Abstract
Epidemiological data and animal models indicate that Helicobacter pylori and dietary NaCl have a synergistic ill effect on gastric maladies. Here we show that the Ferric Uptake Regulator (Fur), which is a crucial regulatory factor required for H. pylori colonization, is essential for growth in the presence of high NaCl concentrations. Moreover, we demonstrate that the transcriptional response induced by sodium chloride stress exhibits similarities to that seen under iron depletion.
Helicobacter pylori can be found in the gastric mucosa of more than half of the human population (Pounder and Ng 1995). Long-term colonization results in gastritis and may lead to gastric or duodenal ulcers (Malaty 2007). Infection with H. pylori is also a risk factor for development of gastric cancer (Matysiak-Budnik and Megraud 2006; Mbulaiteye et al. 2009); to date, H. pylori is the only bacterium classified as a carcinogen (Anon. 1994). For colonized individuals, epidemiological data indicate a synergistic ill effect between diets rich in sodium chloride and H. pylori infection (Tsugane and Sasazuki 2007; Wang et al. 2009). The role of a high salt diet on H. pylori infection was also previously examined in several animal models. In a mouse model of infection, an increase in bacterial load was observed in animals consuming a high salt diet (Rogers et al. 2005). Whereas, in a gastric chemical carcinogenesis model of H. pylori-infected Mongolian gerbils, a high salt diet resulted in the dose-dependent promotion of cancer initiation that was accompanied by a shift in mucin production from the glandular to the surface mucous cells (Kato et al. 2006). Taken together, the epidemiological and animal based evidence suggest dietary mediated increases in the severity of H. pylori-induced disease outcomes.
Based on the above observations, we and others postulated and subsequently showed that salt concentration contributes to changes in expression of H. pylori virulence factors such as cagA (Loh et al. 2007) and vacA (Gancz et al. 2008) in vitro. However, the mechanisms of regulation of the salt stress response and the factors required for salt survival are not yet characterized for this organism.
In order to identify genes that contribute to H. pylori’s ability to withstand salt stress, we took a targeted approach and screened defined H. pylori mutants that were constructed in the G27 strain background and already existed in our strain collection (Table 1). Specifically, we targeted strains containing mutations in known regulatory genes (flgR, fur, luxS, spoT) or known virulence genes (cagA, ureA, vacA). All H. pylori strains were maintained as frozen stocks at 80°C as previously described (Carpenter et al. 2010). Prior to each experiment, strains were revived on horse blood agar (HBA) plates as previously described (Carpenter et al. 2010) and used to inoculate liquid cultures that consisted of Brucella broth (Neogen Corporation) supplemented with 10% fetal bovine serum and 10µg/ml vancomycin (BB). These cultures were grown at 37°C with shaking at 100 rpm. Both liquid and plate cultures were grown under microaerobic conditions (5% O2, 10% CO2, and 85% N2) generated with an Anoxomat gas evacuation and replacement system (Spiral Biotech) in gas evacuation jars. When reviving mutant bacterial strains from the freezer only, plates were supplemented with 8µg/ml chloramphenicol (Cm) (EMD Chemicals, Inc.) and/or 25µg/ml kanamycin (Kan) (Gibco) where appropriate (Table 1).
TABLE 1.
