Abstract
The formation of any tissue involves differentiation, cell dynamics and interactions with adjacent tissues. This paper suggests that the complexity of the system as a whole can be represented as a mathematical graph, that is, a set of connected triples of the general form [term] <relationship> [term]. Computationally, such graphs are widely used for modeling data; visually, they form hierarchies and networks. For morphogenesis, the triples are of the general structure <noun> <verb> <noun>, where nouns cover tissues, molecules and networks and verbs describe processes such as moves, differentiates, grows and apoptoses. The paper considers the general formalism of graphs, where graphs are already used in biology, and how developmental anatomy may be described using this format. Representing morphogenesis as a visual graph is complicated as the formalism has to incorporate tissue types, molecular signals, networks, dynamic processes and some aspects, at least, of tissue geometry. The formation of a capillary sprout is chosen as an example of how this complexity can be represented graphically, with colour used to distinguish tissues and molecules. There are three key benefits, beyond its compactness, in using the graph formalism of morphogenesis to complement experimentation. First, it emphasizes the distributed nature of causality in morphogenesis. Secondly, producing all the triples for the visual graph requires explicit formalization of each aspect of the process, and this, in turn, often exposes gaps in knowledge and so suggests new experiments. Thirdly, once the graph has been formalized, triples can be annotated with associated information or IDs (e.g. cell types, publications, gene-expression data) that link to external online resources that may be regularly updated. Such annotations allow the graph to be viewed as a self-maintaining review. The graph approach sees dynamic processes as the drivers of developmental momentum and, because the same processes are used many times during development, it seems appropriate to view them as modules and their underlying networks as genomic subroutines.
Keywords: mathematical graph, morphogenesis, ontology, regulatory networks, systems biology, vascularization
Introduction
The study of morphogenesis has always been difficult. Traditional experimental approaches based on ‘cut and splice’ and similar techniques have revealed the cellular processes that drive morphogenesis, but it has required modern molecular, mutational and genetic engineering technologies to identify the key proteins and networks that control and produce changes in phenotype. As neither approach can easily study the morphogenetic role of the geometry of the participating tissues and their environment, it has been hard to produce coherent explanations of how tissues form their detailed structures within the complex environment of the embryo.
Various authors have tried to construct general frameworks for understanding morphogenesis. Trinkaus (1883) focused on how the properties of cells that could be studied in vitro participated in organogenesis. Bard (1992) tried to show how a set of individual and cooperative dynamic cell properties, the morphogenetic toolkit, operating within geometric constraints were responsible for building tissues. Davies (2005) has described the molecular basis of these properties, their physical implications and their modular nature. Most recently, Newman & Bhat (2009) have considered how a toolkit of primitive molecular networks (‘dynamical patterning modules’) in conjunction with mesoscale physical processes that predated metazoa could easily evolve to produce a ‘pattern language’ capable of generating all metazoan body plans and organ forms. None of these approaches has provided a full theory of morphogenesis for contemporary animals.
The alternative is to model morphogenesis using top-down systems approaches that integrate the various structural, molecular and cellular components underpinning morphogenesis. One line that has been successful in other areas of developmental biology has been the use of coupled sets of ordinary differential equations to link molecular dynamics to tissue-level events. This approach, which has led to models of the formation of pigment patterns in vertebrate skins (Bard, 1981; Murray, 1981) based on the model of Turing (1952), is currently being used, for example, to model somite patterning (Goldbeter & Pourquié, 2008) and gene regulatory networks (GRN; Ribeiro & Lloyd-Price, 2007). Analysing morphogenetic processes such as cell sorting (Painter, 2009) is, however, much harder and requires the additional sophistication provided by partial differential equations. More may still be needed: Marée & Hogeweg (2001) needed to combine partial differential equation formulations with cellular automata properties to model the transformation of the Dictyostelium discoideum slug into a fruiting body.
Any analysis of a morphogenetic event presupposes that there is a good description of that event. Producing this is often complicated, as morphogenesis involves not only cell, tissue and molecular dynamics, but also the structure of the participating tissues and their environment. Disentangling the facets of the story is not always easy and this paper suggests that a helpful way of formalizing the process is by representing it as a mathematical graph, which is a set of linked triads (Junker & Schreiber, 2008). For morphogenesis, each triad describes a fact and is of the general form:
Such graphs are a key tool of systems biology and are not new (Doi, 1984); indeed, they are now used in biology, informally at least, to describe evolutionary hierarchies, protein networks and ontologies (formal descriptions of an area of knowledge, such as the types of cells).
