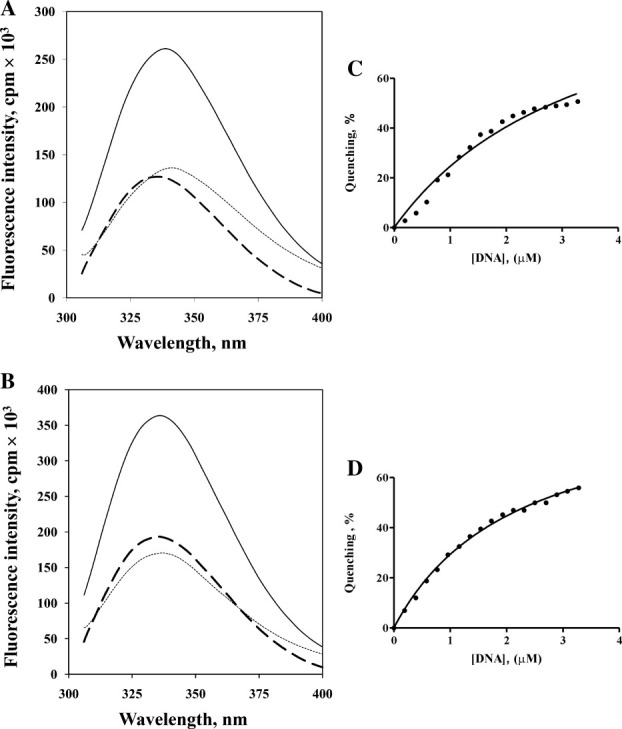
Fluorescence titration of DNA with the monomeric A39P mutants of FOXP1 with excitation at 285 nm. (A) shows the emission spectra of free FOXP1 A39P/C61S (___), the saturated DNA/FOXP1 A39P/C61S complex (••••), and their difference spectrum (----). (C) shows the emission spectra of free FOXP1 A39P/C61Y (___), the saturated DNA/FOXP1 A39P/C61Y complex (••••), and their difference spectrum (----). Binding isotherm for (B) FOXP1 A39P/C61S & DNA (Kd=3.65±0.40 μM) and for (D) FOXP1 A39P/C61Y & DNA (Kd=1.510±.06 μM) was obtained by using standard fluorometric titration analysis described in the method.
NMR assignment of the monomeric and dimeric forms of FOXP1
NMR experiments were performed to compare the structural differences between monomer and swapped dimer of FOXP1. The C61Y and A39P/C61Y mutants of FOXP1 were used to compare the structural differences between monomer and dimer, because C61S and A39P/C61S mutants had solubility and stability problems during the NMR experiments. The 2D 1H-15N-HSQC spectrum of A39P/C61Y mutant showed uniform intensities for most of the resonances at pH 5.5, and only the resonances of residues S11, L12, F41, and K49 cannot be found (Supporting Information Fig. S3A). In contrast, the 2D 1H-15N-HSQC spectrum of C61Y mutant showed that two sets of resonances for the residues I17-E19, E22, Q24, L25, N28, I30, W33, F38, R53, E66, W73, E77, T80-K82, and R84 were identified (Supporting Information Fig. S3B). Superimposition of the 15N-HSQC spectra of the C61Y and A39P/C61Y mutants showed that the set of minor resonances of 15N-HSQC spectra of FOXP1 C61Y was overlapped with the corresponding resonances of 2D 1H-15N-HSQC spectrum of the A39P/C61Y mutant, suggesting that they were the resonances of FOXP1 monomer [Fig. 4(A)]. The intensity ratio of 3.4:1 for the resonances of dimer and monomer was consistent with the calculation based on the dissociation constant of FOXP1 C61Y mutant at a concentration of 2.5 mM. The formation of a three-stranded antiparallel β-sheet was characterized by the CαH-CαH, CαH-NH, and NH-NH NOE patterns of the connecting strands, the slowly exchanging amide protons, and the upfield-shifted Cα chemical shift. The formation of a four-helix-bundle was also confirmed by dNN(i, i+2) and dαN(i, i+3) NOE connectivities and by chemical shift index.
Figure 4.
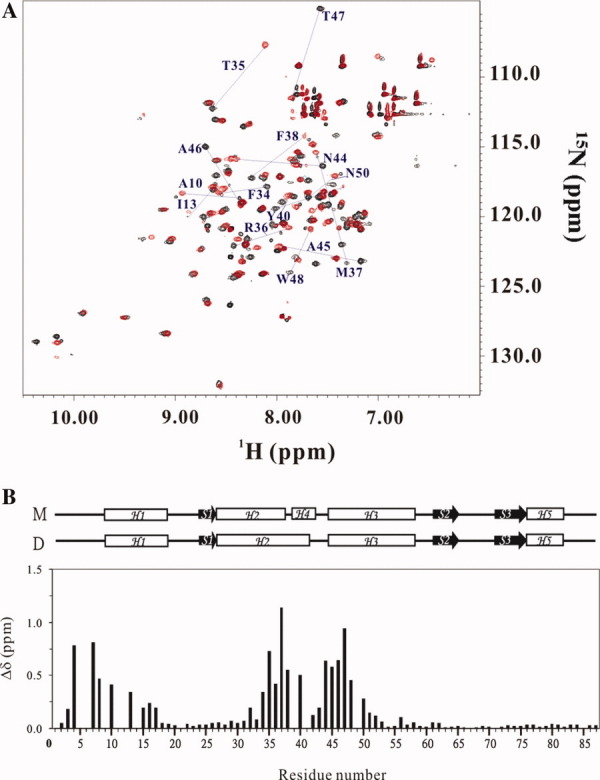
NMR analysis of FOXP1 C61Y and A39P/C61Y mutants. (A) Superimposition of HSQC spectra of FOXP1 C61Y (black) and FOXP1 A39P/C61Y (red). The residues with chemical shift perturbation greater than 0.5 are shown. (B) Chemical shift difference between FOXP1 C61Y and A39P/C61Y mutants. The chemical shift perturbation is calculated according to the equation 1δ={[0.02x(1δN)2+0.5x(1δH)2]1/2}. The secondary structures of monomer (M) and dimer (D) are shown on top of the figure.
Chemical shift perturbation was used to compare structural differences between FOXP1 monomer and dimer. The analysis of 2D 1H-15N-HSQC spectra of FOXP1 A39P/C61Y and C61Y mutants showed that five regions—namely R4-T8, A10-I13, F34-M37, F38-Y40, and N44-N50—exhibited values of chemical shift perturbation greater than 0.25. They were located at N-terminus, H1, H2, the hinge region, and H3 [Fig. 4(B)]. In particular, their largest chemical shift difference was found from the hinge region. This was consistent with the reported X-ray structures of the FOXP2/DNA complex that two short 11-residue H2 and 4-residue H4 of the monomer are replaced with a long 15-residue H2 of the swapped dimer. In contrast, significant reduction is observed in the residues immediately adjacent to A39. This reduction may suggest that A39 and A41-43 did not undergo a significant structural change during the transition from monomer to dimer. The analysis of Cα chemical shift showed that the secondary structures of the A39P/C61Y mutant contained four α-helices (H1, residues 9–18; H2, residues 27–37; H3, residues 45–48; H5, residues 73–76) and three β-strands (S1, residues 24–26; S2, residues 62–65; S3, residues 73–76) [Fig. 5(A)]. The residues from 27 to 40 of FOXP1 C61Y mutant exhibited the downfield-shifted Cα chemical shifts, indicating the formation of a long 15-residue H2 [Fig. 5(B)]. The formation of their secondary structures was also confirmed from the NOE data. These results showed that FOXP1 C61Y mutant existed as a mixture of swapped dimer and monomer in the absence of DNA. In contrast, the A39P/C61Y mutant was a monomer.
Figure 5.
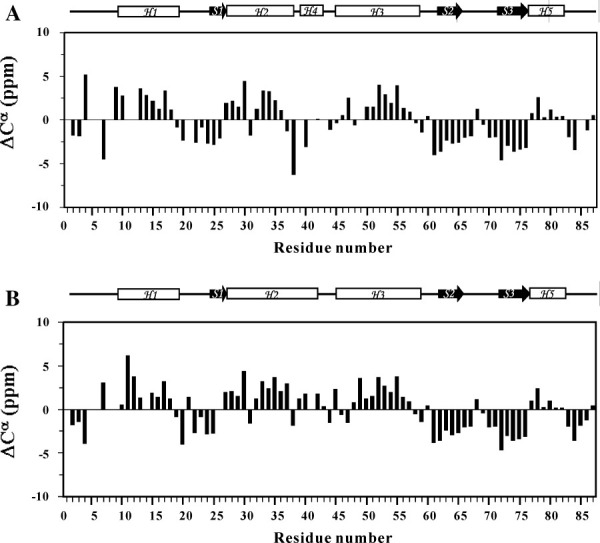
Chemical shift index analysis of FOXP1 C61Y and A39P/C61Y mutants. The chemical shift differences of Cα resonances relative to those of random coil values are calculated for (A) the monomeric form of FOXP1 A39P/C61Y mutant and (B) the dimeric form of FOXP1 C61Y mutant. Schematic representations of the secondary structures are shown above the figure.
