Abstract
Plants perceive light via specialized photoreceptors of which the phytochromes (phyA-E), absorbing far-red (FR) and red light (R) are best understood. Several nuclear and cytoplasmic proteins have been characterized whose deficiencies lead to changes in light-dependent morphological responses and gene expression. However, no plastid protein has yet been identified to play a role in phytochrome signal transduction. We have isolated a new Arabidopsis mutant, laf (long after FR) 6, with reduced responsiveness preferentially toward continuous FR light. The disrupted gene in laf6 encodes a novel plant ATP-binding-cassette (atABC1) protein of 557 amino acids with high homology to ABC-like proteins from lower eukaryotes. In contrast to lower eukaryotic ABCs, however, atABC1 contains an N-terminal transit peptide, which targets it to chloroplasts. atABC1 deficiency in laf6 results in an accumulation of the chlorophyll precursor protoporphyrin IX and in attenuation of FR-regulated gene expression. The long hypocotyl phenotype of laf6 and the accumulation of protoporphyrin IX in the mutant can be recapitulated by treating wild-type (WT) seedlings with flumioxazin, a protoporphyrinogen IX oxidase (PPO) inhibitor. Moreover, protoporphyrin IX accumulation in flumioxazin-treated WT seedlings can be reduced by overexpression of atABC1. Consistent with the notion that ABC proteins are involved in transport, these observations suggest that functional atABC1 is required for the transport and correct distribution of protoporphyrin IX, which may act as a light-specific signaling factor involved in coordinating intercompartmental communication between plastids and the nucleus.
Keywords: signal transduction, phytochrome A, far-red, ABC proteins, plastids
Because of their photosynthetic and nonmotile nature, plants need to be particularly adaptable to their light environment. Light provides both energy for photosynthesis and informational signals to optimize growth and development from seed germination through adult life. The perception of light is accomplished through the use of specialized photoreceptors of which the phytochromes (phy) are best understood (Kendrick and Kronenberg 1994). Phytochromes are encoded by a small gene family (Pratt 1994). In Arabidopsis, five phytochrome genes have been identified (phyA-E) that perceive R and FR light and that exist as photoconvertible R (Pr) and FR (Pfr) forms. Absorption of photons causes phytochrome to undergo a photoreversible conformational change converting the molecule into the biologically active Pfr form (Furuya 1993; Quail et al. 1995; Neff et al. 2000). The activated phytochrome (Pfr) can subsequently relay information and act as an activator of the downstream signaling pathway leading to the appropriate physiological and molecular responses.
Using phytochrome-deficient mutants, the distinct roles of individual phytochromes have been unraveled. PhyA-deficient mutants are impaired in FR light responses (Whitelam et al. 1993), making it evident that phyA, a light-labile phytochrome (Furuya 1989), is responsible for FR sensing. In contrast, the light-stable phytochromes phyB-E have been shown to be involved in different aspects of R light responses.
Although our understanding of phytochrome action is good, insight into how the perceived light signals are transduced leading to morphological responses and altered gene expression patterns remains sparse. Several approaches have been taken to unravel downstream signaling events. Pharmacological approaches have been applied that have suggested an involvement of G-proteins, cGMP, and calcium as mediators of light-regulated gene expression (Neuhaus et al. 1993; Bowler et al. 1994). A second approach has involved the identification of phytochrome interacting proteins. To date, three phytochrome interacting proteins have been identified, PIF3 (Ni et al. 1998), PKS1 (Fankhauser et al. 1999), and NDPK2 (Choi et al. 1999), interacting not only with phyA but also with phyB.
Genetic screens have identified numerous mutants showing light-dependent phenotypes (cf. Fankhauser and Chory 1998; Neff et al. 2000). In terms of phyA-specific downstream signaling mutants, showing effects only toward FR light, fhy1, fhy3, spa1, fin2, far1, pat1, fin219, and eid1 have been identified (Whitelam et al. 1993; Hoecker et al. 1998, 1999; Soh et al. 1998; Hudson et al. 1999; Bolle et al. 2000; Buche et al. 2000; Hsieh et al. 2000). However, to date, only spa1, far1, pat1, and fin219 have been characterized at the molecular level. Moreover, these four mutants have all been shown to be disrupted in early acting components of the phyA signaling pathway. In contrast to the nuclear localization of SPA1 and FAR1, PAT1 and FIN219 are cytoplasmic proteins.
To date, no plastid-localized phytochrome-signaling component has been identified. PhyA signaling components characterized so far at the molecular level have been shown to be nuclear or cytoplasmic and to interact either with the photoreceptors themselves or to act early in the signaling pathway. An important light-dependent developmental pathway involves the transition of nonphotosynthetically active proplastids into photosynthetically active chloroplasts (Kendrick and Kronenberg 1994). This process involves coordinated expression of both plastidic and nuclear genes and subsequent translocation of nuclear-encoded proteins into developing chloroplasts (Chen and Schnell 1999; Bauer et al. 2000). This finely tuned light-regulated developmental program requires precise communication between the nucleus and plastids where the former senses the functional state of the latter and orchestrates subsequent morphological and molecular manifestations (Oelmuller et al. 1986; Oelmuller 1989; Susek et al. 1993; Lopez-Juez et al. 1998; Streatfield et al. 1999). Indeed, it has been shown that the herbicide Norflurazon, which impairs chloroplast functionality, results in reduced expression of the nuclear encoded genes Lhcb (chlorophyll a/b-binding protein) and Rbcs (small subunit of ribulose bisphosphate carboxylase) (Oelmuller 1989; Susek et al. 1993). One class of Arabidopsis mutants has been identified that shows reduced Lhcb expression and defective chloroplast development, suggesting a close connection between plastid- and phytochrome-regulated nuclear gene expression (Lopez-Juez et al. 1998). One of these mutants has now been cloned, and CUE1 encodes a plastid localized phosphoenolpyruvate/phosphate translocator involved in mesophyll cell development (Streatfield et al. 1999). However, it is still unclear what role plastids have in photomorphogenic responses such as hypocotyl elongation and in controlling nuclear gene expression.
To identify new components of the phyA signaling pathway, we screened Arabidopsis mutants for reduced responsiveness toward FR light irradiation. Here we describe the isolation of laf (long after FR) 6, a mutant preferentially affected toward FR light responses. LAF6 encodes a novel plant ATP-binding-cassette (atABC1) protein with high homology to small ABC-like proteins found in lower eukaryotes. The atABC1 transcript is induced by phyA, and the deficiency of atABC1 in laf6 results in a preferential attenuation of phyA-regulated responses. Furthermore, atABC1 localizes to plastids, and one consequence of the mutant lesion is a deficiency in chlorophyll and an accumulation of the chlorophyll precursor protoporphyrin IX. In addition, flumioxazin-treated WT seedlings, showing increased protoporphyrin IX levels, phenocopy the laf6 hypocotyl mutant phenotype. atABC1 represents an example of a plastid-localized component that may be involved in the intercompartmental communication between the nucleus and plastids for light signaling.