This paper starts by summarizing the types of events that underpin the generation of anatomical structures and so need to be included in graphs; the next section describes graphs, their construction and some biological examples. The major part of the paper shows how to describe morphogenetic processes graphically, with the focus being on the processes that drive change, many of which are frequently repeated during development and can be seen as modules (Davies, 2005). The discussion considers the use of the graphical approach and the implications of modules for how large-scale information may be stored in the genome.
The essential features of developmental anatomy
Producing a tissue involves a wide range of developmental phenomena (Figs 1 and 2; for review, see Gilbert, 2010).
Fig. 1.

A graph showing typical responses to signal activation. Here and in other figures, blue boxes represent tissues; green boxes represent molecular events (often abbreviated for simplicity); yellow boxes are processes and grey boxes (with dotted links) are examples. Shh, sonic hedgehog; EGF, epidermal growth factor; FGF, fibroblast growth factor.
Fig. 2.
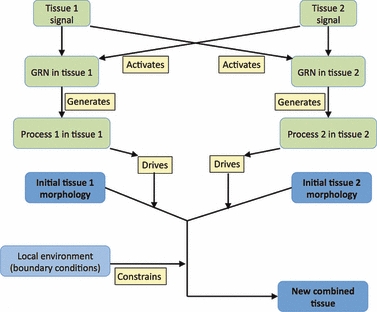
Graphical representation of the morphogenetic interactions between two tissues that lead to the formation of an integrated organ (e.g. the ureteric bud and metanephric mesenchyme combine to give the early metanephros) or the production of a novel feature (when muscle meets bone, an enthesis may form). GRN, gene regulatory network.
Patterning
Morphogenesis is always a late event in tissue development. It requires that the participating tissues be in the right place, with the appropriate molecular apparatus and, sometimes, with any required extracellular pathways (e.g. for contact guidance) already laid down (itself a morphogenetic event). These initial conditions can be viewed as resulting from earlier cell patterning; in pre-molecular days, the tissues would have been described as having become competent. These patterning aspects of morphogenesis will be taken for granted here, even though they are rarely fully understood.
Tissue geometry
Although the geometry of existing structures is important both as the starting point for morphogenesis and as the environment within which morphogenesis is constrained (Bard, 1992; Salazar-Ciudad & Jernvall, 2010), it has not received a great deal of attention. First, all new structures derive from existing ones. Secondly, surrounding structure may facilitate change or provide constraints on growth or movement. Local extracellular matrix, for example, provides a pathway or permissive environment that directs cell migration through contact guidance (e.g. Nakatsuji & Johnson, 1984).
Morphogenetic processes
Table 1 lists the major types of morphogenetic processes that drive development. This list is short because it is restricted to the direct actions of a tissue, defined as a coherent group of histologically similar cells. Thus, movement is not subdivided into contact guidance, chemotaxis, haptotaxis, etc., because these subdivisions are the effect of external constraints on that movement. Of particular interest are changes in differentiation, as these can lead to a change in morphology: for example, one morphogenetic effect of a mesenchyme-to-epithelium transition by a group of cells is that the group acquires a lumen (e.g. the early formation of the nephron; Zeisberg et al. 2005). This is because epithelial cells are polarized and maintain a free apical surface, whereas mesenchymal cells do not. In contrast, such lumens are lost when the reverse transition takes place (e.g. when the medial component of the epithelial somite becomes mesenchymal and forms the sclerotome).
Table 1.
Examples of the processes that drive morphogenesis
| Change in cell property |
| Cell migration initiated or stopped |
| Cell shape change (e.g. columnerisation) |
| Change in cell density |
| Adhesion-protein secretion and metalloproteinase secretion (increase cell density) |
| Adhesion-protein loss and ECM secretion (decrease cell density) |
| Change in cellular organisation |
| Epithelial folding (e.g. to form a duct) Duct branching Convergent extension |
| Cell sorting (e.g. boundary formation through differential expression of cell-surfaceproteins) |
| Change in differentiation |
| Mesenchyme → epithelium transition (polarization and lumen formation) |
| Epithelium → mesenchyme transition polarization (loss of polarization and lumen) |
| Change in cell number |
| Proliferation Apoptosis |
Proliferation and apoptosis
Programmed cell death is used as a shaping mechanism for morphogenesis, and the best-known example is the apoptosis of the mesenchyme in the tetrapod handplate that leads to the separation of the digits (Merino et al. 1999). Cell proliferation can similarly drive morphogenesis, and underpins, for example, the formation of the early limb bud and the provision of its progress-zone cells (Ovchinnikov et al. 2006), while differential growth alters shape. It is also worth noting that enlargement imposes local stresses and strains on the growing tissue and these may induce modifications to the local anatomy.
