Abstract
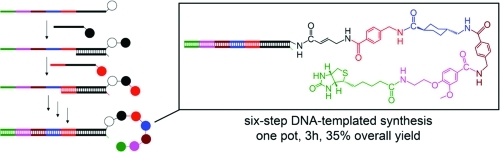
We developed a sequential strand-displacement strategy for multistep DNA-templated synthesis (DTS) and used it to mediate an efficient six-step DTS that proceeded in 35% overall yield (83% average yield per step). The efficiency of this approach and the fact that the final product remains linked to a DNA sequence that fully encodes its reaction history suggests its utility for the translation of DNA sequences into high-complexity synthetic libraries suitable for in vitro selection.
DNA-templated synthesis (DTS), the use of DNA hybridization to dramatically increase the effective molarity of reactants linked to oligonucleotides, is a powerful strategy to control chemical reactivity in a DNA sequence-programmed manner.1−4 Because the reaction products of DTS are encoded by the sequences of the associated DNA templates, they can be subjected to in vitro selection followed by PCR amplification and DNA sequence analysis to enable the discovery of functional small molecules,3,5−7 synthetic polymers,8−10 or novel chemical reactions.11−15 We recently reported the three-step DNA-templated synthesis of a 13,824-membered small-molecule macrocycle library.(16) The library was subjected to in vitro selection for binding affinity to a variety of proteins of biomedical interest, ultimately yielding a new class of macrocyclic kinase inhibitors.(6) Other complementary approaches to generating DNA-encoded libraries have led to the discovery of bioactive small molecules,17−29 including a number of examples in the pharmaceutical industry.5,30,31
Generating DNA-encoded small molecules of significant structural complexity requires multistep DNA-programmed or DNA-tagged synthesis.3,5,23,30,32−37 A number of strategies have been developed to enable multistep DTS. The simplest uses a DNA template strand containing several codons and reagents linked to complementary anticodon oligonucleotides that are added successively.3,32−34 While this approach is conceptually straightforward, it requires several manipulations after each step that increase required time and effort and can decrease overall yields. The relative geometry between reactants on the template and reagent strands also changes after each step in this approach, potentially altering reaction efficiencies.(38) More complex self-assembled DNA structures and devices can also mediate multistep DTS. For example, a DNA three-way junction that contains multiple reagents at the junction has been developed for the construction of DNA-encoded peptides.(5) We developed a DNA mechanical device that moves along a DNA track and mediates autonomous multistep organic synthesis in a single isothermal solution.(35) McKee et al. recently used a DNA strand-exchange strategy to achieve a three-step DTS in which products are swapped between new and old DNA strands with the assistance of a “remover strand” that displaces expended reagent oligonucleotides.(36)
Despite these significant advances and the diversity of approaches to generating multistep DTS products, all multistep DNA-templated small-molecules syntheses reported to date have used only three or fewer DNA-templated steps, and overall yields are generally low (typically <10%). Here we present a new strand-displacement strategy for multistep DTS and its use to mediate a six-step synthesis with an overall yield of 35% (average yield of 83%). By providing products of six-step DNA-programmed reaction sequences in good overall yield, the approach presented here may provide access to high-complexity DNA-templated small-molecule libraries.
Our strategy exploits “toehold displacement,” the known ability of a single-stranded DNA oligonucleotide (AB) to invade an asymmetric DNA duplex (A′B′:B) that contains a single-stranded hybridization site (A′) for the invading strand.(39) Once A:A′ hybridization takes place, base pairing with the invading strand continues, ultimately resulting in strand displacement of the shorter, and therefore less-favorably hybridized, B strand. Displacement results in the formation of a new Watson–Crick complex suitable for DTS. We hypothesized that this approach could represent a highly efficient and very simple way to access products of several consecutive DNA-programmed reactions while preserving the correspondence between DNA sequence and reaction product structure that is required for in vitro selection.