The NOE differences between monomer and the domain-swapped dimer were also used to make comparison. Their apparent differences were NOEs between αH, βH, δH, and γH of F38 residue and the indole NH of W48 residue in the domain-swapped dimer (Supporting Information Fig. S4). In contrast, No NOE was observed between the residues F38 and W48 residue in monomer. This is consistent with the distances of βH of F38 and the indole NH of W48 between monomer and dimer are 9.4 and 3.4 Å, respectively.
Structure determination of the monomeric form of FOXP1
The solution structure of FOXP1 A39P/C61Y mutant was determined by NMR spectroscopy and the hybrid distance geometry-dynamical simulated annealing method. NOE-derived distance restraints were obtained from the 15N- and 13C-edited NOESY data. A total of 1323 experimentally derived restraints, including 1248 NOEs, 34 hydrogen bonds, and 75 dihedral angles, were used for structure calculation. An average of 15.2 restraints per residue was identified (Supporting Information Table I). A stereoview of the 20 best structures from 100 initial structures are shown in Figure 6(A). The backbone and heavy-atom root mean square deviation (RMSD) values for the well-defined regions (residues 9–19, 24–26, 27–37, 45–58, 62–65, 72–76, and 77–82) were 0.68 ± 0.11 and 1.23 ± 0.13 Å, respectively. According to our Ramachandran analysis, all dihedral angles of FOXP1 A39P/C61Y mutant are in the allowed region. A summary of the restraint and structural statistics is presented in Supporting Information Table I. Overall, the secondary structures of the DNA-binding domain of FOXP1 are well defined, and its fold contains a four-helix-bundle packed with a triple-stranded antiparallel β-sheet [Fig. 6(B)]. Superimposition of the backbone atoms of FOXP1 A39P/C61Y mutant and FOXP2 resulted in an RMSD value of 1.13 ± 0.26 Å, indicating no significant structural differences [Fig. 6(B)]. The major difference was found from the region between H2 and H3, where a four-residue helix in FOXP2 and a turn-like structure in FOXP1 A39P/C61Y mutant were formed.
Figure 6.
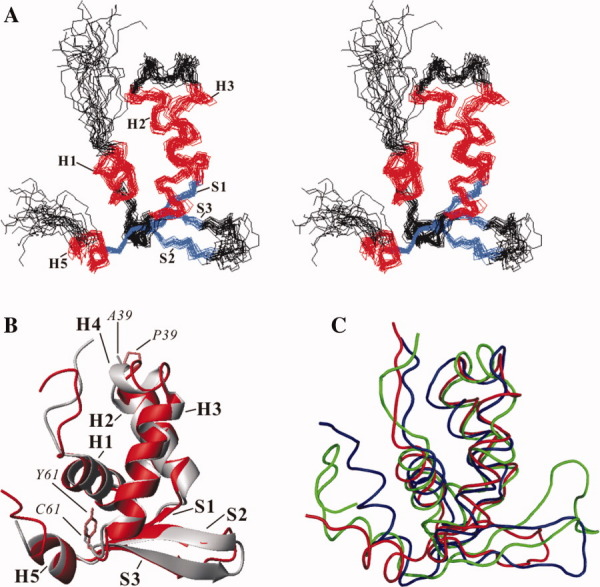
3D structure of FOXP1 (A) Stereoview of 20 NMR structures of FOXP1 A39P/C61Y mutant. The backbone atoms of the secondary structures are superimposed. The α-helix and β-strands are colored in red and blue, respectively. (B) Superimposition of FOXP1 and FOXP2 structures. The FOXP1 and FOXP2 structures are colored in red and gray, respectively. The side chains of two key mutated P39 and Y61 residues of FOXP1 are colored in orange. The side chains of A39 and C61 residues of FOXP2 are colored in gray. (C) Superimposition of FOXP1, FoxD3, and FOXK1a structures. The core regions—including three α-helices and β-sheet—are used for superimposition. The FOXP1, FoxD3, and FOXK1a structures are colored in red, green, and blue, respectively. Their structures are aligned using Theseus (http://www.theseus3d.org/).
Structural comparison with other winged-helix proteins
Many 3D structures of winged helix/forkhead proteins have been determined using NMR spectroscopy and X-ray crystallography.27–34 Structural analysis found that their 3D structures have a similar fold, which consists of three-helix bundle packed with a triple-stranded antiparallel β-sheet.27 35 It was shown that the packing angles between H3, the DNA-recognition helix, and H2 among winged helix/forkhead proteins range from 100° to 150°.35 The analysis showed that the packing angle between H3 and H2 of FOXP1 A39P/C61Y mutant, FoxA3, FOXC2, FoxD3, FOXK1a, FOXO3a, FOXO4, and FOXP2 were 135.5 ± 5.8°, 126.6°, 133.0 ± 2.4°, 114.6 ± 2.8°, 122.4 ± 5.5°, 120.6 ± 0.6°, 123.3 ± 4.4°, and 127.0°, respectively. These results indicate that FOXP1 A39P/C61Y mutant has a canonical winged helix/forkhead fold.
The major structural differences among winged helix/forkhead proteins were the conformation of the connecting region between H2 and H3, the length of the wing 1 region, and the conformation of C-terminal loop.39 We selected the forkhead domain of FOXP1, FoxD3, and FOXK1a for the analysis because they have an α helix in the C-terminal region. Although the forkhead domain of FOXP1 shared 37 and 39% identities with that of FoxD3 and FOXK1a, the superimposition of the backbone atoms of FOXP1 core regions (H1-H3 and S1-S3) with those of FoxD3 and FOXK1a resulted in a maximum likelihood optimization RMSD value of 0.71 Å, respectively [Fig. 6(C)]. The length of the wing 1 region of FOXP1, FoxD3, and FOXK1a contains 6, 11, and 11 amino acids, respectively. Unlike FoxD3 and FOXK1a, the wing 1 region of FOXP1 is the shortest and contains only six amino acid residues with a type I turn structure. Interestingly, their C-terminal α helix, H5, exhibited diverse orientations. The analysis showed that the packing angle between the H1 and H5 of FOXK1a, FoxD3, FOXP2, and FOXP1 was 125.2 ± 5.5°, 138.8 ± 8.6°, 155.9 ± 0.1°, and 162.7 ± 5.7°, respectively. We also found that the packing angle was related to the length of H5. Thus, FOXK1a with longer H5 made more interactions with H1. In contrast, FOXP proteins with shorter H5 made fewer interactions with H1, resulting in a larger packing angle. Because the C-terminal and wing 1 regions are involved in DNA binding, the difference in the orientation of H5 and the length of W1 may result in the modulation of their DNA-binding specificity.
Backbone dynamics of the monomeric and dimeric forms of FOXP1
1H-15N correlated NMR spectroscopy was used to measure 15N R1, 15N R2, and 1H-15N NOE parameters of the monomeric and dimeric forms of FOXP1 (Fig. 7). A total of 71 and 69 backbone amide protons of C61Y and A39P/C61Y mutants were obtained. The trimmed average values of R1, R2, and NOE of FOXP1 monomer were 1.43 s−1, 12.06 s−1, and 0.77. The trimmed average values of R1, R2, and NOE of FOXP1 dimer were 0.96 s−1, 17.11 s−1, and 0.76. The changes in the R1 and R2 relaxation rates were consistent with the changes in the molecular weight between monomer and dimer. Thus, FOXP1 monomer exhibited high R1 and low R2 values in comparison with the values of dimer. In comparison with the R1 and R2 values of individual resonance, the R1 of the residues T25 and A46 as well as the R2 of the Y61 residue in dimer were higher than the average value. In contrast, the R2 value of the A39 was lower than the average value. The trimmed mean R2/R1 ratios of monomer and dimer were 8.4 and 17.8, respectively. These values were larger than expected for a 10.5 kDa of monomer and a 21.0 kDa of dimer. However, they were very similar to the reported values of the hydrated DNA-binding proteins.28 30 In addition, the wing 1, N- and C-terminal regions of FOXP1 monomer, and dimer with low NOE values showed distinctive backbone flexibility, which was consistent with the NMR structure of FOXP1. The average NOE values of monomer and swapped dimer were the same. The hinge region with the residues T35-A46 was the only region that had a higher NOE value on the formation of dimer. It is consistent with the formation of a short α helix in this region.
Figure 7.