Extracellular environment
Morphogenesis can also be driven through changes in extracellular matrix. In the early corneal stroma, for example, mesenchymal cells secrete proteoglycans and the swelling that follows their hydration drives its enlargement; similarly, the secretion of vitreous-humour proteoglycans into the retinal space leads to the anterior retinal epithelium buckling into the ciliary body folds (Bard & Ross, 1982). Perhaps the most dramatic example of environmental activity directing morphogenesis, however, is the force exerted by the movement of blood on the wall of the early cardiac outflow tract: the flow moulds the soft endothelium of the outflow tract (cardiac jelly underlies it) to form the spiral septum (Hove et al. 2003).
The molecular basis of morphogenesis
Like other developmental activities, the initiation of a morphogenetic process usually results from a signal activating a gene regulatory network that will in turn activate the network which drives that process and may also direct the synthesis of some of its proteins (Figs 1 and 3). We now know a great deal about the signals, the receptors and the GRNs that initiate developmental change (for diagrams, see http://www.sabiosciences.com/pathwaycentral.php; for review, see Gilbert, 2010), together with the growth and apoptosis networks. Much is also known about the molecules that drive morphogenetic change directly (adhesions and other membrane-associated molecules, extracellular matrix components, etc.).
Fig. 3.
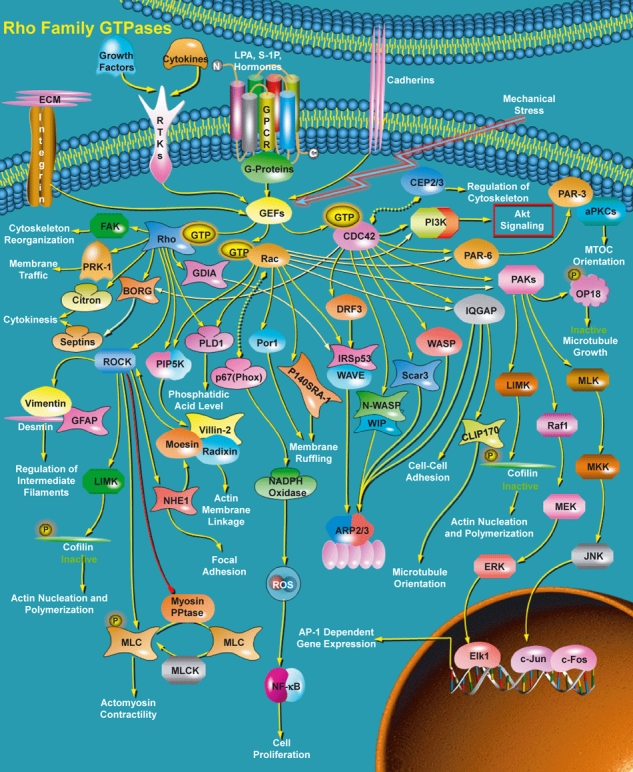
A diagram of the Rho-GTPase network [see text for details; with permission from http://www.sabiosciences.com/pathway.php?sn=Rho_family_GTPase (a Qiagen company)].
We know far less about the process networks activated by the GRNs that produce changes in differentiation and initiate morphogenetic dynamics (Fig. 1). An important exception is the rho-GTPase pathway (Fig. 3, Patwari & Lee, 2008). This network, activated by a wide range of signals, involves rho family members whose activation regulates, through GTP phosphorylation, many of the actin-based morphogenetic properties and cellular architecture features (e.g. migration, traction, folding, convergent extension) of the cell. The rho group of proteins has three subfamilies: there are three rho members, three rac members and two CDC42 members. These activate five, seven and seven subpathways, respectively (and there are two feedback pathways); these in turn mobilize a wide range of processes that mainly involve movement, but include a link to the mitosis pathway. These processes are shown as phrases at the end of pathways in Fig. 3. It is, however, worth noting that this figure is a diagram and not a mathematical graph: there is no annotation of the links between the interacting molecules.
Although elucidating the principal components of this complex network (> 60 proteins) has been a triumph of molecular cell biology, it is still unclear how one or more of the final processes or subpaths in the networks is activated while others are kept silent. In addition, any formal (e.g. differential equation) analysis of the dynamics of the network, either qualitatively or quantitatively is still not feasible because such equations cannot be solved as we lack detailed knowledge of the molecular interactions and the numerical rate constants.
