Abstract
Granulomas, organized aggregates of immune cells, are a hallmark of tuberculosis, and have traditionally been thought to restrict mycobacterial growth. However, analysis of Mycobacterium marinum in zebrafish has shown that the early granuloma facilitates mycobacterial growth; uninfected macrophages are recruited to the granuloma where they are productively infected by M. marinum. Here, we identified the molecular mechanism by which mycobacteria induce granulomas: the bacterial secreted protein ESAT-6, which has long been implicated in virulence, induced matrix metalloproteinase-9 (MMP9) in epithelial cells neighboring infected macrophages. MMP9 enhanced recruitment of macrophages, which contributed to nascent granuloma maturation and bacterial growth. Disruption of MMP9 function attenuated granuloma formation and bacterial growth. Thus, interception of epithelial MMP9 production could hold promise as a host-targeting tuberculosis therapy.
Tuberculous infection begins with recruitment of monocytes to a peripheral infection site where they engulf mycobacteria and migrate to deeper tissues (1, 2). Additional macrophages and other immune cells then aggregate with the infected cells to form granulomas (3). Granulomas, recognized as pathological hallmarks of tuberculosis for over a century, were thought to curtail infection by encasing mycobacteria (4). However, visualization of granuloma formation in transparent zebrafish larvae infected with Mycobacterium marinum (Mm) has revealed that the early granuloma serves to expand bacterial numbers (5, 6). An infected macrophage induces granuloma formation by promoting recruitment of additional phagocytes (6). Upon its death, multiple newly arriving macrophages phagocytose it and thereby become infected. Concerted iteration of these processes makes the early granuloma a site for bacterial expansion (6). Mycobacteria direct these granuloma-forming processes via their RD1 virulence locus that encodes the ESX-1 secretion system (5, 6). The host factors co-opted in RD1-mediated granuloma formation remain unknown.
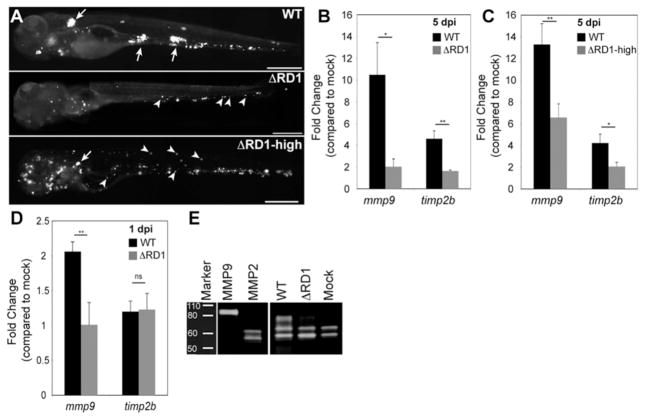
In a host gene expression survey comparing zebrafish larvae infected with wildtype Mm (WT) or RD1-deleted Mm (ΔRD1) (5, 6), we identified matrix metalloproteinase 9 (mmp9) and tissue inhibitor of metalloproteinase 2b (timp2b) as being RD1-induced during granuloma formation at 5 days post infection (5dpi) (Fig. 1A and 1B; tables S1–4; fig. S1A and S1B). To control for ΔRD1’s attenuated infection at 5 dpi (5), we confirmed RD1-dependent gene induction using higher ΔRD1 inoculations that produced similar bacterial burdens at 5 dpi with the expected paucity of ΔRD1 granulomas (5, 6) (Fig. 1A and 1C; fig. S1C). At 1 dpi, only mmp9 was induced, suggesting that timp2b induction at 5 dpi was a compensatory response to increased mmp9 (Fig. 1D). Mmp9 is a gelatinase and gelatin zymography confirmed that RD1-dependent mmp9 mRNA expression resulted in increased Mmp9 gelatinase activity (Fig. 1E). In contrast, mRNA expression and activity of another gelatinase Mmp2 were not altered by infection (fig. S1A and Fig. 1E).
Fig. 1.