Plasmids and strains used in this study
| Plasmid or strain | Description | Reference |
|---|---|---|
| Plasmids | ||
| pDSM142 | pΔHP1027-K7 | (Gancz et al. 2006) |
| pDSM340 | fur, pTM117 complementation vector | (Carpenter et al. 2007) |
| H. pylori strains | ||
| G27 | WT H. pylori | (Covacci et al. 1993) |
| 26695 | WT H. pylori | (Tomb et al. 1997) |
| 43504 | WT H. pylori | (Alamuri et al. 2006) |
| DSM43 | G27 ureB::aphA, Kanr | (Joyce et al. 2001) |
| DSM90 | G27 flgR::cat, Cmr | This study |
| DSM205 | G27 vacA::aphA, Kanr | (Amieva et al. 2002) |
| DSM207 | G27 cagA::cat, Cmr | (Amieva et al. 2002) |
| DSM211 | G27 luxS::aphA, Kanr | Kind gift from J.V. Solnick laboratory |
| DSM300 | G27 Δfur::cat, Cmr | (Carpenter et al. 2007) |
| DSM343 | DSM300 (pDSM340), Kanr Cmr | (Carpenter et al. 2007) |
| DSM357 | 26695 Δfur::cat, Cmr | (Carpenter et al. 2009) |
| DSM388 | 43504 Δfur::aphA, Kanr | This study |
| DSM594 | G27 spoT::aphA, Kanr | (Zhou et al. 2008) |
To determine whether mutation of any of the H. pylori regulatory or virulence genes affected growth in the presence of elevated salt concentrations, we calculated the plating efficiency of each mutant strain and the wildtype G27 control strain by determining the number of colonies obtained after spotting on solidified (1.3% agar final) BB supplemented with various concentrations of NaCl. The normal solidified BB medium contains 5 gr/liter of NaCl (BB5), and we achieved increased salt concentration by use of BB containing 13.5 gr/liter of NaCl (BB13.5). This elevated concentration of sodium chloride is lower than the dramatic growth inhibitory condition we previously described for this strain (BB15) (Gancz et al. 2008). The wildtype strain and each of the defined isogenic mutant strains were grown to mid-exponential phase in liquid media and then O.D. equilibrated to give identical cell densities. Cells were then serially diluted and plated side-by-side on BB5 and BB13.5 plates (Figure 1) as well as on HBA plates (data not shown).
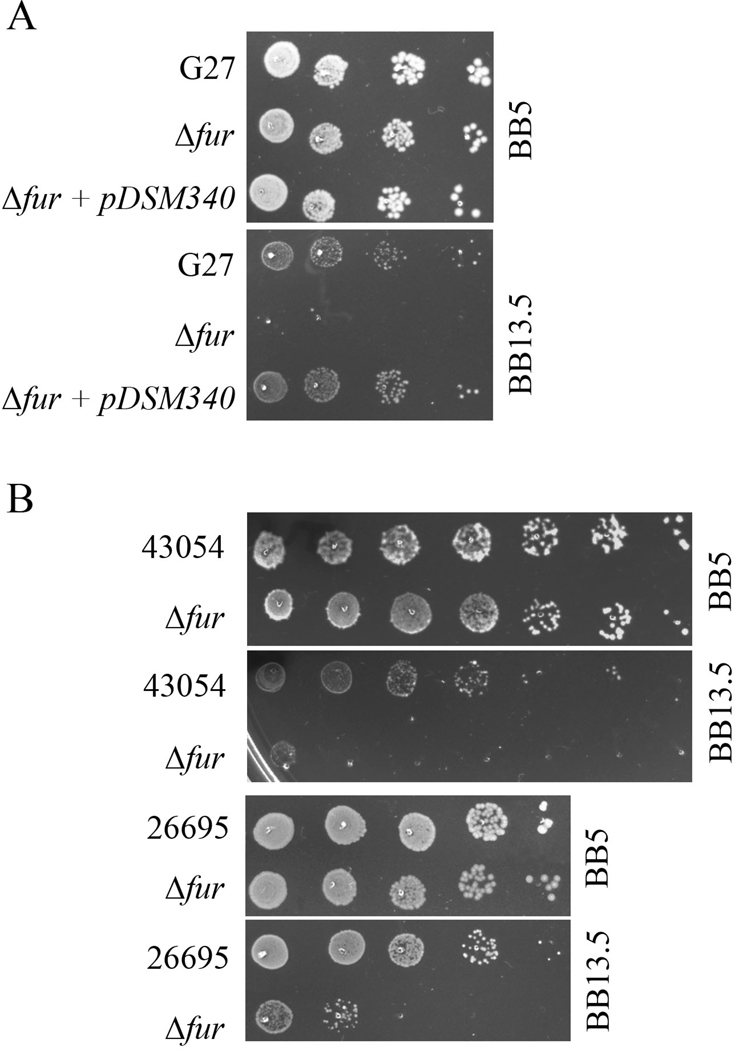
Figure 1.