Results
Mutant screening and isolation
We screened ∼10,000 individual T2-generation Arabidopsis Ds insertion mutants (Sundaresan et al. 1995; Parinov et al. 1999) for elongated hypocotyls after 3 d of FR irradiation (FR-hf; 5.5 μmol m−2; Bolle et al. 2000). Twenty putative mutants displayed a FR-specific long hypocotyl phenotype in the following T3 generation. To determine genetic linkage between the mutant phenotype and the Ds insertion, we back crossed the 20 long-hypocotyl mutants to the isogenic WT (Ler). Segregation analysis and subsequent PCR amplification of the Ds element from F2 seedlings displaying FR-specific long hypocotyls demonstrated that six mutants showed cosegregation of the Ds element with the mutant phenotype. In contrast to the pat mutants (Bolle et al. 2000), these mutants designated as laf, are unable to green in white light (WL) after a pre-FR treatment. Southern blot analysis confirmed that all six mutants contained one single Ds element (data not shown), and one recessive mutant, laf6, was selected for further study.
FR responses are specifically attenuated in laf6
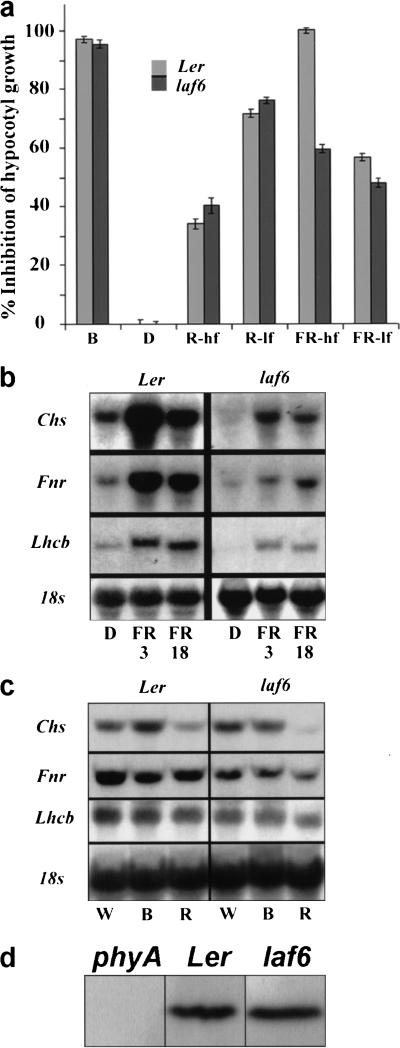
laf6 seedlings displayed an approximately twofold increase in hypocotyl length (Ler ∼ 3 mm, laf6 ∼ 7.5 mm) and reduced cotyledon expansion compared with the isogenic WT after FR-hf light irradiation (Fig. 1a). At low fluences of FR light (FR-lf; 0.34 μmol m−2) laf6 seedlings showed a very similar phenotype to the isogenic WT (Fig. 1a). These results demonstrate a fluence dependency and an attenuation but not an elimination of responsiveness toward FR light in the mutant. The observed fluence dependency suggests that the disrupted protein in laf6 may be rate limiting at high fluences of FR light but is not essential during low fluences. This is in sharp contrast to phyA null mutants, which show a complete loss of responsiveness toward FR light caused by the absence of a functional phyA photoreceptor (Whitelam et al. 1993). The laf6 mutant phenotype is light dependent because no phenotypic differences could be detected between laf6 and WT seedlings in the dark (Fig. 1a).
Figure 1.
Characterization of the laf6 mutant. (a) Percentage inhibition of hypocotyl growth of WT and laf6 after irradiation with B, R (R-hf; high fluence and R-lf; low fluence), and FR (FR-hf; high fluence and FR-lf; low fluence) for 7 d. The hypocotyl length of dark grown (D) seedlings was used as the reference (0% inhibition). A 0% inhibition represents a hypocotyl length of ∼12.5 mm, and a 100% inhibition represents ∼3 mm. (b) Expression of phytochrome-regulated genes in WT and laf6 after 3 and 18 h of FR-hf light irradiation. (c) Northern blot analysis of phytochrome-regulated genes in WT and laf6 after 3 h of white (W; 15 μmol m−2), B, and R light irradiation. (d) Phytochrome A levels in laf6. Western blot showing phytochrome A levels in WT (Ler) compared to laf6. A phytochrome A null mutant (phyA-201) is used as control.
To test the light specificity of the laf6 mutant phenotype, laf6 seedlings were irradiated with either high-fluence red (R) light (R-hf; 6.5 μmol m−2), low-fluence R light (R-lf; 0.88 μmol m−2) or with blue (B) light (8.5 μmol m−2). Figure 1a shows that under these light conditions, only minor differences in hypocotyl length were observed compared to WT, indicating that the laf6 mutant has reduced responsiveness preferentially toward FR light.
The appropriate expression of the light-regulated genes encoding chalcone synthase (Chs), ferredoxin NADP+ oxidoreductase (Fnr), and Lhcb is dependent on a functional phytochrome-signaling pathway. Figure 1b shows that expression of Chs, Fnr, and Lhcb was attenuated in laf6 compared with WT at FR-hf irradiation, while no differences were detected between laf6 and WT at FR-lf light (data not shown). These results extend the fluence dependency observed in terms of laf6 hypocotyl elongation.
To test whether the observed attenuation of Chs, Fnr, and Lhcb expression is specific to FR light irradiation, transcript analysis was performed after 3 h of WL (15 μmol m−2), B light, and R-hf light irradiation (Fig. 1c). Under all the above light conditions, only minor differences were detected between laf6 and WT.
laf6 has normal phyA levels
Previous studies have conclusively demonstrated that the levels of phytochromes have a proportional effect on the sensitivity that seedlings display to light (Whitelam and Devlin 1997). We therefore examined whether the reduced sensitivity of laf6 toward FR light is caused by decreased levels of phyA. Immunoblot analysis showed no difference in phyA protein levels between WT and laf6 seedlings grown in the dark (Fig. 1d). A phyA (phyA-201) null mutant was used as a control. Although immunoblotting provides a way of estimating the levels of total phyA protein, phyA protein accumulation in Arabidopsis is not necessarily linked to the availability of phytochromobilin and, hence, the generation of photoactive phytochrome. To test whether chromophore availability was affected in laf6, we performed feeding experiments where laf6 seedlings were grown in media supplemented with phycocyanobilin, a light-harvesting pigment from cyanobacteria that can substitute for phytochromobilin (Parks and Quail 1991). No physiological effect was observed when laf6 seedlings were treated with FR light irradiation in the presence of this chromophore (data not shown). This demonstrates that the laf6 mutant phenotype is most probably not a result of altered photoactive phyA levels.
laf6 is disrupted in a gene encoding an ABC-like protein
The flanking region of the disrupted gene in laf6 was cloned by inverse PCR. DNA sequencing of the amplified flanking region and subsequent database searches revealed that the Ds insertion site occured 24 bp upstream of the translational initiation site of a gene encoding a novel plant member of the ATP-binding-cassette transporter (atABC1; Accession number AAD03441) superfamily (Holland and Blight 1999) situated on chromosome IV, 21 cM from the RI marker mi306 (Fig. 2a). Analysis of the genomic sequence encompassing atABC1 revealed the presence of two exons and one intron (Fig. 2a). A 1674-bp atABC1 cDNA was cloned by RT–PCR using primers spanning the predicted ORF, and the genomic exon/intron structure was confirmed by DNA sequencing.
Figure 2.
Genomic structure of atABC1 and sequence alignment of small soluble ABC proteins. (a) Genomic structure of atABC1 showing the position of the Ds element insertion. Translation initiation (ATG) and termination (TAA) codons are indicated. (b) Amino acid sequence alignment of small soluble ABC proteins from Arabidopsis (atABC1), Synechocystis sp. strain PCC6803 (synABC), and Guillardia theta (guilABC) using the Clustal method and DNASTAR Megalign 4.0 software. Gaps are inserted to maximize identical sequences. Identical amino acids in at least two of the sequences are shaded in black. The transit peptide is underlined and the putative cleavage site is indicated by an arrow.