In brief, any description of the morphogenesis of a particular tissue has to include the molecular, histological, dynamic and geometric properties of the system – and it is only rarely that we have all of this information.
Graphs
Although we can describe any example of morphogenesis in prose, such a description will be neither simple nor short. Perhaps the key point in this paper is that the mathematical graph (not to be confused with a data graph) can provide a format for describing morphogenesis that is compact, computationally tractable and hence directly linkable to online resources (e.g. literature and gene-expression databases). These graphs are essentially nets of linked triples of the general form:
One advantage of this general formulation is that there is a large body of mathematical theory that can, in principle, be used to help analyse complex graphs (e.g. Junker & Schreiber, 2008). Another is that such triples form the basis of resource description framework (RDF) descriptions, a standard way of modelling information for the semantic web (http://infomesh.net/2001/swintro/), a formalism that has its own benefits (see below). In formal terms, a graph is a set of vertices (or nodes, terms) and edges (or arcs, relationships) where an edge is connected to two vertices and has the form:
with a node being allowed to have more than one link
Informally, and for the cases considered here, a graph triple can be seen as two noun phrases linked by a verb phrase. A simple example is given by the femur, which has two obvious relationships: [femur] <is part of> [leg skeleton] and [femur] <is a> [endochondral bone], and these in turn link to more general terms: thus [leg skeleton] <is part of> [skeleton] and [endochondral bone] <is a> [bone]. Triples thus represent simple facts and sets of connected triples naturally form hierarchies and networks that may well include alternate paths and feedback loops.
The key feature of a triple is the linking relationship and this may be one of two types, directed and undirected. Directed relationships (the direction is indicated by an arrowhead) imply that the relationship is not reciprocal and so indicates a one-way path. Well-known directed relationships in biological graphs include <part of>, <is a>, and <descends from>, with the lineage relationship highlighting the fact that directed relationships can also impose a temporal direction. Undirected relationships imply a symmetric relationship and the convention is that they carry arrowheads at both ends; well known undirected relationships in biological graphs include <next to> and <interacts with>; an obvious undirected relationship in morphogenesis is <forms boundary with>. Undirected relationships allow closed loops or cycles in a graph: an obvious example of this is the map of stations on the London Underground: the triple is of the form [station i] <is next to> [station j] and allows the Circle Line to be a loop. In contrast, directed relationships do not allow loops; a graph with only directed relationships is known as a directed acyclic graph, or DAG.
Graphs have an additional feature that makes them particularly appropriate for formalising biological knowledge. As a triple is essentially a fact, it can be annotated with, for example, a publication reference (e.g. a Pubmed ID). Similarly, nodes can be annotated with associated information: thus an anatomical node can be annotated with IDs of its cell types, tissue type (e.g. endochondral as opposed to membrane bone), tissue name (from an anatomical ontology available from the OBO library, http://obolibrary.org/, see below) and gene-expression data (see http://www.informatics.jax.org/expression.shtml for mouse expression data). A protein can be annotated with Uniprot information, and a molecular network can be linked to a diagram. In addition, edges can be annotated with rate constants and other numbers (Alon, 2007). An annotated graph is a format that allows a lot of information to be collated in an extremely terse way and is essentially a review of the literature.
Because morphogenesis is complicated, and an end result involves many processes and interactions, the complete representation of an event may require hundreds of triples, if all the molecular data is included. Fortunately, one can often make the graph fine-grained where the detail is important and coarse where it is not. If, for instance, during a developmental process EGF activated the EGFR gene regulatory network (Fig. 3) which in turn activated a growth network that resulted in proliferation, and it was just the proliferation rather than the internal molecular interactions that was important, it would clearly be enough to represent it as:
without incorporating all the molecular details of the two networks into the graph, even though this might well be possible. In addition this coarseness of granularity can be used to hide unknown molecular detail, and this is allowable if that detail is not directly relevant.
Biological graphs
There are three obvious areas of biology where knowledge is represented by mathematical graphs, although this term is not usually used: clades, ontologies and molecular networks. The first and simplest example is the evolutionary clade where the nodes are species and the relationship is <evolves from>, a relationship that implies a temporal direction. Ontologies are more or less complex hierarchies that integrate knowledge about, for example, adult and developmental anatomy, cell types and genes (http://www.obofoundry.org). The best known of these is the Gene Ontology (http://www.geneontology.org), a large DAG which incorporates knowledge about the function and cellular location of genes, together with the processes in which they are involved; its relationships are <is a> and <part of>. Anatomical ontologies are hierarchies of tissues where the linking relationships may include <part of> and <is a>, as well as <starts at> and <ends at> if they cover developmental anatomy.