Growth characteristics of H. pylori and isogenic Δfur mutants on normal (BB5) and elevated salt (BB13.5) solidified media. A. Relative growth of G27 wildtype, the isogenic Δfur mutant (DSM300), and the fur complemented strain (DSM343) on BB5 and BB13.5 plates are shown. Ten-fold dilutions of the indicated strain were spotted on solidified Brucella broth based medium containing 5 gr/liter NaCl (BB5) (upper panel) or 13.5 gr/liter NaCl (BB13.5) (lower panel). B. Relative growth of strains 43054 and 26695 and their isogenic Δfur mutant strains on BB5 and BB13.5 plates are shown. Ten-fold dilutions of the indicated strain were spotted on solidified Brucella broth based medium containing 5 gr/liter NaCl (BB5) (upper panel) or 13.5 gr/liter NaCl (BB13.5) (lower panel). The results shown are representative of multiple biological repeats.
Comparison of the plating data revealed that while wildtype G27 showed no decrease in plating efficiency on BB13.5, the resulting colonies were significantly smaller than on non-supplemented media (Figure 1). This fact suggests that the concentration of NaCl found in the BB13.5 plates was sufficient to create stressful conditions under which the bacteria can grow but at a slower rate. Disruption of cagA, flgR, luxS, ureB or vacA all resulted in plating efficiencies that were similar to those of the wildtype (data not shown). However, disruption of the ferric uptake regulator, fur, or spoT resulted in a significant reduction in the plating efficiency of these mutant strains on high salt media (Figure 1 panel A and data not shown). Given that the fur mutant strain showed the most drastic growth defect at elevated salt concentrations and because Fur is known to regulate additional stress responses besides the response to iron ion concentration (Bijlsma et al. 2002; Carpenter et al. 2010), has unique features in H. pylori gene regulation (Delany et al. 2001; Ernst et al. 2005; Miles et al. 2010a) and is the scope of on-going research in our laboratory (Carpenter et al. 2007; Carpenter et al. 2009; Carpenter et al. 2010; Gancz et al. 2006; Miles et al. 2010a; Miles et al. 2010b), we chose to further investigate the role of fur in the response to salt stress.
First, to conclusively demonstrate that the fur mutation was responsible for the demonstrated salt sensitive phenotype, we attempted to complement the sensitivity phenotype using pDSM340, which is a plasmid that carries a wildtype copy of fur (Carpenter et al. 2007). Transformation of this plasmid into the Δfur background resulted in complete restoration of growth to the wildtype phenotype (Figure 1, panel A). Therefore, we conclude that fur is essential for growth under these conditions.
Next, given that H. pylori is considered a panmictic species (Salaun et al. 1998) and since increasing evidence indicates the occurrence of strain specific phenomenon (Gancz et al. 2008), we next investigated the necessity of fur for growth under high salt conditions in two additional strain backgrounds. To this end, we compared the plating efficiency of strains HP26695 and HP43504 to that of their isogenic Δfur mutants, which were constructed as previously described (Carpenter et al. 2009; Gancz et al. 2006). We noticed that although the various wildtype strains differed in their overall salt sensitivity, each of the Δfur strains showed a growth defect in comparison to their respective parental strain (Figure 1, panel B). This indicates that the requirement of Fur for growth under salt stress conditions is not strain dependent. Similar to G27, the other H. pylori strains also exhibited smaller colonies on BB13.5, indicating that they were experiencing stressful conditions (Figure 2, panel B).