The location of the Ds insertion in the atABC1 sequence predicted that atABC1 transcript levels should be severely affected in laf6. Indeed, Northern blot analysis confirmed that the atABC1 transcript in laf6 is reduced to almost undetectable levels compared with WT seedlings (Fig. 3b).
Figure 3.
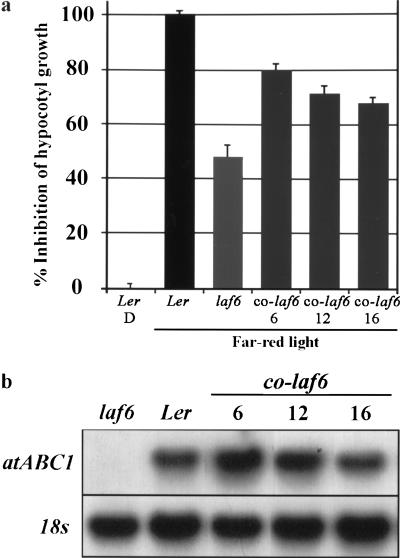
Complementation of laf6 with a full-length genomic copy of atABC1. (a) Percentage inhibition of hypocotyl length of WT (Ler), laf6, and several independent laf6 transgenic lines (homozygous) harboring a genomic copy of atABC1 after FR-hf irradiation for 7 d. The hypocotyl length of dark grown (D) seedlings was used as the reference (0% inhibition). A 0% inhibition represents a hypocotyl length of ∼13 mm, and a 100% inhibition represents ∼3.5 mm. (b) Expression analysis comparing atABC1 transcript levels in the complemented lines. The 18S rRNA was used as a loading control.
To ensure that the disruption of atABC1 in laf6 was the bona fide cause of the mutant phenotype, we transformed laf6 seedlings with a full-length atABC1 genomic copy including ∼1200 bp of the upstream promoter region and ∼80 bp of the 3′ UTR. After FR-hf irradiation, the resulting transgenic T3 seedlings showed near to complete restoration (∼80%) of WT hypocotyl lengths (Fig. 3a), demonstrating that the deficiency of atABC1 is indeed responsible for the laf6 phenotype.
atABC1 encodes a protein of 557 amino acids with high homology (∼80%) to the cyanobacterial Synechocystis sp. strain PCC6803 ABC (Accession number Q55790) and to chloroplast-encoded ABC-like proteins from unicellular organisms such as the cryptophyte alga Guillardia theta (Accession number AF041468.1; Fig. 2b). Although the overall similarity is very high among these ABC-like proteins, atABC1 contains an N-terminal extension absent in soluble ABC proteins of lower eukaryotes (Fig. 2b). The atABC1 N-terminal extension has a high content of serine and threonine residues (∼27%), and computer prediction analysis (http://genome.cbs.dtu.dk) indicated that this region may function as a chloroplast-targeting transit peptide.
By contrast, the overall similarity of atABC1 to classical plant membrane–bound ABC transporter proteins, such as atMRP1 (Lu et al. 1997) and atMRP2 (Lu et al. 1998), is relatively low (∼25%–35%). While classical membrane-bound ABC transporters are typically ∼1600 amino acids in length containing two membrane spanning domains (Lu et al. 1997, 1998), atABC1 is significantly smaller and contains no membrane-spanning regions. Therefore, atABC1 likely belongs to the subfamily of small soluble ABC proteins that require an integral membrane component as an interacting partner to execute their function. Examples of small soluble ABC-like proteins have been found in all species examined to date and are mostly involved in import rather than export reactions (Holland and Blight 1999).
atABC1 is induced by phyA
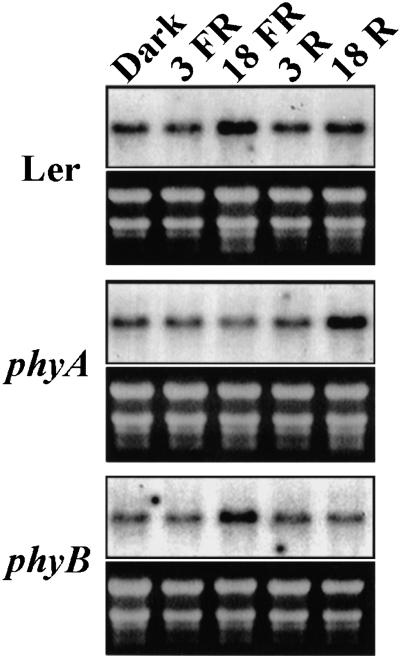
Northern blot analysis revealed that atABC1 is expressed at an equal level in all tissues examined (data not shown). WT seedlings grown in darkness showed a basal level of atABC1 expression (Fig. 4), however, an approximately twofold induction of atABC1 transcript was observed specifically after 18 h of FR-hf irradiation (Fig. 4). There is also a slight induction of atABC1 transcript in response to 18 h of R-hf irradiation. By contrast, no FR-mediated atABC1 induction was observed in a phyA-deficient mutant (Fig. 4; phyA [phyA-201]), demonstrating that the changes in atABC1 levels are induced specifically by phyA. Interestingly, in the phyA-deficient mutant, atABC1 is induced by R-hf light (Fig. 4), suggesting that in the absence of phyA, atABC1 induction can be mediated through phyB. In addition, atABC1 transcript levels were induced after 18 h of FR-hf light irradiation in a phyB-deficient mutant (phyB-5), while no induction above basal levels was observed in response to R-hf light (Fig. 4; phyB [phyB-5]). The observed FR-specific induction in the phyB-deficient mutant corroborates the FR-specific atABC1 induction seen in WT seedlings.
Figure 4.
atABC1 gene expression analysis. Expression profiles of the atABC1 transcript in darkness (4 d) and in response to high-fluence FR-hf and R-hf light irradiation (3 and 18 h) in WT, phyA (phyA-201)-, and phyB (phyB-5)-deficient seedlings.