Ontologies are not meant to be stand-alone items but are intended to provide formal knowledge for use in databases (e.g. the mouse developmental anatomy ontology is linked to gene-expression data in GXD, while the gene ontology links knowledge about protein location, function and process involvement to its associated database of almost 5 × 105 gene products). Links are made with the unique identifiers, and every ontology term has such an ID, which is of the form abc:xyz. Here, abc represents the ontology name and xyz the number for a particular term (e.g. the notochord of the Theiler Stage 16 mouse embryo has ID = EMAP:0001675). One advantage of using standard ontology IDs in an online database is that a search using them can provide instant access to any associated data that is maintained online.
The third type of common biological graph is the molecular network. These extend from small groups of interacting chemicals to large protein networks (e.g. GRNs) and are normally viewed as diagrams with interactions between molecules indicated by simple links or arrows (Fig. 3). They are actually ill-defined graphs, with the nodes being molecules and the edges being one or another sort of interaction. If we had sufficient information about the nature of these interactions, we could give the edges more precise terms such as <binds to>, <activates> and <inhibits>, relationships that may also indicate a temporal direction. The use of such relationships in simple protein networks has been demonstrated by Alon (2007): he used graphing techniques with probability analysis to identify groups of up to five proteins that work cooperatively in bacteria and produce particular functions, such as positive feedback circuits, that are used in many contexts; such functions are known as network motifs.
Key to embedding molecular networks in graphs of, for example, developmental anatomy, is the realization that, for all their internal complexity, their output is one or more processes (Fig. 3); it is this fact that allows a complex network to be represented as a single node. Within a developmental context, such process networks (PN) require to be activated (Fig. 1), and this is the task of a GRN, which may do this by activating transcription factors that in turn direct the synthesis of some of the PN proteins. Thus, for a mesenchyme-to-epithelium transition to take place, two prior events are required: first, a GRN needs to be activated; this in turn activates the PN that will effect differentiation, and may also be responsible for synthesizing some of the PN proteins. For morphogenetic processes, many of the details of how the mechanics of change take place, once the new proteins are in place, are known (for review, see Davies, 2005; Patwari & Lee, 2008). Other than to point out that it is reasonable to assume that an important part of this process is free-energy-driven self-assembly of the components and their localization within the cell, these details will not be further considered here.
Modeling morphogenesis as a graph
There are three basic aspects to making a formal graph: first, all the data has to be collated and organized as triples; second, these triples have to be inspected to ensure that all the links between triples are in place so that the graph is properly formed; third, any annotations (e.g. ID links) have to be added. For the computational representation of the graph, this set of triples is a complete description. Biologists, however, require something more, that the graph be visually comprehensible. Producing the graphical representation of this complexity is much harder than just compiling a list of triples: the events take place at molecular, cellular and tissue levels, and all this information has to be made visually explicit.
Figure 4 shows a general graph of how tissue development is driven by intracellular molecular activity (a less detailed graph for two tissues is shown in Fig. 2) and constrained by the environment. Here, a tissue is used in the sense of a group of coherent cells with the same histological phenotype; hence it usually becomes possible to refer to molecular networks as being within tissues without any loss of precision. Although the molecular activity (green) takes place inside the cells of the tissues (blue), they have been separated for clarity. The nodes and links above the dotted line reflect differentiation, proliferation, apoptosis and any other tissue-autonomous events within the tissue, e.g. [sonic hedgehog] <activates> [the ssh pathway]; [the rho-GTPase pathway] <drives> [migration].
Fig. 4.

A graph showing the processes involved in the morphogenesis of a single tissue. The nodes and processes above the blue dotted line describe tissue-based events (differentiation, growth, movement, etc). The nodes and processes below the dotted line reflect the effects of tissues and matrix external to that tissue and are usually required for describing morphogenesis. ECM, extracellular matrix.
The nodes below the dotted line illustrate how geometric features in the system affect morphogenesis, e.g. [collagen fibrils] <constrain> [cell migration], [swelling of extracellular matrix] <causes> [expansion]. One immediate advantage of this format is that it makes visually explicit the extent to which causality in morphogenesis is distributed across the molecular, cellular and geometric properties of the system. As Noble (2008) has put it in his discussion of causality, ‘there is no preferred level’.