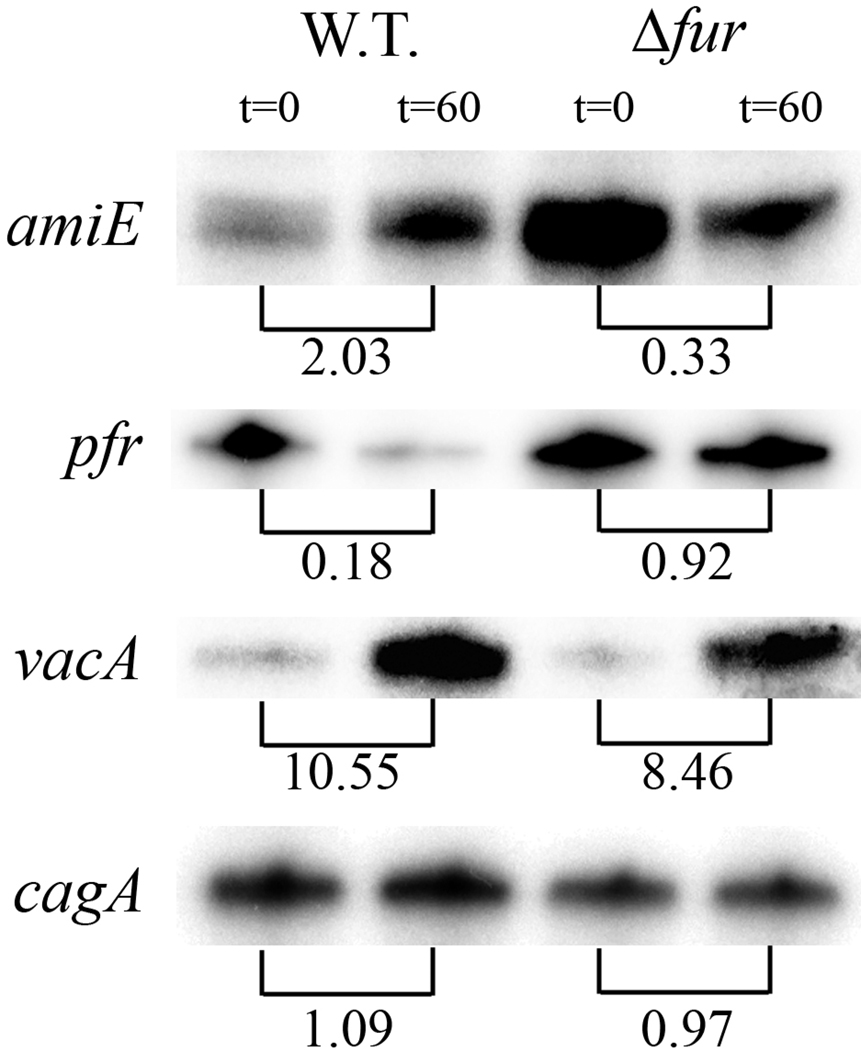
Figure 2.
Transcription of amiE, pfr, vacA and cagA following salt shock as assessed by RNAse protection assay (RPA). Logarithmically growing liquid cultures of G27 and the isogenic Δfur mutant strain were salt shocked by replacing the BB5 growth medium with BB25 medium (containing 25 gr/liter NaCl). Samples were taken immediately after the cells were resuspended in BB25 (t=0) and one hour after the change of the medium (t=60). RNA was harvested, and RPAs were conducted as described in the text. Protected bands for the indicated transcript are shown for each strain. The fold change in expression from the t=0 time point is indicated. These data are representative of three independent biological experiments.
Fur is known to be an important regulatory factor in H. pylori that has been shown to control the expression of a large number of genes (Danielli et al. 2006; Ernst et al. 2005; Gancz et al. 2006; Lee et al. 2004). Included among these genes is amiE, which encodes an aliphatic amidase that is regulated by the iron-bound form of Fur (van Vliet et al. 2003), and pfr, which encodes a nonheme iron-containing ferritin that is regulated by the apo form of Fur (Bereswill et al. 2000). Thus, to determine if Fur regulated processes were taking place during salt stress in H. pylori, we monitored the relative transcription of these two known Fur dependent genes by RNAse protection assay (RPA) as previously described (Carpenter et al. 2007). Additionally, we monitored the Fur-dependent expression of vacA and cagA, two virulence genes whose salt-dependent transcription we previously examined (Gancz et al. 2008). RPAs were conducted with RNA harvested from cultures of the wildtype strain and the isogenic fur mutant strain that were grown in BB5 and then exposed to a salt shock for one hour. To accomplish this, overnight cultures of each strain were harvested by centrifugation and then exposed to salt shock conditions by resuspension of the cells in BB containing 25 gr/liter of NaCl (BB25). RNA was extracted from bacterial cells, the amiE, pfr, cagA and vacA riboprobe templates were generated by PCR, radiolabeled riboprobes were produced, and the RPAs were performed all as previously described (Carpenter et al. 2010; Gancz et al. 2008). The gels were subsequently exposed to phosphor screens, which were scanned using a FLA-5100 scanner (FujiFilm). Data were then analyzed/quantitated using the Multi-Gauge software (version 3.0, FujiFilm).