atABC1 contains a functional transit peptide and localizes to plastids
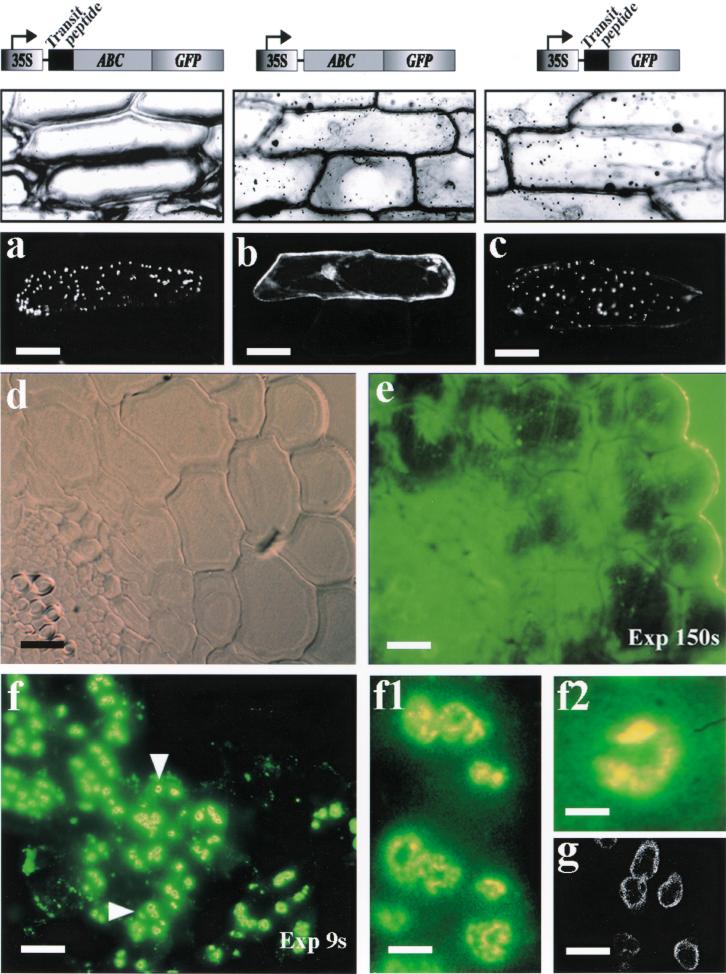
We investigated whether the atABC1 N-terminal extension could function as a chloroplast transit peptide. To this end, fusion genes encoding the full-length atABC1/GFP and atABC1 lacking the 63–amino acid transit peptide/GFP were generated and transiently expressed in onion epidermal cells. Fluorescence analysis demonstrated that full-length atABC1 localizes exclusively to leucoplasts in onion epidermal cells (Fig. 5a). By contrast, the truncated version of atABC1, lacking the transit peptide, remained cytoplasmic (Fig. 5b). To ensure that the plastidic localization of atABC1 represents a genuine translocation event, the 63–amino acid atABC1 transit peptide was fused to the N terminus of GFP, and the fusion protein was found to localize to leucoplasts (Fig. 5c).
Figure 5.
Plastidic localization of atABC1 in transiently transformed onion epidermal cells and in Arabidopsis seedlings. (a–c) Onion epidermal cells. (a) atABC1/GFP, (b) atABC1/GFP lacking the transit peptide (TP), (c) atABC1-TP/GFP. (d–f) Arabidopsis seedlings. (d) Bright-field image of a thin section of a leaf from the laf6 mutant, (e) Epi-fluorescence image (exposure time 150 sec) of (d) incubated with anti-atABC1 antiserum, (f) Epi-fluorescence image (exposure time 9 sec) of a thin section of a leaf from WT seedlings incubated with anti-atABC1 antiserum, (f1–f2) Regions from (f; arrow-heads) at higher magnification. (g) Hypocotyl chloroplasts from transgenic Arabidopsis seedlings overexpressing atABC1/GFP. Bars: a,b,c = 100 μm; d,e,f = 10 μm; f1 = 6 μm; f2 = 3 μm; g = 7 μm.
To further examine the subcellular localization of endogenous atABC1 in chloroplasts, we generated anti-atABC1 antibodies for in situ immunolocalization analysis. Sections 5–8 μm thick of leaves from 4-wk-old WL-grown laf6 and WT seedlings were examined by immunostaining. Specific staining was found in WT chloroplasts (Fig. 5f), while as expected, no staining was detected in laf6 chloroplasts (Fig. 5e). At higher magnifications, it was clear that staining occurred at the periphery of the chloroplasts (Fig. 5f1–f2). In support of this finding, analysis of transgenic Arabidopsis seedlings expressing a full-length atABC1/GFP fusion protein showed peripheral GFP fluorescence in hypocotyl chloroplasts (Fig. 5g). The observation that atABC1 localizes to the periphery of chloroplasts is consistent with the notion that atABC1 likely interacts with an envelope integral membrane component to function as a plastid-localized transport system (Holland and Blight 1999).
The deficiency of atABC1 in laf6 results in an accumulation of protoporphyrin IX
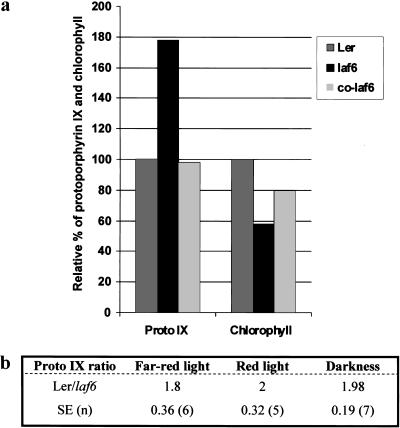
At early stages of development, laf6 seedlings are slightly pale green compared with WT seedlings, and chlorophyll measurements showed a ∼40% decrease in chlorophyll content in laf6 compared to WT (Fig. 6a). This suggested that one consequence of atABC1 deficiency in laf6 is decreased chlorophyll accumulation. In addition, it has been demonstrated that classical membrane-bound plant ABC transporters are involved in transport of various components such as glutathione S-conjugates, bile salts, and chlorophyll catabolites (Lu et al. 1997, 1998). In view of this, we examined whether the deficiency of atABC1 in plastids results in altered transport or distribution characteristics of chlorophyll biosynthetic intermediates. It has also been shown that chlorophyll precursors, such as porphyrin compounds, can prevent the accumulation of light-dependent transcripts in Chlamydomonas (Johanningmeier 1988). The oxidation of the chlorophyll biosynthesis intermediate protoporphyrinogen IX to protoporphyrin IX is catalyzed by the envelope-associated protoporphyrinogen IX oxidase (PPO; Matringe et al. 1992). Subsequent steps, leading ultimately to chlorophyll generation in the thylakoid membrane, occur within the plastid envelope and in the thylakoids, indicating that this spatial separation of events requires both correct distribution and transport of intermediates within the envelope and from the envelope to the thylakoids (Reinbothe and Reinbothe 1996). Using fluorimetric analysis (Lermontova and Grimm 2000) we examined the steady-state protoporphyrin IX levels in laf6 compared with WT seedlings. Figure 6 shows that laf6 seedlings have an approximately twofold increase in protoporphyrin IX levels compared with WT seedlings. Furthermore, spectrophotometric chlorophyll measurements (Gegenheimer 1990) demonstrated almost a twofold reduction in chlorophyll content in laf6 compared with WT (Fig. 6a). As a control, we performed the same analysis on complemented laf6 seedlings (co-laf6), which showed a restoration of WT protoporphyrin IX and chlorophyll levels (Fig. 6a).
Figure 6.
Protoporphyrin IX and chlorophyll levels in WT and laf6 seedlings. (a) Protoporphyrin IX (Proto IX) levels and chlorophyll content of WT (Ler), laf6, and a complemented transgenic line (co-laf6). The bar chart shows a representative experiment under FR-hf (Proto IX) and R-hf light (chlorophyll) conditions. (b) Protoporphyrin IX ratios between WT (Ler) and laf6 seedlings. The standard error (SE) and the number of experiments (n) are given below the ratios.