A case study
While relatively little is known about the molecular details of most morphogenetic events, we have many of the details about how new blood capillaries sprout off existing ones to provide nearby tissues with a blood supply (for review see Karamysheva, 2008), and we can use this as an example of how such events can be represented graphically.
The process is as follows: the eventual target tissue secretes members of the VEGF family (grouped here for simplicity) that diffuse away to form a local concentration gradient. Endothelial cells on a nearby capillary are activated by VEGF and one of them becomes a tip cell, which blocks its neighbours (via notch-delta signaling) from differentiating in this way (Suchting et al. 2008). Instead, they proliferate and, with the tip cell leading, form a sprout that migrates up the VEGF gradient to invade the original target tissue. Migration is facilitated by two additional processes: tip cells secrete proteases that loosen local tissue, while the endothelial cells that will form the sprout break the focal adhesions that originally stabilized them.
The essential features of capillary sprouting can be represented graphically (Fig. 5) with nodes representing tissues, molecules and networks and edges representing processes (indicated by an arrowhead for a directed action, or a blocking symbol for the delta network). For simplicity, and because the additional information is not required here, the molecular details of the networks are not shown. In formal terms, the graph is composed of ∼ 20 triples (those that localize networks to tissues are not detailed but are represented visually by locating the network nodes within the boxes of the tissue nodes). Two additional features make the visual representation of the graph easy to follow: first, different colours are used for tissues (mid- and light blue), molecular components (green) and processes (yellow); and secondly, molecular networks are shown as located within tissues while VEGF signal is in the intervening space. This convention not only maintains a sense of tissue geometry but helps keep the graph compact.
Fig. 5.
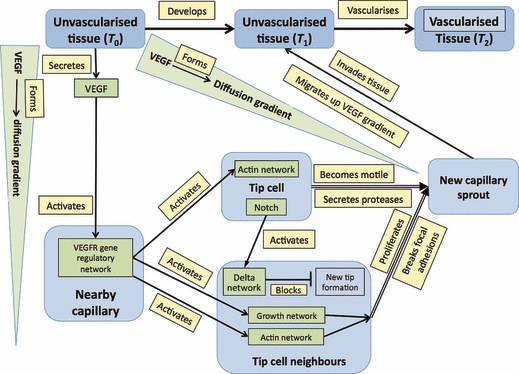
A graph showing the stages in the process of the vascularization of a tissue (blue boxes) from a nearby capillary that produces a sprout composed of a tip cell and its neighbours (light blue boxes). At the molecular level (green boxes) the tissue secretes VEGF that activates networks in nearby endothelial cells. The processes that drive morphogenetic change are in yellow boxes. Networks are shown within tissues (unlike in Figs 1 and 2) to keep the diagram compact.
As mentioned earlier, nodes, processes and triples can be annotated with identifiers that point to external sources such as databases and ontologies. For the graph of capillary formation, many such links are immediately available: Karamysheva (2007) and Suchting et al. (2007) have Pubmed IDs of PMID:18707583 and PMID:17296941, respectively; all proteins have Uniprot IDs, endothelial cells have a cell-type ontology ID of CL:0000071; while the adult mouse capillary system has an ID of MA:0000711 that currently links to a database entry of 10 expressed genes (http://www.informatics.jax.org/expression.shtml). Showing this information in the visualized graph would clutter it unreasonably, but the links can all be readily included if the graph is maintained as a computer file using standard tools (http://java-source.net/open-source/rss-rdf-tools), or as an online diagram where terms are annotated with hyperlinks. Indeed, if every term in a graph had its own ID, the graph would meet current computing requirements for a semantic web application ((http://infomesh.net/2001/swintro/).
Discussion
The purpose of this paper is to suggest that the mathematical graph, widely used outside of development, provides a useful framework for describing our knowledge about a specific morphogenetic or other developmental event. Making such a graph requires an understanding not only of the events at the molecular and cellular level in the tissues undergoing morphogenesis, but also of how features in the local environment constrain the geometry of their development. The key step is to turn this knowledge into small facts each of which can be structured as a triple and that together form a graph describing our knowledge of the phenomenon.
Such graphs can be represented in both computational and visual formats and there are advantages to both. Although less attention has been paid to the former here, it simply requires articulating the developmental information as a set of connected triples annotated with identifiers that will link it to online resources. It is likely that this format will become increasingly important as links to external resources that are regularly updated enable the graph as a whole to keep abreast of new data without additional effort. To help with this representation, there are a range of computational tools for forming, checking, visualizing, and analysing graphs (e.g. http://www.babelgraph.org/links.html).