Examination of the response of the virulence factors vacA and cagA to salt shock by RPAs demonstrated that, as expected for strain G27 (Gancz et al. 2008), upon exposure to increased salt, vacA expression was increased by 10.55 fold, while cagA expression was virtually unchanged (Figure 2). These responses were almost identical in the fur mutant strain (Figure 2), indicating that Fur does not function as a critical regulatory factor for cagA and vacA in these conditions. Conversely, Fur does function as a critical regulator of amiE and pfr in these conditions; in the wildtype strain, amiE expression was increased upon salt shock, while pfr expression was decreased, but these changes were completely lost in the fur mutant strain (Figure 2). Interestingly, these changes in expression closely resemble the previously characterised transcriptional response of these genes to iron depletion (Carpenter et al. 2007; Carpenter et al. 2009). For example, amiE expression in response to salt stress increased 2.03 fold in comparison to 5.8 fold under iron depletion (Carpenter et al. 2007), while the pfr transcript showed a 5.6 decrease under salt shock conditions and a 3.3 fold decrease under iron depletion (Carpenter et al. 2007). Taken together, these data suggest that for H. pylori, salt shock mimics iron depletion and that Fur plays a crucial role in mediating changes in gene expression that allow growth under these conditions.
The involvement of Fur in the response to high concentrations of salt was previously also suggested for other microorganisms: Desulfovibrio vulgaris (Bender et al. 2007; He et al. 2010; Mukhopadhyay et al. 2006), the halophilic bacterium Chromohalobacter salexigens (Argandona et al. 2010), Bacillus subtilis (Hahne et al. 2010; Hoffmann et al. 2002; Steil et al. 2003), and Bacillus cereus (den Besten et al. 2009). For D. vulgaris, microarray analysis demonstrated that many of the genes showing marked changes in transcription following salt exposure belonged to the Fur regulon. Despite this, the wildtype D. vulgaris and the Δfur strain display virtually identical suppressed growth curves under elevated salt conditions. However, the fur mutant strain differs from the wildtype in that while growth of the wildtype in salt can be restored by the addition of glycine betaine as an osmoprotectant, the fur mutant strain is unable to be rescued from salt induced poor growth (Bender et al. 2007). In B. subtilis the connection between Fur and salt stress was inferred from proteomic examination of the response to high salt conditions (Hoffmann et al. 2002) and was further studied by comparative transcriptional profiling in response to iron depletion and salt shock (Hahne et al. 2010; Steil et al. 2003). Interestingly, in this organism addition of iron to the growth media partially reversed the B. subtilis growth defect induced by salt-stress (Hoffmann et al. 2002). Similarly, in the closely related bacterium, B. cereus, salt exposure causes induction of genes involved in iron homeostasis (den Besten et al. 2009). Thus, while it is clear that in H. pylori Fur acts as a global transcriptional regulator that responds to and or mediates the response to an assortment of environmental signals (Bijlsma et al. 2002; Carpenter et al. 2010), we suggest that the link between salt, iron and mediation of the osmotic stress response by Fur crosses bacterial taxa and may prove to be a general one.
Acknowledgments
We thank members of the Merrell lab for useful discussions and Jay Solnick for the G27 luxS mutant. Research in the laboratory of D. Scott Merrell is made possible by grants R073PW from USUHS and AI065529 from the NIAID. Contents of this manuscript are the sole responsibility of the authors and do not necessarily represent the official views of the NIH or the DOD.