A PPO inhibitor can phenocopy laf6
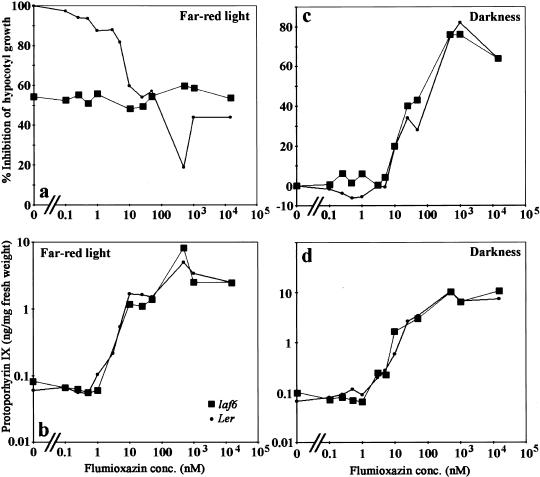
Inhibition of PPO should result in the export of protoporphyrinogen IX to the cytoplasm followed rapidly by cytoplasmic protoporphyrin IX accumulation caused by the nonspecific oxidation of protoporphyrinogen IX by plasma membrane-bound peroxidases (Jacobs and Jacobs 1993; Lee et al. 1993). The accumulation of protoporphyrin IX in laf6 suggests that inhibition of PPO should phenocopy laf6. To test this notion, we treated WT seedlings with a wide range (0.1–105 nM) of the PPO inhibitor flumioxazin (Arnould and Camadro 1998). As expected, we detected an increase in protoporphyrin IX levels with increasing flumioxazin concentration in FR light and in darkness (Fig. 7b,d). In darkness, increased protoporphyrin IX levels had a similar effect on both WT and laf6 seedlings (Fig. 7c) in that the hypocotyl length decreased in a concentration-dependent manner most probably caused by an inhibitory effect on cell elongation. In response to FR-hf light, however, WT seedlings treated with 1–100 nM flumioxazin displayed an increase in hypocotyl length similar to untreated laf6 seedlings (Fig. 7a). This indicates that the hypocotyl phenotype of laf6 can be phenocopied by artificially increasing cytoplasmic protoporphyrin IX levels in WT seedlings. Comparison of Figure 7a and 7b reveals a clear correlation between increased protoporphyrin IX levels and hypocotyl lengths in WT seedlings; as the protoporphyrin IX levels increase, the hypocotyl length increases in a dose-dependent manner. Interestingly, there was little change in hypocotyl lengths of the laf6 mutant (Fig. 7a) toward flumioxazin-induced protoporphyrin IX accumulation (Fig. 7b). This is likely to be because of the fact that laf6 seedlings already have increased protoporphyrin IX levels and that increasing the levels further has no effect on hypocotyl elongation. Taken together, these observations argue that the effect of increased protoporphyrin IX levels on hypocotyl length is FR-light specific, which corroborates the observed FR-specific hypocotyl phenotype of laf6 (Fig. 1a).
Figure 7.
Influence of increasing flumi oxazin concentrations on hypocotyl growth (percentage inhibition) of WT and laf6 seedlings in response to FR-hf light (a) and darkness (c) and on protoporphyrin IX levels (ng/mg fresh weight) in response to FR-hf light (b) and darkness (d). A 100% inhibition in a represents a hypocotyl length of ∼3.5 mm, and a 0% inhibition in (c) represents a hypocotyl length of ∼13 mm. Note that the Y-axes in b and d are given as log scales. Filled circles denote WT and filled squares denote laf6.
Increased atABC1 levels result in reduced sensitivity toward protoporphyrin IX
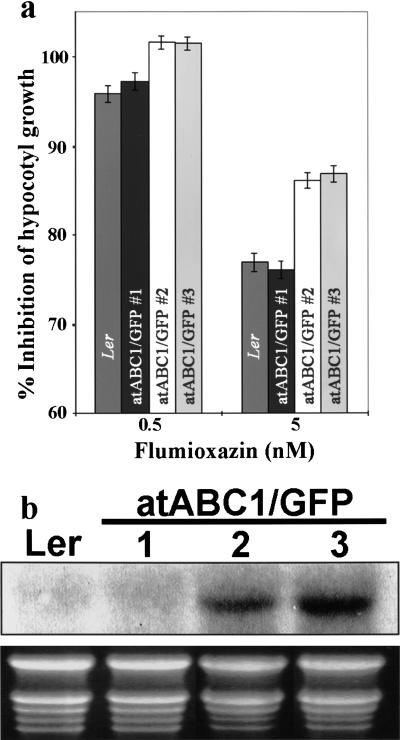
To further substantiate our findings, we examined what effect increased atABC1 levels have on FR-induced hypocotyl length and on protoporphyrin IX levels after flumioxazin treatment. To this end, we generated transgenic Arabidopsis seedlings overexpressing an atABC1/GFP fusion protein. To ensure that the overexpressed version of atABC1 localized to the correct subcellular compartment, we monitored GFP fluorescence in transgenic seedlings and found that the atABC1/GFP fusion protein localizes to the periphery of plastids (Fig. 5g), as was shown for the endogenous atABC1 (Fig. 5f1–f2). WT seedlings and transgenic seedlings overexpressing atABC1 (Fig. 8b; transgenic lines 2 and 3) were irradiated with FR-hf light in the presence of 0.5 and 5 nM flumioxazin (Fig. 8a) to artificially stimulate an increase in protoporphyrin IX levels. Figure 8a shows that transgenic Arabidopsis seedlings with increased atABC1 levels (atABC1/GFP 2 and 3) display a significant reduction in hypocotyl length compared to WT seedlings in response to flumioxazin treatment. Moreover, this effect is more prominent in response to 5 nM flumioxazin, which has been shown to increase protoporphyrin IX levels approximately fivefold compared with untreated seedlings (Fig. 7b). As an additional control, we measured the hypocotyl length in a transgenic line that showed the presence of the transgene (determined by PCR analysis; data not shown) but that did not overexpress the atABC1-GFP transcript (Fig. 8b, transgenic line 1) and found no significant difference in FR-light induced–hypocotyl length elongation compared to WT seedlings (Fig. 8a, atABC1/GFP 1).
Figure 8.
The effect of increased atABC1 levels on hypocotyl growth (percentage inhibition) after FR-hf irradiation in the presence of flumioxazin. (a) Bar chart showing inhibition of hypocotyl growth (percentage inhibition) of WT (Ler), of transgenic line 1 (atABC1/GFP number 1; not overexpressing atABC1/GFP, as shown in b), and of transgenic lines 2 and 3 (atABC1/GFP numbers 2 and 3; overexpressing atABC1/GFP as shown in b) after 0.5 and 5 nM flumioxazin treatment. The average absolute hypocotyl lengths (70–80 seedlings) after 5 mM flumioxazin treatment are: Ler; 5.3 mm, Line 1; 5.4 mm, Line 2; 4.4 mm; Line 3; 3.9 mm. Note that WT seedlings show ∼95% inhibition of hypocotyl growth after 0.5 nM flumioxazin, as shown in Figure 7a. (b) Expression analysis of the atABC1-GFP transcript in WT (Ler) and in transgenic lines 1, 2, and 3 (atABC1/GFP 1–3), using the atABC1 cDNA as a probe. The exposure time shown was not sufficient to detect the endogenous atABC1 transcript. Bottom panel shows stained rRNA profiles.