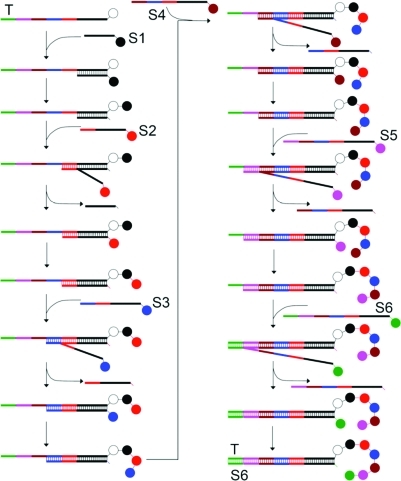
The application of this “toehold displacement” strategy to multistep DTS is summarized in Figure 1. A single-stranded DNA template (T) consists of a 16-base initial reaction site (black) followed by five consecutive 8-base coding segments (colored) that also serve as toeholds to initiate sequential DNA strand displacement. In the first step, substrate DNA S1, which is tethered to the first reactant, hybridizes to T, initiating DTS. If self-cleaving reagents are used, the first reactant group is transferred from S1 to T as a natural consequence of the DNA-templated reaction. After sufficient time to react has elapsed, the second substrate strand S2 is added without requiring product purification or any other manipulation. S2 partially hybridizes with T through the red toehold region and displaces S1 by forming a more stable DNA duplex due to the fact that the oligonucleotide in S2 is eight nucleotides longer than that in S1. As a result, S1 spontaneously leaves the Watson–Crick complex and the second reactant’s group is transferred to the T-linked intermediate through a DNA-templated reaction. Thereafter, an even longer substrate strand S3 is added to displace S2 and initiate a third reaction.
Figure 1.
Toehold displacement–mediated multistep DNA-templated synthesis. In each step, hybridization between the template and substrate oligonucleotides triggers a DNA-templated reaction. The template strand contains multiple toehold regions (colored) that allow successive docking of new substrates and DNA strand displacement of oligonucleotides following each reaction without requiring purification after each step.
So long as unpaired toeholds (colored regions in Figure 1) remain in T, this process can be iterated to generate successively longer or more complex products without requiring any intervention during the reaction sequence beyond the simple addition of successive substrates to the same reaction vessel. Importantly, the effective molarity of the reactants during successive DNA-templated steps in this strategy remains relatively constant since the reactive groups for each step remain at the end of the double-stranded region.(38) As a result, we anticipated that reaction yields would remain higher over the course of many steps than with traditional multistep DTS methods. Crucially, the DNA templates in this strategy retain all product-encoding information throughout the DTS process, thus enabling the multistep products to be subjected to in vitro selections for desired properties, in contrast with a different approach to multistep synthesis that uses DNA strand exchange.(36)
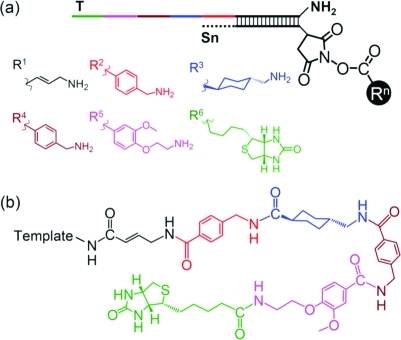
We designed and executed a six-step DNA-templated reaction sequence using this approach. The DNA template (T) contains a 3′ primary amine group, and the first five substrates (S1–S5) contain 5′ N-hydroxysuccinimidyl (NHS) ester-linked amines.32,35,37 The oligonucleotide–reagent bond is automatically cleaved as a result of the amine acylation reaction, thus enabling spontaneous transfer of the amino acid building block from each substrate to the template. The transferred amino acid displays another primary amine group that is poised to further react with the next NHS ester to continue the multistep DTS. The last substrate (S6) contains a biotin NHS ester and lacks a primary amine group (Figure 2).
Figure 2.
(a) Reactant structures; (b) expected six-step DNA-templated reaction product.
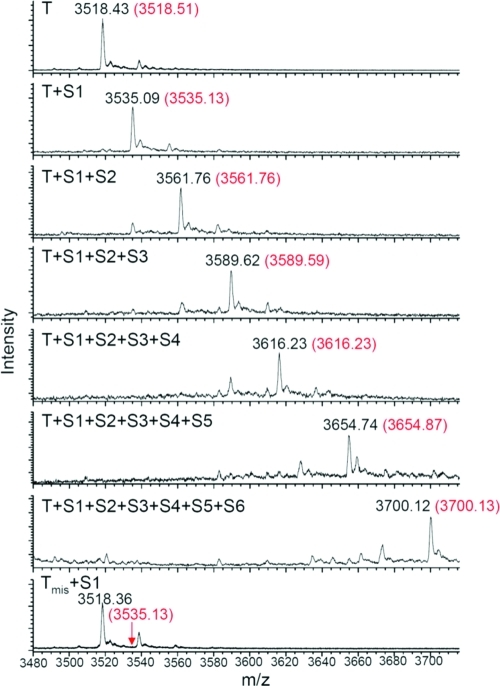
In a typical experiment, T (1 μM) and S1 (1.2 μM) were combined in aqueous buffer containing 50 mM MOPS and 10 mM Mg(OAc)2 at pH 7.5. After 30 min at room temperature, the second substrate S2 was added into the system, followed by another 30-min incubation at room temperature. Substrates S3–S6 were similarly added at successive 30-min intervals. During the entire process, the temperature and buffer conditions were constant. The six-step reaction was therefore carried out in a single vessel at room temperature over 3 h. The crude reaction mixture was desalted by gel filtration and analyzed by high-resolution ESI mass spectrometry. For comparison, reactions containing only T, T+S1, T+S1+S2, T+S1+S2+S3, T+S1+S2+S3+S4, T+S1+S2+S3+S4+S5, and S1+Tmis (a template containing a scrambled DNA sequence) were also subjected to the reaction conditions and analyzed by high-resolution mass spectrometry.