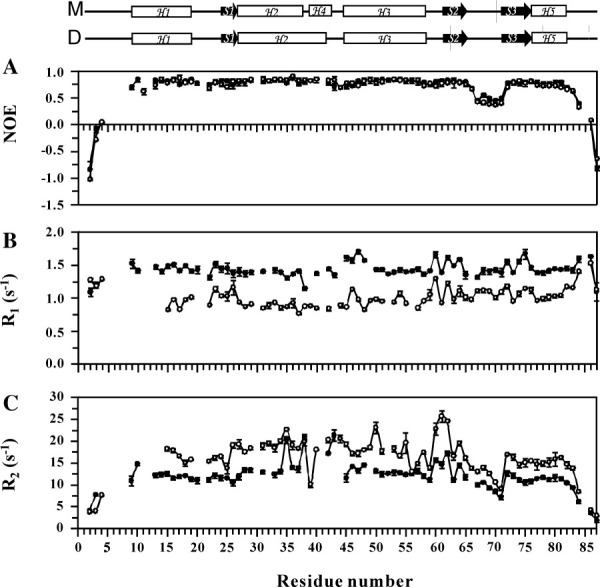
Comparison of the relaxation parameters of FOXP1's two mutants: A39P/C61Y (•) and C61Y (○). (A) 1H-15N steady-state NOE with error. (B) Longitudinal relaxation rates (R1) with error. (C) Transverse relaxation rate (R2) with error. Schematic representations of the secondary structures of the A39P/C61Y and C61Y mutants of FOXP1 are shown at the top. M and D represent the monomer and swapped dimer, respectively.
The square of the generalized order parameter (S2), the effective internal correlation time (τe), and a conformational exchange broadening parameter (Rex) for each backbone amide NH vector were determined using the model-free formalism. The relaxation data of FOXP1 monomer and dimer were analyzed with the isotropic and axially symmetric models, respectively. The optimized values of τm for FOXP1 A39P/C61Y and C61Y mutants were determined to be 8.6 and 14.0 ns. The obtained diffusion tensors of FOXP1 A39P/C61Y and C61Y mutants were symmetric with D///D⊥ = 1.19 and 1.38. These results were consistent with our structural analysis: FOXP1 A39P/C61Y mutant's tertiary fold had a globular shape; FOXP1 C61Y mutant's had an elongated shape.
The optimized values of S2, τe, and Rex for FOXP1 A39P/C61Y and C61Y mutants are shown in Figure 8. The average S2 values of FOXP1 A39P/C61Y and C61Y mutants are 0.88 and 0.85, respectively. The S2 values of N-terminus, wing 1 region, and C-terminus of both FOXP1 conformers were lower than the average value, suggesting that these regions were flexible in both forms of FOXP1 [Fig. 8(A)]. These regions also exhibited the motion on the ps-ns timescale [Fig. 8(B)]. A comparison of the S2 values of FOXP1 monomer and dimer shows that the secondary structure elements of FOXP1 dimer are more flexible than those of monomer (Table II). This is consistent with trends in the temperature factors within the crystal structure of FOXP2 (Supporting Information Fig. S5). We also found that the W1 region had the largest change. The analysis showed that H3, the DNA-recognition helix, and H2 of both FOXP1 conformers exhibited a slow conformational exchange motion on the μs-ms timescale [Fig. 8(C)]. The S2 values of the A39 and Y61 residues of dimer were 0.64 and 0.72, which were lower than the average value of 0.85. In particular, the A39 residue of FOXP1 dimer has a lower parameter with internal motion on the ps-ns timescale. The Y61 residue also exhibited conformational exchange in both conformers. The Rex value of the Y61 residue in dimer was 12.09 s−1, which was much larger than that of monomer with the value of 3.24 s−1. The major difference between monomer and dimer was a slow conformational exchange motion on the μs-ms timescale. The hinge region of the monomer exhibited a slow conformational exchange motion on the μs-ms timescale; and the Rex values of F38 and R43 were 11.58 and 10.28 s−1, respectively [Fig. 8(C)].
Figure 8.
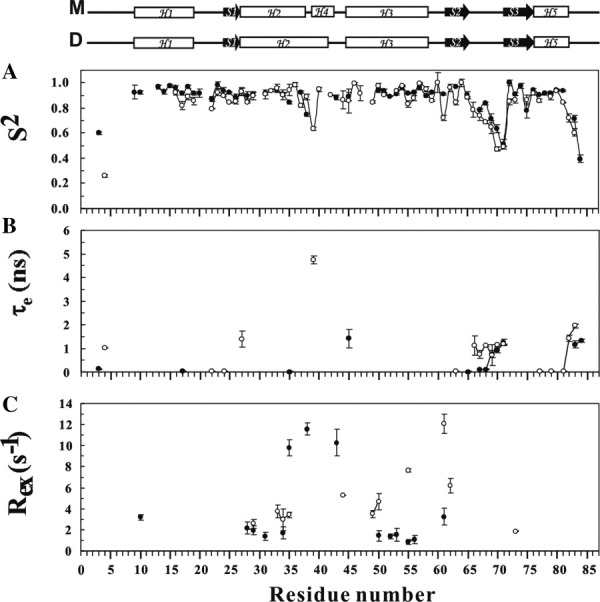
Comparison of model-free parameters of the A39P/C61Y mutant (•) and C61Y mutant (○) of FOXP1. Generalized order parameters S2, τe, and Rex. (A) Generalized order parameter (S2). (B) Effective correlation time (τe). (C) Chemical exchange rate (Rex). Gaps indicate the proline residues. Only some fitting models resulted in a Rex term. Schematic representations of the secondary structures of the A39P/C61Y and C61Y mutants of FOXP1 are shown at the top. M and D represent the monomer and swapped dimer, respectively.
Table II.
Mean Order Parameters of the Secondary Structural Elements of FOXP1 A39P/C61Y and C61Y Mutants
| Secondary structure elements | Position | Number of residuesa (M)c | 〈S2〉 (M)b | Number of residuesa (D)c | 〈S2〉 (D)b |
|---|---|---|---|---|---|
| H1 | 9–18 | 8 | 0.95 ± 0.02 | 3 | 0.88 ± 0.05 |
| T1 | 19–23 | 4 | 0.92 ± 0.05 | 3 | 0.86 ± 0.07 |
| S1 | 24–26 | 3 | 0.92 ± 0.03 | 3 | 0.86 ± 0.03 |
| H2 | 27–37 (27–41)c | 8 | 0.91 ± 0.03 | 13 | 0.89 ± 0.09 |
| H3 | 45–58 | 10 | 0.92 ± 0.03 | 12 | 0.92 ± 0.06 |
| S2 | 62–65 | 2 | 0.94 ± 0.05 | 4 | 0.92 ± 0.07 |
| W1 | 67–71 | 5 | 0.70 ± 0.13 | 5 | 0.61 ± 0.12 |
| S3 | 72–76 | 5 | 0.92 ± 0.09 | 5 | 0.87 ± 0.03 |
| H5 | 77–82 | 5 | 0.92 ± 0.02 | 5 | 0.85 ± 0.08 |
| All | 1–87 | 57 | 0.88 ± 0.12 | 64 | 0.85 ± 0.14 |
Number of residues in each element of secondary structure for which order parameters can be obtained.
M and D represent monomeric (A39P/C61Y mutant) and dimeric (C61Y mutant) forms of FOXP1.
The positions of amino acid sequence of H2 in swapped dimer.
Comparison of dynamical properties of FOXP1 and other winged-helix proteins
Backbone dynamics of many winged helix proteins and a forkhead protein, FoxD3, were determined using NMR spectroscopy.40 Although backbone dynamics of FOXP1 and FoxD3 were very similar, some differences were found. Their N-terminus, wing 1 region, and C-terminus were flexible with low S2 values and with the motion on the ps-ns timescale. The major difference was found from the regions involved in a motion on the μs-ms timescale. Two regions of FoxD3, the linker regions between S1 and H1 and between H2 and H3, show considerable conformational exchange. Three regions of FOXP1—H2, the hinge region between H2 and H3, and H3—exhibited a slow conformational exchange motion on the μs-ms timescale. In particular, the S2 value of the A39 residue of FOXP dimer was 0.64, which was very low. It also exhibited a fast motion on the ps-ns timescale. The analysis of all winged-helix proteins showed that the residues in the hinge region between H2 and H3 exhibited a slow conformational exchange motion on the μs-ms timescale. This finding suggests that the hinge region between H2 and H3 with a slow conformational exchange motion is a general feature for winged-helix proteins.