Of more use to the biologist is the visualized format and it should be emphasized that this is more than an informal diagram. A key requirement here is that the graph be organized in a way that makes it coherent and parsimonious. Doing this is not easy and the careful analysis required for the exercise often reveals gaps in understanding that are not apparent when one merely lists the triples. Filling these gaps always requires further analysis, and often leads to new experimentation, the core purpose of theory. In fact, it is probably impossible to make the full list of triples and associated annotations until the visualization has been done properly!
Modular morphogenesis
The graphical approach captures something of the urgency of embryogenesis because it makes explicit something that is normally implicit: development proceeds because dynamic processes, the output of molecular networks, drive change. These processes extend from molecular interactions at the network level (protein interactions), through the outputs of these networks, e.g. activation of a further network or making a set of new proteins, to major developmental changes (differentiation and morphogenetic processes) and even to the production of new tissues. In the wider context, it is worth emphasizing that these higher-level processes are used many times during development. During the course of, for example, vertebrate embryogenesis, processes such as the various types of differentiation, cell migration, mesenchymal condensation, epithelial folding and apoptosis occur again and again. At a higher level, there are many structures that are repeatedly generated: this is most obvious in the musculo-skeletal system which includes inter alia∼ 100 long bones, the set of vertebrae, ∼ 200 synovial joints and countless links between bone and tendons or ligaments. Other examples include somites (and their immediate derivatives), teeth, hairs, ganglia and blood capillaries.
Davies (2005) has described such repetitions as reflecting modular development and discussed how they might work at the level of the phenotype. It is a reasonable guess that, although the repetitions may not be exact (each vertebra and each tooth has its own detailed morphology), the production of each module can be viewed as the action of a defined set of events. In these, specific groups of cells in specific environments initially undergo an exactly determined series of events to produce early structures that may later be modified by local patterning and growth mechanisms. Such modules are important because they indicate in ways that we do not yet understand, how spatio-temporal information can be encoded in the genome.
There has been discussion in the literature as to whether the genome should be viewed as the computational equivalent of a computer program that directs developmental change or of a database resource to be accessed on demand as required by one or another part of the developing organism (e.g. Werner, 2007; Noble, 2008). Given that developmental processes at both the cellular and tissue levels are often repeated and clearly involve the same networks and outputs, albeit that later growth may be locally specified, it probably makes sense to view such processes as neither a program nor a database, but as subroutines that can be called up on demand.
Graphs of these modules do not of course indicate where or how these sub-routines are located in the genome. The breaking down of the modules into sets of triples is a first step in indicating the separate facets of that module. Such work, combined with knowledge of the relevant process proteins and the location of their underlying sequences in the genome may help unpick how morphogenesis is genetically regulated.
Acknowledgments
I thank Denis Noble, Tom Melham and Eric Werner for discussions, Youichirou Ninomiya for sending me the Doi paper, Gillian Morriss-Kay for commenting on the manuscript and an anonymous referee for helpful criticisms.
References
- Alon U. An Introduction to Systems Biology. Boca Raton: Chapman & Hall/CRC; 2007. [Google Scholar]
- Bard JBL. A model generating aspects of zebra and other mammalian coat patterns. J Theor Biol. 1981;93:363–385. doi: 10.1016/0022-5193(81)90109-0. [DOI] [PubMed] [Google Scholar]
- Bard J. Morphogenesis: the Cellular and Molecular Processes of Developmental Anatomy. 2nd edn. Cambridge: Cambridge University Press; 1992. [Google Scholar]