The unreacted template and all six expected reaction products corresponding to one-, two-, three-, four-, five-, and six-step reaction sequences were observed with high mass accuracy (Figure 3). Each reaction resulted in highly efficient formation of the desired multistep product, such that the predominant template-linked ion observed from each reaction corresponded to the expected one- to six-step reaction product (Figure 3). The observation that in the six-step reaction mixture the template-linked full-length hexaamide product provided the strongest m/z signal among all template-linked ions suggested that the six-step product might be formed in excellent overall yield. The control reaction containing the mismatched template and S1 did not generate detectable product (Figure 3), indicating that hybridization with the template is required to effect product formation under these conditions.
Figure 3.
Mass spectrometric analysis of multistep DNA-templated reaction products generated by sequential strand displacement. Eight reactions using the species shown were separately performed as described in the text. The mass range shown includes all template-linked species. Observed m/z values for ions of −5 charge are shown in black; expected m/z values are shown in red. The arrow in the last spectrum indicates the expected m/z value of the product corresponding to reaction between S1 and a scrambled template (Tmis).
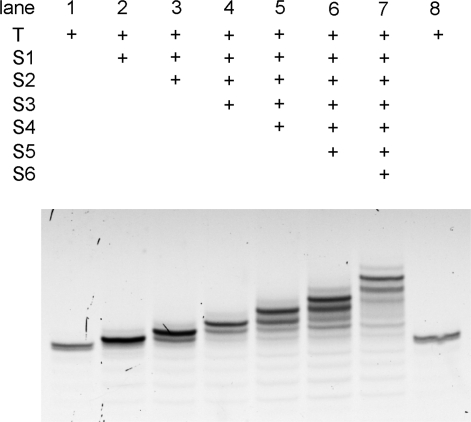
To quantitate reaction yields during all six reaction sequences, we used denaturing polyacrylamide gel electrophoresis (PAGE). Because each building block adds significant mass to the template, the full-length product and all the intermediate products migrate differently during PAGE. To achieve better separation during PAGE, all reaction products were digested with restriction endonuclease DdeI, which cleaves the 56-nt template into a 22-nt 3′ fragment containing the reaction products. To enable accurate quantitation of reaction yields, a single fluorescein group was incorporated into the template (prior to DTS) near the 3′ end, allowing the 22-nt fragments to fluoresce with equal intensity. Densitometry of fluorescein-visualized PAGE gel (Figure 4) revealed that the three-step reaction sequence (lane 4) generated the corresponding three-step product in 55% overall yield, and the six-step reaction sequence (lane 7) produced full-length hexaamide product in 35% overall yield, corresponding to ∼83% average yield per step (see the Supporting Information [SI] for additional details). An independent experiment to determine reaction yields using nonfluoroscein-labeled T in which DdeI-digested reaction products were analyzed by PAGE followed by staining resulted in 38% overall yield of six-step reaction product (see the SI). To our knowledge, these results represent both the highest yielding and longest multistep DNA-templated small-molecule syntheses to date.32,35−37
Figure 4.
Denaturing PAGE analysis of the multistep DTS reactions analyzed in Figure 3. Each template-linked species contained a single fluorescein group to enable uniform visualization. The reaction products were digested with the endonuclease DdeI prior to PAGE analysis to cleave the product-linked oligonucleotides to 22-mers. Lanes 1 and 8: unreacted template; lanes 2–7: DNA template after successive reaction with one, two, three, four, five, or six reagents, respectively.