Discussion
The interchange between protein conformers and the binding of protein to DNA are involved in dynamic process.41–46 To understand the conformational interchange of FOXP1, we used size-exclusion chromatography to determine the monomer–dimer dissociation constants of FOXP1 and its mutants. We also used NMR spectroscopy to determine 3D structure of FOXP1 monomer and backbone dynamics of FOXP1 monomer and domain-swapped dimer. Dynamics analysis of the monomeric and dimeric forms of FOXP1 revealed that slow conformational exchange motion in the hinge region between H2 and H3, particularly in the A39 residue of the dimer containing fast internal motion, are important in domain swapping. The analysis also showed that the residues exhibiting the motions on the ps-ns and μs-ms timescales were located at the DNA-binding surface of FOXP1, indicating the interactions between FOXP1 and DNA are highly dynamic.
In contrast to the formation of a monomer in FOXP1 A39P mutant, we found that the corresponding FOXK1a P39A mutant at any concentration up to 2 mM cannot form a dimer. These findings indicate that both alanines' helix propensity and dynamics properties are important in the formation of a long 15-residue H2 in the swapped dimer. However, we cannot rule out the other possibilities: (1) proline can destabilize the H2 helix formation of the domain-swapped dimer; (2) the decrease of interactions in the dimer can be caused by A39P mutation; and (3) A39P monomer mutant is more stable than C61S dimer mutant. Because FOXP1 and FOXP2 share 88% identity, we plotted the potential DNA-binding residues of FOXP1 using the reported structure of the FOXP2/DNA complex [Fig. 9(A)]. We found that the DNA-binding residues V3 and R83 exhibited τe, and the residues N28, Y31, R43, N50, R53, and S57 contained Rex [Fig. 9(B)]. Thus, the residues in the DNA-binding surface of FOXP1 exhibited the motions on either the ps-ns or μs-ms timescales. However, we cannot find any correlation between backbone dynamics of FOXP1 dimer and its DNA-binding residues (Supporting Information Fig. S6). Thus, most of the DNA-binding residues did not have the motions on the ps-ns and μs-ms timescales. These findings suggest that the dynamics properties of winged-helix proteins may also be important in high-affinity DNA binding.
Figure 9.
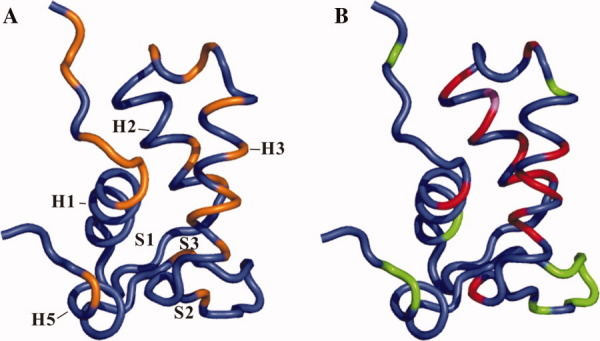
Comparison of the DNA-binding residues and backbone dynamics of FOXP1 monomer. (A) A ribbon illustration of 3D structure of FOXP1 A39P/C61Y mutant highlights the DNA-binding residues, colored in orange. (B) A ribbon illustration of 3D structure of FOXP1 A39P/C61Y mutant highlights the residues with conformational exchange motion (Rex) and internal motion (τe), colored in red and green, respectively. The residues containing both Rex and τe terms are colored in violet.
It is known that three major regions of the DNA-binding domain of forkhead proteins are involved in protein and DNA interactions, including H3, W1, and W2.35 47 48 A sequence comparison of the DNA-binding domain of forkhead proteins shows that the hinge region between H2 and H3, the W1 region, and the C-terminal region are the most diverse regions [Fig. 1(B)]. Although these nonconserved regions make fewer contacts with DNA, they may be important in regulating how the recognition helix makes specific base contacts with DNA. As shown in Figure 1(B), the hinge region of H2 and H3 of FoxA3, FoxD3, and FOXP2 formed short α-helices. In contrast, the region of FOXO3a containing 12 residues and FOXK1a containing seven residues formed coil structures. In particular, FOXO3a and FOXO4 each has an additional five-residue hinge region between H2 and H3 that exhibits, respectively, a coiled structure and a short helix. In this study, we found that the W1 and C-terminal regions are flexible with the motion on the ps-ns timescale, and the hinge region between H2 and H3 shows slow conformational exchange motion on the μs-ms timescale. These findings indicate that not only the conformations, amino acid compositions, and the lengths but also the dynamics properties of these regions affect the affinity and specificity of forkhead proteins.
To date, the FOXP protein family is the only FOX proteins that can form a domain-swapped dimer.36 In this study, we found that the mutation of C61 to Y caused a 12-fold increase in the formation of monomer, suggesting that the hydrophobic interaction may be important. The formation of FOXP1 monomer is stabilized by the hydrophobic interactions among the residues Y9, L12, I13, and I17 in H1; the residues L27, I30, and F34 in H2; the residues W48 and V52 in H3; the residue F62 in S2; and the residue W73 in S3. The packing between H1 and H5 is also stabilized by the hydrophobic interactions between the residues S11 and R14 in H1 and the residues F80 and Q86 in H5. Interestingly, the comparison of chemical shift perturbation between wild-type and C61Y mutant of FOXP1 shows that the H59-Y61, F62, and V75-R83 regions (located at the H3-S2 hinge region), S2, and C-terminal region exhibit large chemical shift perturbation (Supporting Information Fig. S7). These findings show that the hydrophobic interactions among three α-helices, three β-strands, and C-terminal region are important for the formation of monomer. It was reported that the A51T mutation, a disease-causing mutation in FOXP3, used a γ-methyl group packed against Y9 of H1.11 26 This is very similar to the interactions between A10 and Y61, leading to the increase in monomer formation. The disease caused by the A51T mutation may affect not only its DNA binding but also its monomer–dimer equilibrium.
Winged-helix/forkhead proteins have similar binding specificity to the core sequence, 5′-(T/C)(A/C)AA(C/T)A-3′, and the conserved amino acid sequences are found from the DNA-recognition helix.5 6 34 49 This raises an intriguing question about how forkhead proteins use conserved residues to recognize distinct DNA core sequences. Many reports show that the nonconserved elements—including the hinge region between H2 and H3, the W1 region, and the C-terminal region—make fewer contacts with DNA; however, they are important in regulating the interactions of the recognition helix with specific base contacts of DNA. In our study, we found that these regions are very dynamic, leading to the diversity in the conformations of these regions. Thus, both the flexibility and the conformations of these regions may be important in regulating the binding of forkhead proteins with DNA.
In conclusion, we have determined 3D structure of FOXP1 monomer and backbone dynamics of FOXP1 monomer and dimer. Gel filtration analysis shows that FOXP1 existed as a mixture of monomer and dimer with a monomer–dimer dissociation constant of 27.3 μM. Dynamics analysis of FOXP1 monomer and dimer shows that slow conformational exchange motion in the hinge region between H2 and H3—particularly internal motion on the ps-ns timescale in the A39 residue of the dimer—are important in domain swapping. The analysis also showed the residues exhibiting the motions on the ps-ns and μs-ms timescales were located at the DNA-binding surface of FOXP1, indicating the binding of FOXP1 to DNA is highly dynamic. The H2-H3 hinge region, the W1 region, and the C-terminal region are the most dynamic regions regarding the diversity in conformations and amino acid compositions. We postulate that not only the conformations, amino acid compositions, and the lengths but also the dynamics properties of these regions affect the affinity and specificity of forkhead proteins–DNA interactions. This study provides a new insight into how the dynamic properties of the nonconserved regions of forkhead proteins may regulate their DNA-binding specificities.
Materials and Methods
Protein expression and purification
A synthetic gene coding the forkhead domain of FOXP1 was made by eight primers with an overlapping oligonucleotide strategy using polymerase chain reaction (PCR). It was amplified by PCR with sense primer 5′-CATGCCATGGAAGTTAGACCACCATTTACATATGCATCT-3′ with NcoI recognition and antisense primer 5′-CCCAAGCTTCTAGTAGGCGTGGC TGCTCTGCAT GTTTTTAAT-3′ with HindIII recognition. No additional amino acid residues were added into recombinant FOXP1 with these primers. “We designated the residue A462 of FOXP1 as A1 for purposes of comparison.” The gene was then used to produce the DNA fragment encoding the residues A462-K548 of the forkhead domain, and its C61S, C61Y, A39P/C61S, and A39P/C61S mutations were produced using overlap extension PCR, and they were expressed to increase the stability of FOXP1.50 These genes were cloned into the pET21d(+) vector (Novagen, San Diego, CA). The recombinant plasmids were transformed into the E. coli BL21(DE3)pLyS strain, and the system was inducibly expressed under the control of a strong T7 promoter. The cells were grown to A600 = 0.5–0.8 at 37°C in 2 L of M9 minimal medium, and the cells were then incubated in fermentor with 1 L of fresh M9 minimal medium. Protein was induced with 1 mM of isopropyl-1-thio-β-d-galactopyranoside (IPTG) for 3–5 h. The cells were harvested by centrifugation and lysed by liquid shear with a French press to obtain the extract. The proteins were purified by SP-cation sepharose (Amersham Biosciences, Piscataway, NJ) and further purified by reverse-phase C18 high-performance liquid chromatography (HPLC) with a gradient of 30–40% acetonitrile. The solution of protein was lyophilized and kept at −80°C before use.