We also determined the effect of increased atABC1 levels on protoporphyrin IX accumulation. WT seedlings, transgenic line 1 (atABC1/GFP 1; not overexpressing atABC1/GFP) and transgenic lines 2 and 3 (atABC1/GFP 2 and 3; overexpressing atABC1/GFP) were treated with 100 nM flumioxazin. This concentration was chosen because we have shown that 100 nM flumioxazin leads to a large increase in protoporphyrin IX levels (Fig. 7b). Under these conditions, transgenic lines overexpressing atABC1–GFP (atABC1/GFP 2 and 3; Fig. 8b) had a significant reduction in protoporphyrin IX levels (2.2 ng/mg fresh weight and 2.3 ng/mg fresh weight) compared with WT seedlings (2.8 ng/mg fresh weight) and transgenic line 1 (2.7 ng/mg fresh weight; atABC1/GFP 1; Fig. 8b). The observed reduction of protoporphyrin IX levels in the transgenic lines overexpressing atABC1-GFP represent a significant effect, as this reflects a decrease of approximately two to three times the endogenous protoporphyrin IX amount in untreated seedlings. Taken together, these findings indicate that increased atABC1 levels result in decreased sensitivity of hypocotyl elongation toward flumioxazin, most probably because of decreased protoporphyrin IX accumulation.
Discussion
Arabidopsis mutants impaired in their responsiveness toward specific light conditions provide invaluable tools for the dissection of downstream components of light-signaling pathways. The disruption of atABC1 in laf6 results in a preferential insensitivity toward FR light, demonstrating that atABC1 serves as a component of the phyA signaling pathway. Several findings advocate this notion. First, laf6 seedlings show reduced responsiveness toward FR-hf light irradiation with an approximately twofold increase in hypocotyl length compared with WT seedlings and with a clear fluence dependency (Fig. 1a). The observed fluence dependency suggests that atABC1 is only rate limiting at high fluences of FR. Furthermore, laf6 seedlings show no detectable physiological differences compared with WT seedlings when grown in darkness or under R or B light (Fig. 1a). Second, laf6 seedlings have phyA levels similar to WT seedlings (Fig. 1d), and feeding experiments with chromophore showed no effect on the laf6 mutant phenotype. Third, the laf6 mutant phenotype can be complemented by introducing a genomic copy of atABC1 into the laf6 mutant, restoring the FR-inhibition of hypocotyl elongation characteristics of WT (Fig. 3). From these findings, we conclude that atABC1 acts as a regulatory component of phyA signaling and that the disruption of atABC1 in laf6 is responsible for the observed FR-specific mutant phenotype.
Expression of the light-regulated genes Chs, Fnr, and Lhcb is attenuated but not abolished in laf6 in response to FR-hf (Fig. 1b). Although the effect on Chs, Fnr, and Lhcb expression is greatest under FR-hf, there is also a decrease in gene expression, albeit small, in response to R-hf light (Fig. 1c), suggesting a minor effect on the phyB signaling pathway. Consistent with this notion, we found that in the absence of phyA, atABC1 can be induced by R light through phyB (Fig. 4).
Plant ABC transporters characterized to date belong to the classical membrane-bound ABC transporters comprising two integral membrane domains (MD-subunit) and two ATP-hydrolyzing domains (ABC-subunit; Lu et al. 1997, 1998; Holland and Blight 1999). In eukaryotes, the MD- and ABC-subunits normally exist as a single polypeptide chain, while in prokaryotes and lower eukaryotes, the two subunits are encoded by different genes (Schneider and Hunke 1998). On the basis of sequence homology, atABC1 encodes a novel plant ABC protein with no membrane-spanning domains but with high homology to ABC-like proteins from lower eukaryotes (Fig. 2b). The existence of a functional atABC1 transit peptide and the strikingly high amino acid sequence homology to ABC proteins from photosynthetic bacteria strongly suggests that the gene encoding atABC1 most probably relocated to the nucleus during an endosymbiotic event (McFadden 1999). This further suggests that the biological role of atABC1 may serve a conserved plastidic function.
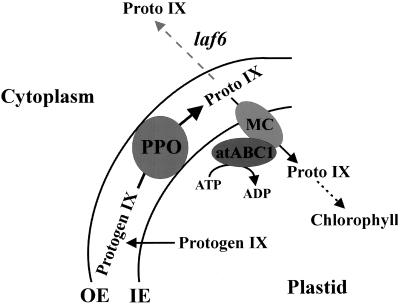
ABC proteins can either function as importers or exporters and are often promiscuous in terms of substrate preference (Holland and Blight 1999). In contrast to export systems, the ABC-subunit of most import systems is encoded as a separate protein, which in turn interacts with an integral membrane component to execute its function. Because atABC1 lacks membrane-spanning domains, it is likely involved in an import mechanism. There are two general models illustrating how ABC import systems may function (Holland and Blight 1999). In the first model, the ABC-subunit itself binds a compound to be imported then docks to its membrane component partner(s) followed by ATP hydrolysis and subsequent transport. In the second model, the compound to be imported binds to a membrane component followed by interaction with the ABC-subunit, which acts as an energy generator to fuel the transport process. During chlorophyll biosynthesis, protoporphyrinogen IX is relocated from the stroma to the plastid envelope, where it is oxidized by the envelope-bound PPO to form protoporphyrin IX (Fig. 9; Reinbothe and Reinbothe 1996). The generated protoporphyrin IX is then imported back into the stroma by yet an unknown mechanism, ultimately leading to the generation of chlorophyll (Fig. 9; Reinbothe and Reinbothe 1996). We show that atABC1 localizes to the envelope region of chloroplasts (Fig. 5) and that increased amounts of protoporphyrin IX in laf6 is a result of atABC1 deficiency (Fig. 6a). These findings indicate that atABC1 may be involved in the protoporphyrin IX reimport process or in the correct distribution of protoporphyrin IX within the plastid envelope (Fig. 9).
Figure 9.
A hypothetical model showing possible involvement of atABC1 in protoporphyrin IX transport and distribution. Protoporphyrinogen IX (Protogen IX) is transported to the plastid envelope, where it is oxidized by the membrane-bound protoporphyrinogen IX oxidase (PPO) to form protoporphyrin IX (Proto IX). Proto IX must then be reimported back into the stroma for chlorophyll biosynthesis to proceed. One possible mode of atABC1 action is to interact with an integral membrane component (MC) or a membrane complex and to fuel the import of Proto IX via ATP-dependent hydrolysis. In laf6, this import mechanism is impaired, leading to cytoplasmic accumulation of Proto IX as shown by the gray dotted arrow. The physiology of the laf6 mutant suggests that the increase in protoporphyrin IX levels can attenuate light responses. OE denotes outer envelope and IO, denotes inner envelope.
To see whether increasing the intracellular concentration of protoporphyrin IX in WT would phenocopy laf6, we treated WT seedlings with the herbicide flumioxazin, an inhibitor of PPO (Jacobs and Jacobs 1993; Lee et al. 1993; Arnould and Camadro 1998). We found that flumioxazin could indeed induce protoporphyrin IX accumulation in WT seedlings in a dose-dependent manner, and this was accompanied by a loss of hypocotyl growth inhibition in response to FR-hf light (Fig. 7a,b). Moreover, the laf6 mutant is not affected in terms of hypocotyl growth by flumioxazin-induced protoporphyrin IX accumulation (Fig. 7a,b). The observation that increased protoporphyrin IX levels has a similar effect on both WT and laf6 seedlings in darkness indicates that protoporphyrin IX can only exert its effect on hypocotyl elongation in the presence of light.