RD1-dependent Mmp9 induction. (A) Representative fluorescence images of 5 dpi embryos used for gene expression studies in (B and C). Embryos in top and middle panels injected with similar doses of WT and ΔRD1, respectively (WT dose 193±36 and ΔRD1 dose 217±63), where ΔRD1 bacterial burdens are lower than WT at 5dpi. Embryo in bottom panel injected with ~ 5-fold more ΔRD1 (ΔRD1- high) to achieve similar bacterial burdens to WT at 5 dpi (5 dpi bacterial burdens were 1601±1071 for WT and 1531±1011 for ΔRD1-high, ns). Arrows, granulomas; arrowheads, single infected macrophages. Scale bars, 400 μm. (B-C) Relative gene expression levels (mean ± SEM of at least 3 biological replicates) of 5 dpi (B) WT- and ΔRD1-infected embryos and (C) WT and ΔRD1- high-infected embryos. Although there appears to be a dose-dependent induction of mmp9 by ΔRD1 (compare induction in panels B and C), the difference is not significant (p=0.2). (D) Relative gene expression levels (mean ± SEM of 3 biological replicates) 1 day after injection with 721±39 WT or 484±147 ΔRD1 (ns). *p < 0.05, **p < 0.01, ns, not significant (Student’s t test). (E) Gelatin zymography of embryos 5 dpi with 200 WT or 700 ΔRD1, or mock-infected. Controls are purified human MMP9 and MMP2.
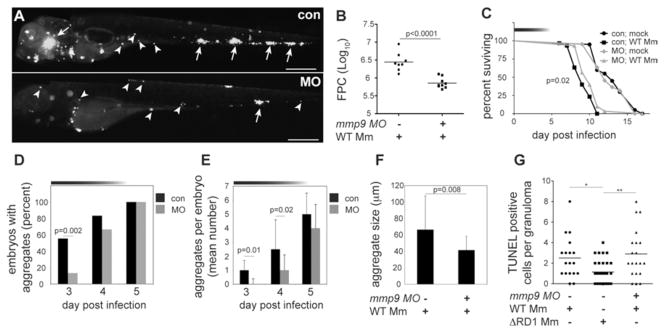
MMP9 is implicated in the pathogenesis of several inflammatory conditions (7, 8) and is highly expressed in human tuberculosis as well as in the mouse model of tuberculosis (9–12)(table S1). In mice, MMP9 activity correlates to increased macrophage migration and granuloma formation; however it is reported to be a host resistance factor, perhaps because its expression is associated with variable effects on infection in different genetic backgrounds (10). In humans, MMP9 clearly mediates susceptibility as its increased activity is correlated with worse outcomes (9). To test mmp9’s role in promoting granuloma formation and virulence, we knocked down its expression transiently with three modified antisense oligonucleotides (morpholinos) (1, 13) (fig. S2). The morpholinos, singly or in combination, reduced gelatinase activity reliably up to 4 dpi with activity returning to control levels by 5 dpi (fig. S2). WT infection of morpholino-injected embryos (morphants) resulted in attenuated infection sharing several features of ΔRD1 infection of control embryos. First, morphants displayed reduced bacteria and granulomas, as well as increased host survival (Fig. 2A-C). Second, kinetic analyses of granuloma formation in the morphants confirmed a specific granuloma-forming deficit (Fig. 2D-F). We found a dynamic link between Mmp9 activity, granuloma formation and bacterial expansion: bacterial burdens and granuloma formation differed only up to 4 dpi, returning to control levels by 5dpi contemporaneous with restoration of Mmp9 activity (Fig. 2A-E; fig. S2). Finally, while the RD1 locus promotes macrophage recruitment to nascent granulomas, it is not required for initial phagocyte migration to infecting bacteria when they are still extracellular (5, 6). Similarly, mmp9 morphants displayed normal macrophage migration to extracellular bacteria when injected into the hindbrain ventricle (fig. S3).
Fig. 2.