The forkhead domain of FOXP1 from A462 to Y565 was expressed with pET21 vector in E. coli. However, the protein was formed inclusion body and cannot be dissolved using a high salt buffer. In contrast, FOXP1's residues from A462 to K548 existed as a soluble form. Recombinant FOXP1 and its mutants were precipitated at the concentration of 0.5 mM in phosphate buffer at pH 7.4. It was reported that the addition of arginine and glutamate can be used to solublize proteins in solution.37 We also found that recombinant FOXP1 was soluble up to the concentration of 2.5 mM with a buffer system containing 50 mM NaCl, 25 mM MgCl2, 50 mM arginine, 50 mM glutamate, and 20 mM phosphate buffer at pH 5.5.
Mass spectrometric measurements
The molecular weights of proteins were confirmed using an API 365 triple quadruple mass spectrometer equipped with a TurboIonSpray source (PE Sciex, Thornill, Canada). Infusion of protein solutions (1∼10 μM in 50∼90% methanol or acetonitrile with 0.1% formic acid) into the mass spectrometer was performed using a syringe pump (Harvard Apparatus, South Natick, MA) at a flow rate of 12∼20 μL/min to acquire full scan mass spectra. The electrospray voltage at the spraying needle was optimized at 5000∼5300 V. The molecular weights of proteins were calculated by a computer software provided with the API 365 mass spectrometer. The experimental molecular weights of the forkhead domain of FOXP1 and its mutant were determined with the deviations less than 1 Da when compared with theoretical values. For example, the experimental molecular weight of wild-type FOXP1 was 10440.1, which is similar to the calculated value of 10441.0.
NMR sample preparation
M9 minimal media was used to produce 15N- and 15N/13C-labeled FOXP1 proteins. One gram/liter 15NH4Cl (98% 15N) and/or two grams/Liter [13C]-glucose (99% 13C) were substituted for the unlabeled compounds in the growth media. Selective [α-15N]-A-, -V-, or -K-labeled proteins were prepared using the protocol described by McIntosh et al.51 NMR samples were prepared in 20 mM sodium phosphate buffer at pH 5.5 containing 50 mM NaCl, 25 mM MgCl2, 50 mM arginine, and 50 mM glutamate with 10 or 100% D2O. NMR experiments were recorded with 1.3–2.5 mM of C61Y mutant and 0.7–2.5 mM of A39P/C61Y mutant. Monomer and dimer of C67Y mutant were separated by Bio-Gel P-30 size-exclusion chromatography. The separated dimer fractions were collected for NMR analysis.
Size-exclusion chromatography analysis
The relative ratios of FOXP1 monomer and dimer were analyzed by Bio-Gel P-30 size-exclusion chromatography (Bio-Rad, Madrid, Spain). The lyophilized FOXP1 proteins were dissolved in a solution of 50 mM NaCl, 25 mM MgCl2, 50 mM arginine, 50 mM glutamate, and 20 mM phosphate buffer at pH 5.5 to avoid protein solubility and stability problem.37 The zymogen form of streptopain (40.3 kDa), mature form of streptopain (27.5 kDa), and cytochrome c (12.3 kDa) were used as molecular weight standards for the analysis. The concentrations of 0.0171–0.171 mM of FOXP1, 0.0128–0.9465 mM of C61S mutant, 0.0635–1.208 mM of C61Y mutant, 0.5–2 mM of A39P/C61S mutant, and 0.5–2 mM of A39P/C61S mutant were incubated at 37°C before the analysis. A total of 750 μL protein solution was applied to the column. The monomer–dimer dissociation constants of FOXP1 and its mutant proteins were calculated using the formula: Kd=[monomer]2/[dimer]. We also carried out the analysis at pH 7.4, and the results were the same.
Differential scanning calorimetry
Calorimetric measurements were carried out with a VP-DSC (Microcal, LLC DSC, GE Healthcare Bio-Sciences). Temperature scans of wild-type FOXP1 and its C61S, C61Y, A39P/C61S, and A39P/C61Y mutants were performed from 15 to 85°C at a scan rate of 60°C/h. The protein concentration was 0.1 mM with a buffer containing 20 mM phosphate, 50 mM NaCl, 25 mM MgCl2, and 5 mM (tris(2-carboxyethyl)phosphine) at pH 5.5. The protein samples were incubated 0, 7, or 15 days before each run. At least three repetitions per experiments were taken, and two experiments were repeated twice. Data analysis was performed with ORIGIN software (version 7) from Origin Lab. The heat capacity of the protein in the initial state was subtracted from the raw signal and corrected for the buffer contribution to obtain the thermograms, Cp − Cp,N versus T curves.
Fluorescence measurements
The interactions between FOXP1 proteins and DNA were analyzed using tryptophan and tyrosine fluorescence of FOXP1, and the spectra were recorded on a Perkin Elmer Luminescence spectrometer LS 45. Binding reactions were performed at 25°C in a total volume of 1 mL in 20 mM phosphate buffer, 100 mM NaCl, 10 mM MgCl2, and 1 mM DTT at pH 7. A concentration of 2 μM FOXP1 A39P proteins was titrated with a stock solution of 12 μM 5′-AACTATGAAACAAATTTCCT-3′ DNA duplex. The samples were excited at 285 nm and were measured from 300 to 500 nm. The maximum fluorescence at 367 nm was used to determine the dissociation constant. Maximum fluorescence (Qmax) was defined as the unliganded fluorophore and observed fluorescence (Qobs) was defined as the liganded fluorophore. Percentage decrease from maximum [(1 − (Qobs/Qmax)) × 100%] was calculated from corrected data52 and further normalized by defining the final saturated titration fluorescence as 100% quenching. Binding affinity was calculated by using the Langmuir binding equation, Y = (Bmax× X)/(Kd + X) and the GraphPad Prizm 3.0 software, where Bmax is maximal binding, and X is total DNA concentration in molarity.53
NMR spectroscopy
All NMR spectra were acquired at 27°C on a Bruker Avance 600 spectrometer equipped with pulse field gradients and xyz-gradient triple resonance probes. Experiments of HNCA, HN(CO)CA, HNCACB, CBCA(CO)NH and 15N-edited TOCSY-HSQC and NOESY-HSQC were carried out for the purpose of proton, carbon, and nitrogen resonance assignments.54–56 2D TOCSY and NOESY experiments with an unlabeled sample in D2O provided the basis for aromatic proton assignments and hydrogen bond determination.57 Distance restraints were obtained from 3D 15N-edited and 13C-edited NOESY-HSQC with mixing times of 100 and 120 ms. HNHA experiment was performed to measure 3JNHα coupling constants and obtain dihedral angles.58 The φ and θ backbone dihedral angles were also calculated by TALOS program.59 Data were processed and analyzed using the Bruker TOPSPIN 1.3 and Aurelia programs.
Chemical shift perturbation was used to determine the difference between FOXP1 C61Y and A39P/C61Y mutants. The weighted average change in the chemical shift of the amide nitrogen and proton atoms was measured through 1H-15N HSQC spectra and treated according to the equation60:
where ΔδN and ΔδH represent respectively the changes in nitrogen and proton chemical shifts (in parts per million).