It is clear from Figures 6 and 7 that a several-fold increase in flumioxazin-induced protoporphyrin IX accumulation is required in WT seedlings, compared with laf6, to exert an effect on hypocotyl elongation. There are several possible reasons for this: first, inhibition of PPO results in protoporphyrin IX accumulation at the plasma membrane caused by the oxidation of protoporphyrinogen IX by plasma membrane-bound peroxidases (Jacobs and Jacobs 1993; Lee et al. 1993), while we suggest that protoporphyrin IX accumulation in laf6 is caused by incorrect transport or interenvelope distribution because of atABC1 deficiency. The differences in localization may, therefore, account for the differences in protoporphyrin IX levels required to exert the same biological effect. Second, although inhibition of PPO results in increased protoporphyrin IX levels, WT seedlings do have intact atABC1, which may lead to the reimport of accumulated protoporphyrin IX during PPO inhibition. This may, in effect, require a higher cytoplasmic protoporphyrin IX pool size to exert an effect on hypocotyl elongation. Finally, laf6 seedlings may be sensitized with respect to protoporphyrin IX levels compared with WT seedlings.
Herbicides, as do most inhibitors, may have the disadvantage of exerting unspecific effects. Although flumioxazin treatment clearly leads to increased protoporphyrin IX levels, there may be secondary toxic side-effects such as the observed decrease in hypocotyl length in dark-grown seedlings (Fig. 7c,d). To more firmly pinpoint the relationship between atABC1, hypocotyl length, and protoporphyrin IX levels, we generated transgenic plants overexpressing atABC1 and monitored the effect of increased atABC1 levels on FR-induced hypocotyl elongation and on protoporphyrin IX accumulation. According to our hypothesis, we expected that by increasing atABC1 levels there would be a concurrent decrease in the sensitivity toward elevated protoporphyrin IX levels. Using two different flumioxazin concentrations (0.5 and 5 nM) to artificially stimulate cytoplasmic accumulation of protoporphyrin IX, we compared hypocotyl elongation in WT seedlings to transgenic seedlings overexpressing atABC1 after FR-hf light irradiation. This analysis showed that transgenic seedlings with increased atABC1 levels show increased hypocotyl growth inhibition compared with WT (Fig. 8a). In addition, we have demonstrated that increased atABC1 levels result in decreased protoporphyrin IX accumulation. Taken together, these findings suggest that the availability and, moreover, the abundance of atABC1, determines the physiological sensitivity toward elevated protoporphyrin IX levels, which in turn regulates hypocotyl lengths in FR light.
The coordinated expression of both plastidic and nuclear genes requires a finely tuned light-regulated developmental program, which is dependent on precise communication between the nucleus and plastids. For instance, functional chloroplasts are required for the appropriate expression of several nuclear genes, and many studies have shown that the nucleus can sense the functional state of plastids and orchestrate subsequent morphological manifestations (Oelmuller et al. 1986; Mayfield 1990; Lopez-Juez et al. 1998; Streatfield et al. 1999). Indeed, herbicides that impair chloroplast development, such as Norflurazon, have been shown to repress the expression of Lhcb and Rbcs (Oelmuller 1989; Susek et al. 1993). Furthermore, chlorophyll precursors, such as porphyrin compounds, can prevent the accumulation of light-regulated transcripts in Chlamydomonas (Johanningmeier 1988). These observations led us to suggest that the increased protoporphyrin IX levels in laf6 may be responsible for the observed attenuation of light-regulated gene expression (Fig. 1b,c). Protoporphyrin IX may act as a plastid-derived signaling factor coordinating certain aspects of nuclear gene expression in response to the light environment. In the absence of plastid-localized atABC1, the increased protoporphyrin IX levels may act as a negative signal to attenuate the expression of nuclear encoded light-regulated genes.
Several models can be envisaged to explain the modes of action of atABC1 in terms of protoporphyrin IX transport and distribution. Based on the presence of a functional transit peptide, atABC1 is most probably located on the stromal side of the inner plastid envelope interacting with one or several integral membrane component(s) or with a membrane protein complex (Fig. 9). Soluble ABC proteins involved in import reactions can also insert themselves across membranes through proteinaceous complexes (Holland and Blight 1999). If this is the case for atABC1, exposure to the intermembrane space may allow atABC1 to bind protoporphyrin IX directly, facilitating its import via ATP-dependent translocation. Alternatively, atABC1 may not bind protoporphyrin IX and may simply serve as an ATP-driven energy generator for the import process (Fig. 9). Which of these hypotheses is correct remains to be determined.
Although one clear effect of atABC1 deficiency in laf6 is increased protoporphyrin IX accumulation, it is possible, based on the promiscuous nature of ABCs, that atABC1 is also involved in the transport of other compounds. In addition, other related transport systems are probably involved in protoporphyrin IX transport, as laf6 seedlings are able to accumulate chlorophyll, albeit at a lower efficiency.
In conclusion, atABC1 represents the first example of a plastid-localized protein involved in phyA signal transduction. One consequence of atABC1 deficiency is elevated protoporphyrin IX levels, which may be involved in the intercompartmental communication between plastids and the nucleus. The preferential specificity of laf6 toward FR light and the lack of physiological effects in darkness suggest further that elevated protoporphyrin IX levels most probably interact with the cytosolic light-signaling pathway or trigger a secondary pathway ultimately coordinating nuclear gene expression patterns. This interaction results in decreased inhibition of hypocotyl elongation in response to FR light and attenuated expression of light-regulated genes. Determining how atABC1 precisely coordinates the interplay between plastids and the nucleus, ultimately modifying gene expression affecting hypocotyl elongation, will be an exciting future challenge.
Materials and methods
Plant material and growth conditions
Ds insertion mutants were generated in Arabidopsis thaliana ecotype Landsberg erecta (Ler; Sundaresan et al. 1995). A phyA-deficient mutant (phyA-201) in ecotype Ler was used as a control in all experiments described. Unless otherwise stated, plant growth conditions and light sources were as described by Bolle et al. (2000).
Mutant screening and genetic analysis
On MS plates, 10,000 T2 Ds insertion Arabidopsis mutants were grown individually under high-fluence FR light (FR-hf; 5.5 μmol m−2) for 4 d and screened (Bolle et al. 2000) for reduction of hypocotyl growth inhibition compared with the isogenic WT. Twenty putative mutants were rescreened at the T3 generation and back-crossed to WT. F2 seedlings were then examined for cosegregation of kanamycin resistance (Ds-element associated) with the mutant phenotype (laf), and seedlings with a mutant phenotype were subjected to PCR amplification of the β-glucuronidase gene (Ds-element associated) using primers GUS1 (5′CCATTTG AAGCCGATGTCACGCCG-3′) and GUS2 (5′-CCAGTTGCAACCACCTGTTGATCC GG-3′) followed by Southern blotting using the entire Ds element as a probe. laf mutants have long hypocotyls after FR irradiation but, in contrast to pat mutants (Bolle et al. 2000), are unable to green in subsequent WL. Six laf mutants showed clear cosegregation of the Ds element with the mutant phenotype, and laf6 was selected for this study.
Characterization of the laf6 mutant
WT and laf6 mutant seedlings were grown on MS plates for 7 d at 22°C in darkness and under FR (high fluence 5.5 μmol m−2; low fluence 0.34 μmol m−2), R (high fluence 6.5 μmol m−2; low fluence 0.88 μmol m−2), and B light (8.5 μmol m−2 and 2 μmol m−2) followed by hypocotyl measurements. Each sample contained 60–80 seedlings.