Mmp9 promotes granuloma formation and virulence. (A) Fluorescence images of representative control (con), and mmp9 morphant (MO) embryos 4 dpi with 116 WT. Arrows, granulomas; arrowheads, single infected macrophages. Scale bars, 400 μm (B) Bacterial burdens of all 4dpi embryos determined by fluorescence pixel counts (FPC) (see supplementary methods). (C) Survival of con and MO embryos (n=30 each) infected with 150 CFU WT or mock-infected (n=20 each). Median survival 10 days for infected MO and 9 days for infected con (p=0.02; Log-rank test) and no different for uninfected MO and con. Top horizontal bar denotes duration of MO activity (see fig. S2). (D to F) Kinetics of granuloma formation in con and MO embryos infected with 101 WT. Data in (D) analyzed by Fisher’s exact test of a contingency table. Bars in (E) and (F) represent mean ± SEM (Student’s t test). (G) Median number of TUNEL-positive cells per con or MO granuloma 4dpi with 37 CFU WT and con granulomas 4 dpi with 585 ΔRD1. (ANOVA; One-Way Analysis of Variance p=0.003, with Dunnet’s Multiple Comparison Test).
RD1 probably contributes to granuloma expansion through pleiotropic effects including inducing apoptosis of infected macrophages and recruitment of new uninfected macrophages (5, 6, 14, 15). In contrast, Mmp9 was not required for RD1-induced cell death; morphant and control granulomas in WT infection contained similar numbers of TUNEL-positive cells, whereas control granulomas in ΔRD1 infection exhibited the expected reduction (13) (Fig. 2G). Thus RD1-induced apoptosis is Mmp9-independent and cannot mediate bacterial expansion in the absence of new macrophage recruitment, and Mmp9-mediated acceleration of macrophage recruitment to granulomas is an independent mediator of pathogenesis.
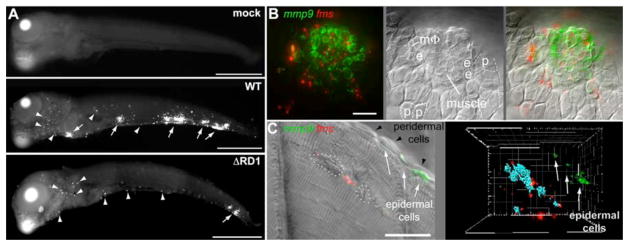
Multiple cell types express MMP9 in many inflammatory conditions (7). In the context of tuberculosis, it is induced in cultured Mtb-infected monocytes (9, 16–18) and in epithelial cells (19). In advanced human tuberculosis, induced expression is reported in some monocytes and multinucleated giant cells abutting necrotic centers of lymph node granulomas (17, 20) and in epithelial cells proximal to lung granulomas (19). To understand how the RD1-Mmp9 axis mediates granuloma formation we assessed localization of mmp9 expression during this process. Fluorescence whole mount in situ hybridization (FISH) (1) revealed RD1-dependent mmp9 induction in 5 dpi embryos in cells associated with granulomas as well as in distal single cells (Fig. 3A). Multiplex FISH combining the mmp9 and macrophage-specific fms probes, or mmp9 and neutrophil-specific mpo probes (1), showed that the distal single cells consisted largely of neutrophils with a minor macrophage contribution (fig. S4 and S5). However, mmp9 expression by neutrophils and macrophages was unlikely to be relevant for granuloma formation because their mmp9 expression induced by infection was RD1-independent, and most granulomas contained very few, if any, of these cells (Fig. 3B, fig. S4 and S5).