Structure calculations
The software X-PLOR Version 3.851 was used to calculate the solution structure of FOXP A39P/C61Y mutant using the hybrid distance geometry-dynamic simulated annealing method.61 62 The cross-peak intensities of resonance assignments were classified into strong, medium, and weak. These intensities were further converted into distance restraints of 1.8–2.8, 1.8–3.6, 1.8–5, and 2.3–6 Å, respectively. Pseudoatoms of methylene, methyl, and aromatic protons were corrected by adding a further 0.5 Å to the upper limit distances involving methyl protons. The dihedral angles φ were determined from the 3JNHα coupling constants.58 For 3JNHα values less than 5 Hz, φ values were restricted from –25° to –95°; and for 3JNHα values greater than 9 Hz, φ values were restricted from –100° to –170°. Backbone hydrogen bonds within the antiparallel β-sheets were identified according to the criteria previously laid out.63 Two restraints were used for each NH–CO backbone hydrogen bond with dN–O restricted to 2.4–3.3 Å and dH–O to 1.7–2.3 Å. A total of 100 structures were generated using NOE distance, dihedral angle, and hydrogen bond restraints. During the first phase of dynamics at 2000 K, the value of the force constant of the NOE term was kept constant at 50 kcal mol−1 Å−2. The repulsion term was gradually increased from 0.03 to 4.0 kcal mol−1 Å−2, and the torsion angle term from 5 to 200 kcal mol−1 rad−2. The simulated annealing refinement consisted of a 9-ps cooling dynamics and then 200 cycles of Powell minimization. The twenty lowest-energy structures were accepted based on violations of distance restraints smaller than 0.5 Å, dihedral angle restraints smaller than 5°, a van der Waals energy cut-off value of 35 kcal/mol, and an NOE energy cut-off value of 55 kcal/mol. The structure figures were prepared using the MOLMOL and PyMOL programs.64 65
NMR relaxation measurements
Backbone dynamics of FOXP1 C61Y and A39P/C61Y mutants were determined by two-dimensional proton-detected heteronuclear NMR spectroscopy. NMR relaxation data were collected at 300 K. The 15N-R1 and 15N-R2 relaxation rates and steady-state 1H-15N NOEs were performed as described by Farrow et al.66 67 A relaxation delay of 6 s was used for all relaxation experiments, and the spectra were recorded as 512 × 2 K complex matrices with 16 scans per F1 experiment. Spectral width of 1946 and 7184 Hz were used in F1 and F2, respectively. A total of 10 data sets were collected to measure R1 of FOXP1 A39P/C61Y mutant with the delay values of: 30, 100, 300, 500, 650, 800, 900, 1000, 1500, and 3000 ms and R1 of C61Y mutant with the delay values of 2, 120, 240, 360, 480, 600, 720, 900, 1020, and 1290 ms. A total of 10 data sets were collected to measure R2 of FOXP1 A39P/C61Y mutant with delay values of 20, 40, 60, 80, 100, 120, 150, 200, 300, and 250 ms and R2 of C61Y mutant with the delay values of 2, 20, 35, 50, 70, 85, 100, 120, 135, and 150 ms. The longitudinal and transverse relaxation rate constants, R1 and R2, were obtained from exponential fits of the peak height data using least-squares fit software SigmaPlot (Jandel Scientific Software, San Rafael, CA). In NOE experiment, two spectra (one with the NOE and one without) were collected. The NOE effect was calculated as the ratio of peak heights in spectra collected with and without NOE. The reported NOE value was the average value from three pairs of NOE experiments. The reported Ri values are the mean values of at least two independent data sets. A relaxation delay of 6 s was used, and 256 complex t1 increments of 32 scans were acquired.
The model-free analysis
The heteronuclear 15N relaxation rate constants, R1 and R2, and the 1H-15N steady state NOE values were analyzed using the FASTModelfree program.68 This program is based on the model-free formalism that was pioneered by Lipari and Szabo.69 70 In this approach, the overall and internal molecular motions were assumed to be independent, and the spectral density function for a molecule undergoing isotropic tumbling was calculated using the appropriate expression,
where 1/τ = 1/τm + 1/τe and 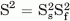 , τm is overall rotational correlation time of the molecule, τe is the effective correlation time for the motions on the slower of the two time scale, S2 is the square of the generalized order parameter, and
, τm is overall rotational correlation time of the molecule, τe is the effective correlation time for the motions on the slower of the two time scale, S2 is the square of the generalized order parameter, and 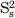 and
and 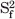 are the squares of order parameters for the motion on the fast and slow time scales, respectively.
are the squares of order parameters for the motion on the fast and slow time scales, respectively.
The 15N relaxation parameters were analyzed according to the following criteria: the residues with large amplitudes in molecular motions were excluded in the diffusion tensor calculations, and the residues with significant internal motions on ps-ns time scales were not included in calculations when the residues exhibited 1H-15N NOE less than 0.65. The program models of each 15N–H bond, R1, R2, and NOE parameters were available, consist of the following extended model-free parameters: (1) S2; (2) S2, τe = τf; (3) S2, Rex; (4) S2, τe = τf, Rex; and (5) 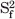 ,
, 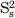 , τe = τs. The dynamic model describing internal motion was selected in a residue-specific manner, and the numerical optimization procedure described by Mandel et al. was used to estimate the involved parameters for the model.71 The program QUADRIC DIFFUSION was used to calculate the diffusion tenors of C61Y mutant for axially symmetric motional models from experimental 15N spin relaxation data.72 All optimization procedures were involved in the minimization of the χ2 function.73
, τe = τs. The dynamic model describing internal motion was selected in a residue-specific manner, and the numerical optimization procedure described by Mandel et al. was used to estimate the involved parameters for the model.71 The program QUADRIC DIFFUSION was used to calculate the diffusion tenors of C61Y mutant for axially symmetric motional models from experimental 15N spin relaxation data.72 All optimization procedures were involved in the minimization of the χ2 function.73
Protein Data Bank accession number
The coordinates of 20 calculated structures of FOXP1 A39P/C61Y mutant have been deposited in the Protein Data Bank under the accession number 2kiu.
Acknowledgments
NMR spectra were obtained at National Cheng Kung University or the High-Field Biomacromolecular NMR Core Facility supported by the National Research Program for Genomic Medicine.
Glossary
Abbreviations:
- HSQC
heteronuclear single quantum correlation
- NMR
nuclear magnetic resonance
- NOE
nuclear Overhauser effect
- TOCSY
Total correlated spectroscopy.
Supplementary material
References
- 1.Katoh M. Human FOX gene family (Review) Int J Oncol. 2004;25:1495–1500. [PubMed] [Google Scholar]
- 2.Weigel D, Jackle H. The fork head domaa novel DNA binding motif of eukaryotic transcription factors? Cell. 1990;63:455–456. doi: 10.1016/0092-8674(90)90439-l. [DOI] [PubMed] [Google Scholar]
- 3.Kaestner KH, Knochel W, Martinez DE. Unified nomenclature for the winged helix/forkhead transcription factors. Genes Dev. 2000;14:142–146. [PubMed] [Google Scholar]
- 4.Mazet F, Yu JK, Liberles DA, Holland LZ, Shimeld SM. Phylogenetic relationships of the Fox (Forkhead) gene family in the Bilateria. Gene. 2003;316:79–89. doi: 10.1016/s0378-1119(03)00741-8. [DOI] [PubMed] [Google Scholar]
- 5.Kaufmann E, Knochel W. Five years on the wings of fork head. Mech Dev. 1996;57:3–20. doi: 10.1016/0925-4773(96)00539-4. [DOI] [PubMed] [Google Scholar]
- 6.Carlsson P, Mahlapuu M. Forkhead transcription factors: key players in development and metabolism. Dev Biol. 2002;250:1–23. doi: 10.1006/dbio.2002.0780. [DOI] [PubMed] [Google Scholar]