Applying this strategy for future multistep DNA-templated library syntheses will require that reagent-linked oligonucleotides each be prepared as mixtures containing all possible anticodons for prior DNA-encoded steps. The recent availability of inexpensive custom oligonucleotide pools from chip-based parallel synthesis40−43 provides practical access to these materials. In addition, the fact that the specificity-determining step of each reaction is hybridization of the 8-base toehold, which encodes the current reagent, implies that mismatch hybridization elsewhere in the template:reagent oligonucleotide duplex will not affect the fidelity of the multistep DTS process. Moreover, longer DNA templates could conceivably give rise to products of more than six DNA-programmed reactions. Indeed, we have observed that a 64-base, 80-base, and 96-base DNA strand in a duplex, sufficient to encode up to ∼10 DNA-templated reactions, can be effectively displaced using a 72-mer, 88-mer, or 104-mer (see the SI and Figure S3).
In summary, we developed a rapid, one-pot, highly efficient multistep DNA-templated synthesis strategy that is driven by successive toehold DNA strand displacement. The strategy was used to generate a six-step DTS reaction product in 35% overall yield (83% average yield per step), such that the desired product was the major observed template-linked species after six steps with no purification or reaction manipulation required beyond addition of each substrate every 30 min. Since the reaction history of the products remains encoded by their associate templates, this strategy may enable future efforts to generate high-complexity (e.g., n6) DNA-templated libraries suitable for in vitro selection.
Acknowledgments
This work was supported by the Howard Hughes Medical Institute and NIH/NIGMS (R01GM065865). We also thank Jia Niu for experimental assistance and David Gorin for helpful discussions.
Supporting Information Available
Complete ref (30), experimental methods, and additional experimental data. This material is available free of charge via the Internet at http://pubs.acs.org.
Funding Statement
National Institutes of Health, United States
Supplementary Material
References
- Gartner Z. J.; Liu D. R. J. Am. Chem. Soc. 2001, 123, 6961–6963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gartner Z. J.; Kanan M. W.; Liu D. R. Angew. Chem., Int. Ed. 2002, 41, 1796–1800. [DOI] [PubMed] [Google Scholar]
- Gartner Z. J.; Tse B. N.; Grubina R.; Doyon J. B.; Snyder T. M.; Liu D. R. Science 2004, 305, 1601–1605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X. Y.; Liu D. R. Angew. Chem., Int. Ed. 2004, 43, 4848–4870. [DOI] [PubMed] [Google Scholar]
- Hansen M. H.; Blakskjaer P.; Petersen L. K.; Hansen T. H.; Hojfeldt J. W.; Gothelf K. V.; Hansen N. J. V. J. Am. Chem. Soc. 2009, 131, 1322–1327. [DOI] [PubMed] [Google Scholar]
- Kleiner R. E.; Dumelin C. E.; Tiu G. C.; Sakurai K.; Liu D. R. J. Am. Chem. Soc. 2010, 132, 11779–11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Silverman S. K. Angew. Chem., Int. Ed. 2010, 49, 7180–7201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleiner R. E.; Brudno Y.; Birnbaum M. E.; Liu D. R. J. Am. Chem. Soc. 2008, 130, 4646–4659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenbaum D. M.; Liu D. R. J. Am. Chem. Soc. 2003, 125, 13924–13925. [DOI] [PubMed] [Google Scholar]
- Brudno Y.; Birnbaum M. E.; Kleiner R. E.; Liu D. R. Nat. Chem. Biol. 2010, 6, 148–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanan M. W.; Rozenman M. M.; Sakurai K.; Snyder T. M.; Liu D. R. Nature 2004, 431, 545–549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozenman M. M.; Kanan M. W.; Liu D. R. J. Am. Chem. Soc. 2007, 129, 14933–14938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momiyama N.; Kanan M. W.; Liu D. R. J. Am. Chem. Soc. 2007, 129, 2230–2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y.; Kamlet A. S.; Steinman J. B.; Liu D. R. Nat. Chem. 2011, 3, 146–153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gorin D. J.; Kamlet A. S.; Liu D. R. J. Am. Chem. Soc. 2009, 131, 9189–9191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tse B. N.; Snyder T. M.; Shen Y. H.; Liu D. R. J. Am. Chem. Soc. 2008, 130, 15611–15626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dumelin C. E.; Scheuermann J.; Melkko S.; Neri D. Bioconjugate Chem. 2006, 17, 366–370. [DOI] [PubMed] [Google Scholar]