Fig. 3.
mmp9 is selectively induced in epithelial cells neighboring infected macrophages. (A) mmp9 FISH images of embryos 5 days after mock infection or infection with 78 CFU WT or 130 CFU ΔRD1. Arrows, mmp9 expression corresponding to granulomas, arrowheads, single mmp9-expressing cells. Scale bars, 400μm. (B and C) Images of WT granulomas after dual mmp9 and fms FISH. (B) Fluorescence (left), DIC (middle), and overlay (right) images. e, epidermal cell; p, peridermal cell; MΦ, macrophage. Scale bar, 20μm. Also see movie S1. (C) Fluorescence and DIC overlay of nascent WT muscle granuloma (left panel). Dotted white circles outline bacterial clusters discerned by DIC microscopy. Fluorescence data has been deconvolved. Right panel represents 3D reconstruction from fluorescence image of same lesion with bacterial locations pseudocolored blue, showing complete absence of mmp9 expression in adjacent muscle, and strong mmp9 expression in nearest neighboring epidermal cells. Scale bar, 20μm. Also see movie S2.
Differential Interference Contrast (DIC) and confocal microscopy revealed that granuloma-associated mmp9 expression was localized to epithelial cells proximate to infected macrophages (21) (Fig. 3B, movie S1). Expression was restricted to specific epithelial cell types: epidermal cells adjacent to the granuloma expressed mmp9 whereas immediately overlying peridermal cells did not (21) (movie S1). Epidermal cell-specific expression was highlighted in granulomas forming in muscle where mmp9 was expressed not by the immediately surrounding myocytes but by their closest epidermal neighbors (Fig. 3C, movie S2). Every granuloma analyzed had proximate mmp9-expressing epithelial cells (n=35 granulomas in 9 fish), including the smallest identifiable macrophage aggregates (fig. S6 and movie S3). Thus Mmp9 induction is critical for granuloma formation from the very earliest stages and probably in later stages as well, given RD1’s continued influence on granuloma structure in chronic tuberculous infection (5, 22).
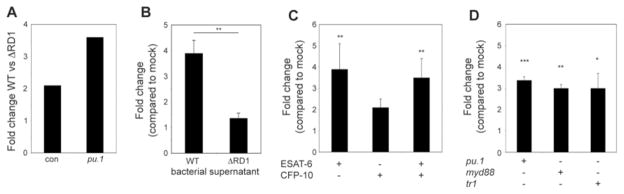
Bacteria residing in macrophages could induce epithelial cell mmp9 in two ways: (i) RD1 might induce macrophage signals such as secreted cytokines (23, 24) that in turn elicit mmp9 secretion by epithelial cells, or (ii) bacteria (25, 26) or bacterial products (27) released from macrophages might interact directly with epithelial cells. To distinguish between these mechanisms, we assessed mmp9 induction in pu.1 morphants that lack macrophages and in which infection results in extracellular mycobacterial growth (1). pu.1 morphants exhibited RD1-dependent mmp9 induction, suggesting that bacteria or their products interact directly with epithelial cells to induce mmp9 by a macrophage-independent mechanism (Fig. 4A).
Fig. 4.
Mycobacterial ESAT-6 is sufficient to induce mmp9 in epithelial cells independent of Myd88 and TNF signaling. (A-D) Relative mmp9 expression analyzed by qRT-PCR of (A) con or pu.1 morphant embryos 3 dpi with 84 WT or 126 ΔRD1 (represents one biological replicate), (B) 34 hpf embryos 4 hours post-injection with WT or ΔRD1 bacterial supernatant. Bars represent means ± SEM of three biological replicates. (C) 34 hpf con embryos 4 hours post-injection with 4.8×10−17 moles of purified ESAT-6 or CFP-10, or 4.9×10−17 moles of ESAT-6 plus 5.0×10−17 moles of CFP-10. Bars represent means (± SEM) of five biological replicates. (D) 34 hpf con embryos, myd88 morphants, or tr1 morphants 4 hours post-injection with 5.7×10−17 moles of purified ESAT-6. Bars represent means ± SEM of four biological replicates (pu.1 morphant), or three biological replicates (myd88 and tr1 morphants).