- 7.Lehmann OJ, Sowden JC, Carlsson P, Jordan T, Bhattacharya SS. Fox's in development and disease. Trends Genet. 2003;19:339–344. doi: 10.1016/S0168-9525(03)00111-2. [DOI] [PubMed] [Google Scholar]
- 8.Hannenhalli S, Kaestner KH. The evolution of Fox genes and their role in development and disease. Nat Rev Genet. 2009;10:233–240. doi: 10.1038/nrg2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Banham AH, Beasley N, Campo E, Fernandez PL, Fidler C, Gatter K, Jones M, Mason DY, Prime JE, Trougouboff P, Wood K, Cordell JL. The FOXP1 winged helix transcription factor is a novel candidate tumor suppressor gene on chromosome 3p. Cancer Res. 2001;61:8820–8829. [PubMed] [Google Scholar]
- 10.Lai CS, Fisher SE, Hurst JA, Vargha-Khadem F, Monaco AP. A forkhead-domain gene is mutated in a severe speech and language disorder. Nature. 2001;413:519–523. doi: 10.1038/35097076. [DOI] [PubMed] [Google Scholar]
- 11.Bennett CL, Christie J, Ramsdell F, Brunkow ME, Ferguson PJ, Whitesell L, Kelly TE, Saulsbury FT, Chance PF, Ochs HD. The immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome (IPEX) is caused by mutations of FOXP3. Nat Genet. 2001;27:20–21. doi: 10.1038/83713. [DOI] [PubMed] [Google Scholar]
- 12.Lu MM, Li S, Yang H, Morrisey EE. Foxp4: a novel member of the Foxp subfamily of winged-helix genes co-expressed with Foxp1 and Foxp2 in pulmonary and gut tissues. Mech Dev. 2002;119(Suppl 1):S197–S202. doi: 10.1016/s0925-4773(03)00116-3. [DOI] [PubMed] [Google Scholar]
- 13.Wang B, Lin D, Li C, Tucker P. Multiple domains define the expression and regulatory properties of Foxp1 forkhead transcriptional repressors. J Biol Chem. 2003;278:24259–24268. doi: 10.1074/jbc.M207174200. [DOI] [PubMed] [Google Scholar]
- 14.Fetterman CD, Rannala B, Walter MA. Identification and analysis of evolutionary selection pressures acting at the molecular level in five forkhead subfamilies. BMC Evol Biol. 2008;8:261–273. doi: 10.1186/1471-2148-8-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li C, Tucker PW. DNA-binding properties and secondary structural model of the hepatocyte nuclear factor 3/fork head domain. Proc Natl Acad Sci USA. 1993;90:11583–11587. doi: 10.1073/pnas.90.24.11583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shu W, Yang H, Zhang L, Lu MM, Morrisey EE. Characterization of a new subfamily of winged-helix/forkhead (Fox) genes that are expressed in the lung and act as transcriptional repressors. J Biol Chem. 2001;276:27488–27497. doi: 10.1074/jbc.M100636200. [DOI] [PubMed] [Google Scholar]
- 17.Bates GJ, Fox SB, Han C, Launchbury R, Leek RD, Harris AL, Banham AH. Expression of the forkhead transcription factor FOXP1 is associated with that of estrogen receptorbeta in primary invasive breast carcinomas. Breast Cancer Res Treat. 2008;111:453–459. doi: 10.1007/s10549-007-9812-4. [DOI] [PubMed] [Google Scholar]
- 18.Fox SB, Brown P, Han C, Ashe S, Leek RD, Harris AL, Banham AH. Expression of the forkhead transcription factor FOXP1 is associated with estrogen receptor alpha and improved survival in primary human breast carcinomas. Clin Cancer Res. 2004;10:3521–3527. doi: 10.1158/1078-0432.CCR-03-0461. [DOI] [PubMed] [Google Scholar]
- 19.Koon HB, Ippolito GC, Banham AH, Tucker PW. FOXP1: a potential therapeutic target in cancer. Expert Opin Ther Targets. 2007;11:955–965. doi: 10.1517/14728222.11.7.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barrans SL, Fenton JA, Banham A, Owen RG, Jack AS. Strong expression of FOXP1 identifies a distinct subset of diffuse large B-cell lymphoma (DLBCL) patients with poor outcome. Blood. 2004;104:2933–2935. doi: 10.1182/blood-2004-03-1209. [DOI] [PubMed] [Google Scholar]
- 21.Fenton JA, Schuuring E, Barrans SL, Banham AH, Rollinson SJ, Morgan GJ, Jack AS, van Krieken JH, Kluin PM. t(3;14)(p14;q32) results in aberrant expression of FOXP1 in a case of diffuse large B-cell lymphoma. Genes Chromosomes Cancer. 2006;45:164–168. doi: 10.1002/gcc.20278. [DOI] [PubMed] [Google Scholar]
- 22.Streubel B, Vinatzer U, Lamprecht A, Raderer M, Chott A. T(3;14)(p14.1;q32) involving IGH and FOXP1 is a novel recurrent chromosomal aberration in MALT lymphoma. Leukemia. 2005;19:652–658. doi: 10.1038/sj.leu.2403644. [DOI] [PubMed] [Google Scholar]
- 23.Sagaert X, de Paepe P, Libbrecht L, Vanhentenrijk V, Verhoef G, Thomas J, Wlodarska I, De Wolf-Peeters C. Forkhead box protein P1 expression in mucosa-associated lymphoid tissue lymphomas predicts poor prognosis and transformation to diffuse large B-cell lymphoma. J Clin Oncol. 2006;24:2490–2497. doi: 10.1200/JCO.2006.05.6150. [DOI] [PubMed] [Google Scholar]
- 24.Goatly A, Bacon CM, Nakamura S, Ye H, Kim I, Brown PJ, Ruskone-Fourmestraux A, Cervera P, Streubel B, Banham AH, Du MQ. FOXP1 abnormalities in lymphoma: translocation breakpoint mapping reveals insights into deregulated transcriptional control. Mod Pathol. 2008;21:902–911. doi: 10.1038/modpathol.2008.74. [DOI] [PubMed] [Google Scholar]
- 25.Enard W, Przeworski M, Fisher SE, Lai CS, Wiebe V, Kitano T, Monaco AP, Paabo S. Molecular evolution of FOXP2, a gene involved in speech and language. Nature. 2002;418:869–872. doi: 10.1038/nature01025. [DOI] [PubMed] [Google Scholar]
- 26.Bennett CL, Ochs HD. IPEX is a unique X-linked syndrome characterized by immune dysfunction, polyendocrinopathy, enteropathy, and a variety of autoimmune phenomena. Curr Opin Pediatr. 2001;13:533–538. doi: 10.1097/00008480-200112000-00007. [DOI] [PubMed] [Google Scholar]
- 27.Clark KL, Halay ED, Lai E, Burley SK. Co-crystal structure of the HNF-3/fork head DNA-recognition motif resembles histone H5. Nature. 1993;364:412–420. doi: 10.1038/364412a0. [DOI] [PubMed] [Google Scholar]
- 28.van Dongen MJ, Cederberg A, Carlsson P, Enerback S, Wikstrom M. Solution structure and dynamics of the DNA-binding domain of the adipocyte-transcription factor FREAC-11. J Mol Biol. 2000;296:351–359. doi: 10.1006/jmbi.1999.3476. [DOI] [PubMed] [Google Scholar]
- 29.Marsden I, Jin C, Liao X. Structural changes in the region directly adjacent to the DNA-binding helix highlight a possible mechanism to explain the observed changes in the sequence-specific binding of winged helix proteins. J Mol Biol. 1998;278:293–299. doi: 10.1006/jmbi.1998.1703. [DOI] [PubMed] [Google Scholar]
- 30.Jin C, Liao X. Backbone dynamics of a winged helix protein and its DNA complex at different temperatures: changes of internal motions in genesis upon binding to DNA. J Mol Biol. 1999;292:641–651. doi: 10.1006/jmbi.1999.3106. [DOI] [PubMed] [Google Scholar]
- 31.Tsai KL, Huang CY, Chang CH, Sun YJ, Chuang WJ, Hsiao CD. Crystal structure of the human FOXK1a-DNA complex and its implications on the diverse binding specificity of winged helix/forkhead proteins. J Biol Chem. 2006;281:17400–17409. doi: 10.1074/jbc.M600478200. [DOI] [PubMed] [Google Scholar]
- 32.Liu PP, Chen YC, Li C, Hsieh YH, Chen SW, Chen SH, Jeng WY, Chuang WJ. Solution structure of the DNA-binding domain of interleukin enhancer binding factor 1 (FOXK1a) Proteins. 2002;49:543–553. doi: 10.1002/prot.10227. [DOI] [PubMed] [Google Scholar]
- 33.Tsai KL, Sun YJ, Huang CY, Yang JY, Hung MC, Hsiao CD. Crystal structure of the human FOXO3a-DBD/DNA complex suggests the effects of post-translational modification. Nucleic Acids Res. 2007;35:6984–6994. doi: 10.1093/nar/gkm703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weigelt J, Climent I, Dahlman-Wright K, Wikstrom M. Solution structure of the DNA binding domain of the human forkhead transcription factor AFX (FOXO4) Biochemistry. 2001;40:5861–5869. doi: 10.1021/bi001663w. [DOI] [PubMed] [Google Scholar]
- 35.Gajiwala KS, Burley SK. Winged helix proteins. Curr Opin Struct Biol. 2000;10:110–116. doi: 10.1016/s0959-440x(99)00057-3. [DOI] [PubMed] [Google Scholar]
- 36.Stroud JC, Wu Y, Bates DL, Han A, Nowick K, Paabo S, Tong H, Chen L. Structure of the forkhead domain of FOXP2 bound to DNA. Structure. 2006;14:159–166. doi: 10.1016/j.str.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 37.Golovanov AP, Hautbergue GM, Wilson SA, Lian LY. A simple method for improving protein solubility and long-term stability. J Am Chem Soc. 2004;126:8933–8939. doi: 10.1021/ja049297h. [DOI] [PubMed] [Google Scholar]