REFERENCES
- 1.Schistosomes, liver flukes and Helicobacter pylori. IARC Working Group on the Evaluation of Carcinogenic Risks to Humans. Lyon. Lyon, 7–14 June 1994. IARC Monogr Eval.Carcinog.Risks Hum. 1994;61:1–241. [PMC free article] [PubMed] [Google Scholar]
- 2.Alamuri P, Mehta N, Burk A, Maier RJ. Regulation of the Helicobacter pylori Fe-S cluster synthesis protein NifS by iron, oxidative stress conditions, and fur. J.Bacteriol. 2006;188:5325–5330. doi: 10.1128/JB.00104-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Amieva MR, Salama NR, Tompkins LS, Falkow S. Helicobacter pylori enter and survive within multivesicular vacuoles of epithelial cells. Cell Microbiol. 2002;4:677–690. doi: 10.1046/j.1462-5822.2002.00222.x. [DOI] [PubMed] [Google Scholar]
- 4.Argandona M, Nieto JJ, Iglesias-Guerra F, Calderon MI, Garcia-Estepa R, Vargas C. Interplay between iron homeostasis and the osmotic stress response in the halophilic bacterium Chromohalobacter salexigens. Appl.Environ.Microbiol. 2010;76:3575–3589. doi: 10.1128/AEM.03136-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bender KS, Yen HC, Hemme CL, Yang Z, He Z, He Q, Zhou J, Huang KH, Alm EJ, Hazen TC, Arkin AP, Wall JD. Analysis of a ferric uptake regulator (Fur) mutant of Desulfovibrio vulgaris Hildenborough. Appl.Environ.Microbiol. 2007;73:5389–5400. doi: 10.1128/AEM.00276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bereswill S, Greiner S, van Vliet AH, Waidner B, Fassbinder F, Schiltz E, Kusters JG, Kist M. Regulation of ferritin-mediated cytoplasmic iron storage by the ferric uptake regulator homolog (Fur) of Helicobacter pylori. J.Bacteriol. 2000;182:5948–5953. doi: 10.1128/jb.182.21.5948-5953.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bijlsma JJ, Waidner B, Vliet AH, Hughes NJ, Hag S, Bereswill S, Kelly DJ, Vandenbroucke-Grauls CM, Kist M, Kusters JG. The Helicobacter pylori homologue of the ferric uptake regulator is involved in acid resistance. Infect.Immun. 2002;70:606–611. doi: 10.1128/iai.70.2.606-611.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carpenter BM, Gancz H, Benoit SL, Evans S, Olsen CH, Michel SL, Maier RJ, Merrell DS. Mutagenesis of Conserved Amino Acids of Helicobacter pylori Fur Reveals Residues Important for Function. J.Bacteriol. 2010;192:5037–5052. doi: 10.1128/JB.00198-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carpenter BM, Gancz H, Gonzalez-Nieves RP, West AL, Whitmire JM, Michel SL, Merrell DS. A single nucleotide change affects fur-dependent regulation of sodB in H. pylori. PLoS.One. 2009;4:e5369. doi: 10.1371/journal.pone.0005369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Carpenter BM, McDaniel TK, Whitmire JM, Gancz H, Guidotti S, Censini S, Merrell DS. Expanding the Helicobacter pylori genetic toolbox, modification of an endogenous plasmid for use as a transcriptional reporter and complementation vector. Appl.Environ.Microbiol. 2007;73:7506–7514. doi: 10.1128/AEM.01084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Covacci A, Censini S, Bugnoli M, Petracca R, Burroni D, Macchia G, Massone A, Papini E, Xiang Z, Figura N. Molecular characterization of the 128-kDa immunodominant antigen of Helicobacter pylori associated with cytotoxicity and duodenal ulcer. Proc.Natl.Acad.Sci.U.S.A. 1993;90:5791–5795. doi: 10.1073/pnas.90.12.5791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Danielli A, Roncarati D, Delany I, Chiarini V, Rappuoli R, Scarlato V. In vivo dissection of the Helicobacter pylori Fur regulatory circuit by genome-wide location analysis. J.Bacteriol. 2006;188:4654–4662. doi: 10.1128/JB.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Delany I, Spohn G, Rappuoli R, Scarlato V. The Fur repressor controls transcription of iron-activated and -repressed genes in Helicobacter pylori. Mol.Microbiol. 2001;42:1297–1309. doi: 10.1046/j.1365-2958.2001.02696.x. [DOI] [PubMed] [Google Scholar]