The observation that uninfected epithelial cell mmp9 induction can occur distant from infection foci (Fig. 3C and movie S3) implicated an RD1-dependent secreted determinant rather than direct bacterial contact with epithelial cells. Indeed, injection of WT but not ΔRD1 bacterial supernatants rapidly induced mmp9 expression (Fig. 4B). The ESX-1 secretion system secretes five proteins that are all mutually co-dependent for secretion, so distinguishing their individual roles in virulence has been difficult (14, 15). We pursued ESAT-6 as the lead candidate for inducing mmp9 for two reasons: ESAT-6 mediates virulence independent of secretion (15) and its pore-forming activity (28, 29) could allow it direct access to epithelial cells. Injection of 4.8×10−17 moles of purified ESAT-6 was sufficient to induce mmp9 within 4 hours (Fig. 4C). In contrast, 5.0×10−17 moles CFP-10, thought to bind ESAT-6 and serve as its chaperone (15), failed to induce mmp9 significantly (Fig. 4C). Moreover, co-injection of CFP-10 and ESAT-6 did not augment the induction observed with ESAT-6 alone, confirming an ESAT-6-specific effect (Fig. 4C). Finally, similar to RD1-competent bacteria (Fig. 4A), ESAT-6 induced mmp9 in pu.1 morphants (Fig. 4D), consistent with a direct interaction with epithelial cells. We next asked if epithelial cell mmp9 induction was dependent on MyD88 and TNF signaling, as each can enhance mycobacterial induction of mmp9 in cultured cells under certain conditions (18, 19). ESAT-6 induced mmp9 in myd88 and tnf-receptor 1(tr1) morphants (Fig. 4D) suggesting a novel pathway for this epithelial cell-specific interaction. Moreover, TNF-independent induction of mmp9 is consistent with the finding that TNF does not mediate granuloma formation either in the presence or absence of bacterial RD1 (13).
Thus ESAT-6 functions in virulence by promoting granuloma formation via interaction with epithelial cells, previously regarded as bystanders in the pathogenesis of tuberculosis (fig. S7). The co-option of epithelial cells may offer mycobacteria a means of amplifying MMP9 secretion in the vicinity of a single infected macrophage to establish the granuloma niche. In addition, the differential induction of inflammatory programs in macrophages and epithelial cells may generate a hospitable growth niche in macrophages while harnessing epithelial cells to facilitate the chemotaxis of additional macrophages for niche expansion (6) (fig. S7). Our work provides a mechanistic explanation for the implication of MMP9 in human susceptibility to tuberculosis (9, 11, 12) and suggests targeted inhibition of its expression as a host-directed antituberculous therapy. Because, increased MMP9 is detrimental in both tuberculosis and a variety of noninfectious inflammatory conditions (7), interception of this pathway may have broad utility in treating a variety of inflammatory conditions in addition to tuberculosis.
Supplementary Material
Acknowledgments
We thank J.I. Gordon, W. Parks, D. Raible, D. Sherman, K. Urdahl, and P. Elkington for advice and discussion, D. Beery and R. Kim for help with microinjections, L. Swaim. H. Wiedenhoft and J. Cameron for fish facility maintenance. K. Winglee for developing FPC analysis methods, R. Burmeister for graphic design and D. Tobin, B. Cormack, W. Parks, D. Stetson, R. Berg and F. Chu for review of the manuscript. This work was supported by the Burroughs Wellcome Fund (LR), the Pew Scholars Program (JFR), the National Institutes of Health (LR and JFR), an American Heart Association predoctoral fellowship (HEV), a Pediatric Infectious Diseases Society postdoctoral award, the Children’s Health Research Center new investigator award and a National Institutes of Health diversity supplement (TCP) and a National Defense Science and Engineering predoctoral fellowship (JMD).