- Bard JBL, Ross ASA. The morphogenesis of the ciliary body of the avian eye. II: Differential enlargement causes an epithelium to buckle. Dev Biol. 1982;92:87–96. doi: 10.1016/0012-1606(82)90153-1. [DOI] [PubMed] [Google Scholar]
- Davies JA. Mechanisms of Morphogenesis. San Diego: Elsevier Academic Press; 2005. [Google Scholar]
- Doi H. Graph-theoretical analysis of cleavage pattern: graph developmental system and its application to cleavage pattern of ascidian egg. Develop Growth Differ. 1984;26:49–60. doi: 10.1111/j.1440-169X.1984.00049.x. [DOI] [PubMed] [Google Scholar]
- Gilbert S. Developmental Biology. 10th edn. Sunderland: Sinauer Press; 2010. [Google Scholar]
- Goldbeter A, Pourquié O. Modeling the segmentation clock as a network of coupled oscillations in the Notch, Wnt and FGF signaling pathways. J Theor Biol. 2008;252:574–585. doi: 10.1016/j.jtbi.2008.01.006. [DOI] [PubMed] [Google Scholar]
- Hove JR, Köster RW, Forouhar AS, et al. Intracardiac fluid forces are an essential epigenetic factor for embryonic cardiogenesis. Nature. 2003;421:172–177. doi: 10.1038/nature01282. [DOI] [PubMed] [Google Scholar]
- Junker BH, Schreiber F. Analysis of Biological Networks. Hoboken: John Wiley & Sons, Inc; 2008. [Google Scholar]
- Karamysheva AF. Mechanisms of angiogenesis. Biochemistry (Mosc) 2008;73:751–762. doi: 10.1134/s0006297908070031. [DOI] [PubMed] [Google Scholar]
- Marée AFN, Hogeweg P. How amoeboids self-organize into a fruiting body: multicellular coordination in Dictyostelium discoideum. Proc Natl Acad Sci U S A. 2001;98:3879–3883. doi: 10.1073/pnas.061535198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino R, Rodriguez-Leon J, Macias D, et al. The BMP antagonist Gremlin regulates outgrowth, chondrogenesis and programmed cell death in the developing limb. Development. 1999;126:5515–5522. doi: 10.1242/dev.126.23.5515. [DOI] [PubMed] [Google Scholar]
- Murray JD. On pattern formation mechanisms for lepidopteran wing patterns and mammalian coat markings. Phil Trans R Soc Lond. 1981;295:473–496. doi: 10.1098/rstb.1981.0155. [DOI] [PubMed] [Google Scholar]
- Nakatsuji N, Johnson KE. Experimental manipulation of a contact guidance system in amphibian gastrulation by mechanical tension. Nature. 1984;307:453–455. doi: 10.1038/307453a0. [DOI] [PubMed] [Google Scholar]
- Newman SA, Bhat R. Dynamical patterning modules: a ‘pattern language’ for development and evolution of multicellular form. Int J Dev Biol. 2009;53:693–705. doi: 10.1387/ijdb.072481sn. [DOI] [PubMed] [Google Scholar]
- Noble D. Genes and causation. Philos Trans R Soc. 2008;366:3001–3015. doi: 10.1098/rsta.2008.0086. [DOI] [PubMed] [Google Scholar]
- Ovchinnikov DA, Selever J, Wang Y, et al. BMP receptor type IA in limb bud mesenchyme regulates distal outgrowth and patterning. Dev Biol. 2006;295:103–115. doi: 10.1016/j.ydbio.2006.03.013. [DOI] [PubMed] [Google Scholar]
- Painter KJ. Continuous models for cell migration in tissues and applications to cell sorting via differential chemotaxis. Bull Math Biol. 2009;71:1117–1147. doi: 10.1007/s11538-009-9396-8. [DOI] [PubMed] [Google Scholar]
- Patwari P, Lee RT. Mechanical control of tissue morphogenesis. Circ Res. 2008;103:234–243. doi: 10.1161/CIRCRESAHA.108.175331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro AS, Lloyd-Price J. SGN Sim, a stochastic genetic networks simulator. Bioinformatics. 2007;23:777–779. doi: 10.1093/bioinformatics/btm004. [DOI] [PubMed] [Google Scholar]
- Salazar-Ciudad I, Jernvall J. A computational model of teeth and the developmental origins of morphological variation. Nature. 2010;464:583–586. doi: 10.1038/nature08838. [DOI] [PubMed] [Google Scholar]
- Suchting S, Freitas C, le Noble F, et al. The Notch ligand Delta-like 4 negatively regulates endothelial tip cell formation and vessel branching. Proc Natl Acad Sci. 2007;104:3225–3230. doi: 10.1073/pnas.0611177104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trinkaus JP. Cells into Organs. The Forces that Shape the Embryo. Englewood Cliffs, NJ: Prentice-Hall; 1983. [Google Scholar]
- Turing AM. The chemical theory of morphogenesis. Philos Trans R Soc Lond B Biol Sci. 1952;237B:37–72. [Google Scholar]
- Werner E. How central is the genome? Science. 2007;317:753–754. [Google Scholar]
- Zeisberg M, Shah AA, Kalluri R. Bone morphogenic protein-7 induces mesenchymal to epithelial transition in adult renal fibroblasts and facilitates regeneration of injured kidney. J Biol Chem. 2005;280:8094–8100. doi: 10.1074/jbc.M413102200. [DOI] [PubMed] [Google Scholar]