- 14.den Besten HM, Mols M, Moezelaar R, Zwietering MH, Abee T. Phenotypic and transcriptomic analyses of mildly and severely salt-stressed Bacillus cereus ATCC 14579 cells. Appl.Environ.Microbiol. 2009;75:4111–4119. doi: 10.1128/AEM.02891-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ernst FD, Bereswill S, Waidner B, Stoof J, Mader U, Kusters JG, Kuipers EJ, Kist M, van Vliet AH, Homuth G. Transcriptional profiling of Helicobacter pylori Fur- and iron-regulated gene expression. Microbiology. 2005;151:533–546. doi: 10.1099/mic.0.27404-0. [DOI] [PubMed] [Google Scholar]
- 16.Ernst FD, Homuth G, Stoof J, Mader U, Waidner B, Kuipers EJ, Kist M, Kusters JG, Bereswill S, van Vliet AH. Iron-responsive regulation of the Helicobacter pylori iron-cofactored superoxide dismutase SodB is mediated by Fur. J.Bacteriol. 2005;187:3687–3692. doi: 10.1128/JB.187.11.3687-3692.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gancz H, Censini S, Merrell DS. Iron and pH homeostasis intersect at the level of Fur regulation in the gastric pathogen Helicobacter pylori. Infect.Immun. 2006;74:602–614. doi: 10.1128/IAI.74.1.602-614.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gancz H, Jones KR, Merrell DS. Sodium chloride affects Helicobacter pylori growth and gene expression. J.Bacteriol. 2008;190:4100–4105. doi: 10.1128/JB.01728-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hahne H, Mader U, Otto A, Bonn F, Steil L, Bremer E, Hecker M, Becher D. A comprehensive proteomics and transcriptomics analysis of Bacillus subtilis salt stress adaptation. J.Bacteriol. 2010;192:870–882. doi: 10.1128/JB.01106-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.He Z, Zhou A, Baidoo E, He Q, Joachimiak MP, Benke P, Phan R, Mukhopadhyay A, Hemme CL, Huang K, Alm EJ, Fields MW, Wall J, Stahl D, Hazen TC, Keasling JD, Arkin AP, Zhou J. Global transcriptional, physiological, and metabolite analyses of the responses of Desulfovibrio vulgaris hildenborough to salt adaptation. Appl.Environ.Microbiol. 2010;76:1574–1586. doi: 10.1128/AEM.02141-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffmann T, Schutz A, Brosius M, Volker A, Volker U, Bremer E. High-salinity-induced iron limitation in Bacillus subtilis. J.Bacteriol. 2002;184:718–727. doi: 10.1128/JB.184.3.718-727.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Joyce EA, Gilbert JV, Eaton KA, Plaut A, Wright A. Differential gene expression from two transcriptional units in the cag pathogenicity island of Helicobacter pylori. Infect.Immun. 2001;69:4202–4209. doi: 10.1128/IAI.69.7.4202-4209.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kato S, Tsukamoto T, Mizoshita T, Tanaka H, Kumagai T, Ota H, Katsuyama T, Asaka M, Tatematsu M. High salt diets dose-dependently promote gastric chemical carcinogenesis in Helicobacter pylori-infected Mongolian gerbils associated with a shift in mucin production from glandular to surface mucous cells. Int.J.Cancer. 2006;119:1558–1566. doi: 10.1002/ijc.21810. [DOI] [PubMed] [Google Scholar]
- 24.Lee HW, Choe YH, Kim DK, Jung SY, Lee NG. Proteomic analysis of a ferric uptake regulator mutant of Helicobacter pylori, regulation of Helicobacter pylori gene expression by ferric uptake regulator and iron. Proteomics. 2004;4:2014–2027. doi: 10.1002/pmic.200300740. [DOI] [PubMed] [Google Scholar]
- 25.Loh JT, Torres VJ, Cover TL. Regulation of Helicobacter pylori cagA expression in response to salt. Cancer Res. 2007;67:4709–4715. doi: 10.1158/0008-5472.CAN-06-4746. [DOI] [PubMed] [Google Scholar]
- 26.Malaty HM. Epidemiology of Helicobacter pylori infection. Best.Pract.Res.Clin.Gastroenterol. 2007;21:205–214. doi: 10.1016/j.bpg.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 27.Matysiak-Budnik T, Megraud F. Helicobacter pylori infection and gastric cancer. Eur.J.Cancer. 2006;42:708–716. doi: 10.1016/j.ejca.2006.01.020. [DOI] [PubMed] [Google Scholar]