Footnotes
Materials and Methods
Figs. S1 to S7
Tables S1 to S4
Movies S1 to S3
References
References and Notes
- 1.Clay H, et al. Cell Host & Microbe. 2007;2:29. doi: 10.1016/j.chom.2007.06.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wolf AJ, et al. J Immunol. 2007;179:2509. doi: 10.4049/jimmunol.179.4.2509. [DOI] [PubMed] [Google Scholar]
- 3.Adams DO. Am J Pathol. 1976;84:164. [PMC free article] [PubMed] [Google Scholar]
- 4.Ulrichs T, Kaufmann SH. J Pathol. 2006;208:261. doi: 10.1002/path.1906. [DOI] [PubMed] [Google Scholar]
- 5.Volkman HE, et al. PLoS Biol. 2004;2:e367. doi: 10.1371/journal.pbio.0020367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Davis JM, Ramakrishnan L. Cell. 2009;136:37. doi: 10.1016/j.cell.2008.11.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Van den Steen PE, et al. Crit Rev Biochem Mol Biol. 2002;37:375. doi: 10.1080/10409230290771546. [DOI] [PubMed] [Google Scholar]
- 8.Greenlee KJ, Werb Z, Kheradmand F. Physiol Rev. 2007;87:69. doi: 10.1152/physrev.00022.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Price NM, et al. J Immunol. 2001;166:4223. doi: 10.4049/jimmunol.166.6.4223. [DOI] [PubMed] [Google Scholar]
- 10.Taylor JL, et al. Infect Immun. 2006;74:6135. doi: 10.1128/IAI.02048-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Park KJ, et al. Respiration. 2005;72:166. doi: 10.1159/000084048. [DOI] [PubMed] [Google Scholar]
- 12.Sheen P, et al. Eur Respir J. 2009;33:134. doi: 10.1183/09031936.00127807. [DOI] [PubMed] [Google Scholar]
- 13.Clay H, Volkman HE, Ramakrishnan L. Immunity. 2008;29:283. doi: 10.1016/j.immuni.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.DiGiuseppe Champion PA, Cox JS. Cell Microbiol. 2007;9:1376. doi: 10.1111/j.1462-5822.2007.00943.x. [DOI] [PubMed] [Google Scholar]
- 15.Simeone R, Bottai D, Brosch R. Curr Opin Microbiol. 2009;12:4. doi: 10.1016/j.mib.2008.11.003. [DOI] [PubMed] [Google Scholar]
- 16.Chang JC, et al. Thorax. 1996;51:306. doi: 10.1136/thx.51.3.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Price NM, Gilman RH, Uddin J, Recavarren S, Friedland JS. J Immunol. 2003;171:5579. doi: 10.4049/jimmunol.171.10.5579. [DOI] [PubMed] [Google Scholar]
- 18.Shi S, et al. J Exp Med. 2003;198:987. doi: 10.1084/jem.20030603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Elkington PT, et al. Am J Respir Cell Mol Biol. 2007 [Google Scholar]
- 20.Zhu XW, Price NM, Gilman RH, Recarvarren S, Friedland JS. J Infect Dis. 2007;196:1076. doi: 10.1086/521030. [DOI] [PubMed] [Google Scholar]
- 21.Le Guellec D, Morvan-Dubois G, Sire J. Int J Dev Biol. 2004;48:217. doi: 10.1387/ijdb.15272388. [DOI] [PubMed] [Google Scholar]
- 22.Sherman DR, et al. J Infect Dis. 2004;190:123. doi: 10.1086/421472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Koo IC, et al. Cell Microbiol. 2008;10:1866. doi: 10.1111/j.1462-5822.2008.01177.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stanley SA, Raghavan S, Hwang WW, Cox JS. Proc Natl Acad Sci U S A. 2003;100:13001. doi: 10.1073/pnas.2235593100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stamm LM, et al. J Exp Med. 2003;198:1361. doi: 10.1084/jem.20031072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagedorn M, Rohde KH, Russell DG, Soldati T. Science. 2009;323:1729. doi: 10.1126/science.1169381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Russell DG. Nat Rev Microbiol. 2007;5:39. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- 28.Hsu T, et al. Proc Natl Acad Sci U S A. 2003;100:12420. doi: 10.1073/pnas.1635213100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Jonge MI, et al. J Bacteriol. 2007;189:6028. doi: 10.1128/JB.00469-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.