- Halpin D. R.; Harbury P. B. PLoS Biol. 2004, 2, 1015–1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin D. R.; Harbury P. B. PLoS Biol. 2004, 2, 1022–1030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halpin D. R.; Lee J. A.; Wrenn S. J.; Harbury P. B. PLoS Biol. 2004, 2, 1031–1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melkko S.; Zhang Y.; Dumelin C. E.; Scheuermann J.; Neri D. Angew. Chem., Int. Ed. 2007, 46, 4671–4674. [DOI] [PubMed] [Google Scholar]
- Melkko S.; Mannocci L.; Dumelin C. E.; Villa A.; Sommavilla R.; Zhang Y. X.; Grutter M. G.; Keller N.; Jermutus L.; Jackson R. H.; Scheuermann J.; Neri D. ChemMedChem 2010, 5, 584–590. [DOI] [PubMed] [Google Scholar]
- Melkko S.; Scheuermann J.; Dumelin C. E.; Neri D. Nat. Biotechnol. 2004, 22, 568–574. [DOI] [PubMed] [Google Scholar]
- Scheuermann J.; Dumelin C. E.; Melkko S.; Zhang Y. X.; Mannocci L.; Jaggi M.; Sobek J.; Neri D. Bioconjugate Chem. 2008, 19, 778–785. [DOI] [PubMed] [Google Scholar]
- Mannocci L.; Melkko S.; Buller F.; Molnar I.; Bianke J. P. G.; Dumelin C. E.; Scheuermann J.; Neri D. Bioconjugate Chem. 2010, 21, 1836–1841. [DOI] [PubMed] [Google Scholar]
- Buller F.; Steiner M.; Scheuermann J.; Mannocci L.; Nissen I.; Kohler M.; Beisel C.; Neri D. Bioorg. Med. Chem. Lett. 2010, 20, 4188–4192. [DOI] [PubMed] [Google Scholar]
- Buller F.; Zhang Y. X.; Scheuermann J.; Schafer J.; Buhlmann P.; Neri D. Chem. Biol. 2009, 16, 1075–1086. [DOI] [PubMed] [Google Scholar]
- Mannocci L.; Zhang Y. X.; Scheuermann J.; Leimbacher M.; De Bellis G.; Rizzi E.; Dumelin C.; Melkko S.; Neri D. Proc. Natl. Acad. Sci. U.S.A. 2008, 105, 17670–17675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wrenn S. J.; Harbury P. B. Annu. Rev. Biochem. 2007, 76, 331–349. [DOI] [PubMed] [Google Scholar]
- Clark M. A.; et al. Nat. Chem. Biol. 2009, 5, 647–654. [DOI] [PubMed] [Google Scholar]
- Driggers E. M.; Favaloro F.; Li X.; Li G.; Benton B.; Fraley A.; Bittker J.; Morales A.; Harbeson S.; Coull J.. 232nd ACS National Meeting, San Francisco, CA, United States, Sept. ffa10–14, 2006, pp 10–14,
- Gartner Z. J.; Kanan M. W.; Liu D. R. J. Am. Chem. Soc. 2002, 124, 10304–10306. [DOI] [PubMed] [Google Scholar]
- Li X. Y.; Gartner Z. J.; Tse B. N.; Liu D. R. J. Am. Chem. Soc. 2004, 126, 5090–5092. [DOI] [PubMed] [Google Scholar]
- Calderone C. T.; Liu D. R. Angew. Chem., Int. Ed. 2005, 44, 7383–7386. [DOI] [PubMed] [Google Scholar]
- He Y.; Liu D. R. Nat. Nanotechnol. 2010, 5, 778–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKee M. L.; Milnes P. J.; Bath J.; Stulz E.; Turberfield A. J.; O’Reilly R. K. Angew. Chem., Int. Ed. 2010, 49, 7948–7951. [DOI] [PubMed] [Google Scholar]
- Snyder T. M.; Liu D. R. Angew. Chem., Int. Ed. 2005, 44, 7379–7382. [DOI] [PubMed] [Google Scholar]
- Gartner Z. J.; Grubina R.; Calderone C. T.; Liu D. R. Angew. Chem., Int. Ed. 2003, 42, 1370–1375. [DOI] [PubMed] [Google Scholar]
- Green C.; Tibbetts C. Nucleic Acids Res. 1981, 9, 1905–1918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J. D.; Gong H.; Sheng N. J.; Zhou X. C.; Gulari E.; Gao X. L.; Church G. Nature 2004, 432, 1050–1054. [DOI] [PubMed] [Google Scholar]
- Lee C. C.; Snyder T. M.; Quake S. R. Nucleic Acids Res. 2010, 38, 2514–2521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian J. D.; Ma K. S.; Saaem I. Mol. Biosyst. 2009, 5, 714–722. [DOI] [PubMed] [Google Scholar]
- Saaem I.; Ma K. S.; Marchi A. N.; LaBean T. H.; Tian J. D. ACS Appl. Mater. Interfaces 2010, 2, 491–497. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.