- 38.Wu Y, Borde M, Heissmeyer V, Feuerer M, Lapan AD, Stroud JC, Bates DL, Guo L, Han A, Ziegler SF, Mathis D, Benoist C, Chen L, Rao A. FOXP3 controls regulatory T cell function through cooperation with NFAT. Cell. 2006;126:375–387. doi: 10.1016/j.cell.2006.05.042. [DOI] [PubMed] [Google Scholar]
- 39.Obsil T, Obsilova V. Structure/function relationships underlying regulation of FOXO transcription factors. Oncogene. 2008;27:2263–2275. doi: 10.1038/onc.2008.20. [DOI] [PubMed] [Google Scholar]
- 40.Jin C, Marsden I, Chen X, Liao X. Sequence specific collective motions in a winged helix DNA binding domain detected by 15N relaxation NMR. Biochemistry. 1998;37:6179–6187. doi: 10.1021/bi980031v. [DOI] [PubMed] [Google Scholar]
- 41.Bracken C, Carr PA, Cavanagh J, Palmer AG., III Temperature dependence of intramolecular dynamics of the basic leucine zipper of GCN4: implications for the entropy of association with DNA. J Mol Biol. 1999;285:2133–2146. doi: 10.1006/jmbi.1998.2429. [DOI] [PubMed] [Google Scholar]
- 42.Miloushev VZ, Bahna F, Ciatto C, Ahlsen G, Honig B, Shapiro L, Palmer AG., III Dynamic properties of a type II cadherin adhesive domaimplications for the mechanism of strand-swapping of classical cadherins. Structure. 2008;16:1195–1205. doi: 10.1016/j.str.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yuan Y, Simplaceanu V, Lukin JA, Ho C. NMR investigation of the dynamics of tryptophan side-chains in hemoglobins. J Mol Biol. 2002;321:863–878. doi: 10.1016/s0022-2836(02)00704-0. [DOI] [PubMed] [Google Scholar]
- 44.Jin C, Marsden I, Chen X, Liao X. Dynamic DNA contacts observed in the NMR structure of winged helix protein-DNA complex. J Mol Biol. 1999;289:683–690. doi: 10.1006/jmbi.1999.2819. [DOI] [PubMed] [Google Scholar]
- 45.Yan H, Liao X. Amino acid substitutions in a long flexible sequence influence thermodynamics and internal dynamic properties of winged helix protein genesis and its DNA complex. Biophys J. 2003;85:3248–3254. doi: 10.1016/S0006-3495(03)74742-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhu L, Hu J, Lin D, Whitson R, Itakura K, Chen Y. Dynamics of the Mrf-2 DNA-binding domain free and in complex with DNA. Biochemistry. 2001;40:9142–9150. doi: 10.1021/bi010476a. [DOI] [PubMed] [Google Scholar]
- 47.Wolberger C, Campbell R. New perch for the winged helix. Nat Struct Biol. 2000;7:261–262. doi: 10.1038/74004. [DOI] [PubMed] [Google Scholar]
- 48.Kenney LJ. Structure/function relationships in OmpR and other winged-helix transcription factors. Curr Opin Microbiol. 2002;5:135–141. doi: 10.1016/s1369-5274(02)00310-7. [DOI] [PubMed] [Google Scholar]
- 49.Overdier DG, Porcella A, Costa RH. The DNA-binding specificity of the hepatocyte nuclear factor 3/forkhead domain is influenced by amino-acid residues adjacent to the recognition helix. Mol Cell Biol. 1994;14:2755–2766. doi: 10.1128/mcb.14.4.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schochetman G, Ou CY, Jones WK. Polymerase chain reaction. J Infect Dis. 1988;158:1154–1157. doi: 10.1093/infdis/158.6.1154. [DOI] [PubMed] [Google Scholar]
- 51.McIntosh LP, Wand AJ, Lowry DF, Redfield AG, Dahlquist FW. Assignment of the backbone 1H and 15N NMR resonances of bacteriophage T4 lysozyme. Biochemistry. 1990;29:6341–6362. doi: 10.1021/bi00479a003. [DOI] [PubMed] [Google Scholar]
- 52.Landon LA, Harden W, Illy C, Deutscher SL. High-throughput fluorescence spectroscopic analysis of affinity of peptides displayed on bacteriophage. Anal Biochem. 2004;331:60–67. doi: 10.1016/j.ab.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 53.Kim C, Paulus BF, Wold MS. Interactions of human replication protein A with oligonucleotides. Biochemistry. 1994;33:14197–14206. doi: 10.1021/bi00251a031. [DOI] [PubMed] [Google Scholar]
- 54.Clore GM, Gronenborn AM. Determining the structures of large proteins and protein complexes by NMR. Trends Biotechnol. 1998;16:22–34. doi: 10.1016/S0167-7799(97)01135-9. [DOI] [PubMed] [Google Scholar]
- 55.Grzesiek SaB A. Improved 3D triple-resonance NMR techniques applied to a 31 kDa protein. J Magn Reson. 1992;96:432–440. [Google Scholar]
- 56.Bax A. Two-dimensional NMR and protein structure. Annu Rev Biochem. 1989;58:223–256. doi: 10.1146/annurev.bi.58.070189.001255. [DOI] [PubMed] [Google Scholar]
- 57.Wüthrich K. NMR of Proteins and Nucleic Acid. New York: John Wiley & Sons; 1986. [Google Scholar]
- 58.Vuister GW, Bax A. Measurement of four-bond HN-H alpha J-couplings in staphylococcal nuclease. J Biomol NMR. 1994;4:193–200. doi: 10.1007/BF00175247. [DOI] [PubMed] [Google Scholar]
- 59.Cornilescu G, Delaglio F, Bax A. Protein backbone angle restraints from searching a database for chemical shift and sequence homology. J Biomol NMR. 1999;13:289–302. doi: 10.1023/a:1008392405740. [DOI] [PubMed] [Google Scholar]
- 60.Foster MP, Wuttke DS, Clemens KR, Jahnke W, Radhakrishnan I, Tennant L, Reymond M, Chung J, Wright PE. Chemical shift as a probe of molecular interfaces: NMR studies of DNA binding by the three amino-terminal zinc finger domains from transcription factor IIIA. J Biomol NMR. 1998;12:51–71. doi: 10.1023/a:1008290631575. [DOI] [PubMed] [Google Scholar]
- 61.Brunger AT. X-PLOR Version 3.1: a system for X-ray crystallography and NMR. New Haven, CT: Yale University Press; 1992. [Google Scholar]
- 62.Nilges M, Clore GM, Gronenborn AM. Determination of three-dimensional structures of proteins from interproton distance data by dynamical simulated annealing from a random array of atoms. Circumventing problems associated with folding. FEBS Lett. 1988;239:129–136. doi: 10.1016/0014-5793(88)80559-3. [DOI] [PubMed] [Google Scholar]
- 63.Wagner G, Braun W, Havel TF, Schaumann T, Go N, Wuthrich K. Protein structures in solution by nuclear magnetic resonance and distance geometry. The polypeptide fold of the basic pancreatic trypsin inhibitor determined using two different algorithms, DISGEO and DISMAN. J Mol Biol. 1987;196:611–639. doi: 10.1016/0022-2836(87)90037-4. [DOI] [PubMed] [Google Scholar]
- 64.DeLano WL. The PyMol molecular graphics system. Sam Carlos, CA, USA: DeLano Scientific; 2002. [Google Scholar]
- 65.Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. J Mol Graph. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. 29–32. [DOI] [PubMed] [Google Scholar]
- 66.Farrow NA, Muhandiram R, Singer AU, Pascal SM, Kay CM, Gish G, Shoelson SE, Pawson T, Forman-Kay JD, Kay LE. Backbone dynamics of a free and phosphopeptide-complexed Src homology 2 domain studied by 15N NMR relaxation. Biochemistry. 1994;33:5984–6003. doi: 10.1021/bi00185a040. [DOI] [PubMed] [Google Scholar]
- 67.Farrow NA, Zhang O, Forman-Kay JD, Kay LE. Comparison of the backbone dynamics of a folded and an unfolded SH3 domain existing in equilibrium in aqueous buffer. Biochemistry. 1995;34:868–878. doi: 10.1021/bi00003a021. [DOI] [PubMed] [Google Scholar]
- 68.Cole R, Loria JP. FAST-Modelfree: a program for rapid automated analysis of solution NMR spin-relaxation data. J Biomol NMR. 2003;26:203–213. doi: 10.1023/a:1023808801134. [DOI] [PubMed] [Google Scholar]
- 69.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 2. Analysis of experimental results. J Am Chem Soc. 1982;104:4559–4570. [Google Scholar]
- 70.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104:4546–4559. [Google Scholar]
- 71.Mandel AM, Akke M, Palmer AG., III Backbone dynamics of Escherichia coli ribonuclease HI: correlations with structure and function in an active enzyme. J Mol Biol. 1995;246:144–163. doi: 10.1006/jmbi.1994.0073. [DOI] [PubMed] [Google Scholar]
- 72.Sarsa A, Boronat J, Casulleras J. Quadratic diffusion Monte Carlo and pure estimators for atoms. J Chem Phys. 2002;116:5956–5962. [Google Scholar]
- 73.Paquin Rl, Ferrage F, Mulder FAA, Akke M, Bodenhausen G. Multiple-timescale dynamics of side-chain carboxyl and carbonyl groups in proteins by 13C nuclear spin relaxation. J Am Chem Soc. 2008;130:15805–15807. doi: 10.1021/ja803794g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.