- 28.Mbulaiteye SM, Hisada M, El-Omar EM. Helicobacter Pylori associated global gastric cancer burden. Front Biosci. 2009;14:1490–1504. doi: 10.2741/3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miles S, Carpenter BM, Gancz H, Merrel DS. Helicobacter pylori apo-Fur regulation appears unconserved across species. J.Microbiol. 2010;48:378–386. doi: 10.1007/s12275-010-0022-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Miles S, Piazuelo MB, Semino-Mora C, Washington MK, Dubois A, Peek RM, Jr, Correa P, Merrell DS. Detailed in vivo analysis of the role of Helicobacter pylori Fur in colonization and disease. Infect.Immun. 2010;78:3073–3082. doi: 10.1128/IAI.00190-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Mukhopadhyay A, He Z, Alm EJ, Arkin AP, Baidoo EE, Borglin SC, Chen W, Hazen TC, He Q, Holman HY, Huang K, Huang R, Joyner DC, Katz N, Keller M, Oeller P, Redding A, Sun J, Wall J, Wei J, Yang Z, Yen HC, Zhou J, Keasling JD. Salt stress in Desulfovibrio vulgaris Hildenborough, an integrated genomics approach. J.Bacteriol. 2006;188:4068–4078. doi: 10.1128/JB.01921-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pounder RE, Ng D. The prevalence of Helicobacter pylori infection in different countries. Aliment.Pharmacol.Ther. 1995;(9 Suppl 2):33–39. [PubMed] [Google Scholar]
- 33.Rogers AB, Taylor NS, Whary MT, Stefanich ED, Wang TC, Fox JG. Helicobacter pylori but not high salt induces gastric intraepithelial neoplasia in B6129 mice. Cancer Res. 2005;65:10709–10715. doi: 10.1158/0008-5472.CAN-05-1846. [DOI] [PubMed] [Google Scholar]
- 34.Salaun L, Audibert C, Le LG, Burucoa C, Fauchere JL, Picard B. Panmictic structure of Helicobacter pylori demonstrated by the comparative study of six genetic markers. FEMS Microbiol.Lett. 1998;161:231–239. doi: 10.1111/j.1574-6968.1998.tb12953.x. [DOI] [PubMed] [Google Scholar]
- 35.Steil L, Hoffmann T, Budde I, Volker U, Bremer E. Genome-wide transcriptional profiling analysis of adaptation of Bacillus subtilis to high salinity. J.Bacteriol. 2003;185:6358–6370. doi: 10.1128/JB.185.21.6358-6370.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tomb JF, White O, Kerlavage AR, Clayton RA, Sutton GG, Fleischmann RD, Ketchum KA, Klenk HP, Gill S, Dougherty BA, Nelson K, Quackenbush J, Zhou L, Kirkness EF, Peterson S, Loftus B, Richardson D, Dodson R, Khalak HG, Glodek A, McKenney K, Fitzegerald LM, Lee N, Adams MD, Hickey EK, Berg DE, Gocayne JD, Utterback TR, Peterson JD, Kelley JM, Cotton MD, Weidman JM, Fujii C, Bowman C, Watthey L, Wallin E, Hayes WS, Borodovsky M, Karp PD, Smith HO, Fraser CM, Venter JC. The complete genome sequence of the gastric pathogen Helicobacter pylori. Nature. 1997;388:539–547. doi: 10.1038/41483. [DOI] [PubMed] [Google Scholar]
- 37.Tsugane S, Sasazuki S. Diet and the risk of gastric cancer, review of epidemiological evidence. Gastric.Cancer. 2007;10:75–83. doi: 10.1007/s10120-007-0420-0. [DOI] [PubMed] [Google Scholar]
- 38.van Vliet AH, Stoof J, Poppelaars SW, Bereswill S, Homuth G, Kist M, Kuipers EJ, Kusters JG. Differential regulation of amidase- and formamidase-mediated ammonia production by the Helicobacter pylori fur repressor. J.Biol.Chem. 2003;278:9052–9057. doi: 10.1074/jbc.M207542200. [DOI] [PubMed] [Google Scholar]
- 39.Wang XQ, Terry PD, Yan H. Review of salt consumption and stomach cancer risk, epidemiological and biological evidence. World J.Gastroenterol. 2009;15:2204–2213. doi: 10.3748/wjg.15.2204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhou YN, Coleman WG, Jr, Yang Z, Yang Y, Hodgson N, Chen F, Jin DJ. Regulation of cell growth during serum starvation and bacterial survival in macrophages by the bifunctional enzyme SpoT in Helicobacter pylori. J.Bacteriol. 2008;190:8025–8032. doi: 10.1128/JB.01134-08. [DOI] [PMC free article] [PubMed] [Google Scholar]