Northern blot analysis
Total RNA was extracted using the RNeasy Plant mini kit (QIAGEN). Northern blots containing 15 μg of total RNA from WT and laf6 seedlings were hybridized to random primed [32P] cDNA probes encoding chalcone synthase (Chs; accession number AF112086), chlorophyll a/b binding protein (Lhcb; accession number X03908), and ferredoxin NADP+-oxidoreductase (Fnr; PCR amplified from genomic DNA) and atABC1. For analysis of light-induced expression profiles of atABC1, WT, phyA-deficient (phyA-201) and phyB-deficient (phyB-5) mutant seedlings were grown as described above under FR-hf and R-hf light.
PhyA extraction, immunoblotting and phycocyanobilin feeding
WT, laf6, and phyA-deficient (phyA-201) mutant seedlings were grown in darkness for 4 d, followed by total protein extraction, protein determination, SDS–PAGE analysis, and Western blotting as described by Kunkel et al. (1993). Phycocyanobilin extraction was performed as described by Kunkel et al. (1993), and feeding experiments were performed as described by Parks and Quail (1991).
Isolation of atABC1 and sequence analysis
Genomic DNA was extracted from laf6 seedlings using the NUCLEON DNA extraction kit (Vector labs). Inverse PCR was then performed using two primers designed within the left border of the Ds element (Ds3–1; 5′-CGATTACCGTATTTATC CCGTTCG-3′ and Ds3–2 5′; 5′-CGATCCGGTCGGGTTAAA GTCG-3′), enabling amplification of the genomic DNA flanking the Ds element. Reamplification was performed using a nested primer pair (Ds3–2; 5′-CCGGTATATCCCGTTTTCG-3′ and Ds3–1 5′; 5′-GGGAACCGGTATTTTTGTTCGG-3′), and the resulting 230-bp amplification product was cloned into pCRScript (Stratagene). Following DNA sequence analysis, database searches (U.S. National Center for Biotechnology Information), and BLAST searches (Altschul et al. 1990), we found that the Ds element was inserted 24 bp upstream of the translational start site of an ABC-like protein (atABC1; BAC T4B21; accession number AAD03441). Amino acid alignments were performed using the DNASTAR Megalign v. 4.0 software package. A full-length atABC1 1674-bp cDNA was isolated by RT–PCR (ProStar, Stratagene; ABC/1; 5′-TACTCGAGATGGCGT CTCTTCTCGCAAACGG-3′ and ABC/2; 5′-AAGTCGACC TCTAGATTAACCCACTGATCCTTCAAGC-3′) using Pwo Polymerase (Boehringer Mannheim).
To ensure that the cloned genomic flanking region truly represented the region adjacent to the Ds element, a combination of primers was designed inside the Ds element (β-glucuronidase and neomycin phosphotransferase II gene) and the cloned genomic flanking region. PCR amplification of four fragments spanning the two border regions (left-border and right-border) between the Ds element and the genomic flanking regions were cloned into pCRScript (Stratagene) and subjected to DNA sequencing.
Complementation analysis
A full-length genomic copy of atABC1 including ∼1200 bp of the upstream promoter region and ∼80 bp of the 3′UTR was amplified (ABC/3; 5′-AACCATGGCAGTAAACGCGGATTTGTG GTTAATCCCGG-3′ and ABC/4; 5′-CCTCTAGATTTATTGA GAAGAAGCACAAAGACCACCCC-3′), cloned into a promoterless version of the binary vector pBA002 (Kost et al. 1998), and transformed into laf6 seedlings by vacuum infiltration (Clough and Bent 1998). T1 transformants were selected, and T2 seedlings were analyzed by Northern blotting for the presence of the atABC1 transcript. Homozygous T3 seedlings showing the presence of the atABC1 transcript were grown under FR-hf light conditions, as for the initial screening procedure.
Heterologous expression of atABC1 and production of antisera
The full-length ORF of atABC1 was amplified by RT–PCR (Prostar, Stratagene; ABC/5; 5′-TAGGATCCATGGCGTCTC TTCTCGCAAACGG-3′ and ABC/6; 5′-AAGTCGAGCTCT AGATTAACCCACTGATCCTTCAAGC-3′) using Pwo polymerase (Boehringer Mannheim) and cloned into pQE30-1 (QIAGEN). The resulting construct was subjected to DNA sequencing and transformed into Escherichia coli BL21 (Novagen). Expression of full-length recombinant atABC1 was induced by 0.4 mM IPTG for 3 h, followed by affinity purification under denaturing conditions using Ni2+-agarose resin following standard procedures (QIAGEN). Anti-atABC1 antibodies were generated in white New Zealand rabbits following standard immunization protocols.
Analysis of atABC1 localization
The full-length atABC1 cDNA (atABC1), atABC1 lacking the transit sequence (atABC1–), and the 189-bp atABC1 transit sequence (atABC1/TP) were PCR amplified, removing the stop codon using Pwo polymerase (Boehringer Mannheim) using the following primer combinations (atABC1: ABC/7; 5′-TACTCG AGATGGCGTCTCTTCTCGCAAACGG-3′ and ABC/8; 5′ATGGTACCACCCACTGATCCTTCAAGC-3′, atABC1-: ABC/9; 5′-TACTCGAGATGGCTTCTGAATCATCATCATCAGG-3′ and ABC/8, atABC1/TP: ABC/7 and ABC/10; ATGGTACCTC CGATGGGACGAGAATCGATTCC-3′). The amplified fragments were cloned into pGFP2 (Kost et al. 1998) under the control of the CaMV35S promoter as translational fusions to the N terminus of GFP. The resulting constructs were transfected into onion epidermal cells by particle bombardment (Kost et al. 1998) and analyzed for GFP expression using a Zeiss LSM410 inverted confocal microscope. For Arabidopsis transformation, the entire CaMV35S/atABC1/GFP cassette was subcloned into a promoterless version of the binary vector pBA002 (Kost et al. 1998) and transformed into WT Arabidopsis seedlings as described earlier. Individual T2 seedlings were analyzed by Northern blotting for the presence of the atABC1/GFP fusion transcript using the both the atABC1 and GFP cDNAs as probes. Positive transgenic lines were subsequently analyzed for GFP fluorescence using a Zeiss LSM410 inverted confocal microscope.
In situ immunolocalization of atABC1 in WT and laf6 mutant leaves was performed in 5–8-μm thin sections after fixation in 2% paraformaldehyde and 2.5% glutaraldehyde and embedding in Historesin (Reichert and Jung). For immunofluorescence, atABC1 antiserum was diluted 1 : 200, and staining was performed using biotinylated anti-rabbit antibodies and fluorescein isothiocyanate (FITC)-labeled avidin (Vector labs). FITC fluorescence was viewed using an Axioskop microscope (Zeiss).
Flumioxazin treatment and analysis of protoporphyrin IX and chlorophyll levels
Stock solutions of flumioxazin were dissolved in DMSO. For herbicide experiments, flumioxazin was added to MS agar plates at 60°C to give final concentrations between 0.1 nM and 30 μM. Flumioxazin-containing plates were stored at 4°C in darkness. For each flumioxazin concentration, the hypocotyl lengths of 60 and 80 seedlings and the corresponding protoporphyrin IX amounts of four independent samples were measured. Protoporphyrin IX levels were determined as described by Lermontova and Grimm (2000) with slight modifications (based on emissions of 650 nm and calculated using standard protoporphyrin IX solutions). Note that the measured protoporphyrin IX levels presented vary slightly between independent experiments. Chlorophyll measurements were performed as described by Gegenheimer (1990).
Acknowledgments
We are very grateful to Venkatesan Sundaresan for providing Ds tagged lines generated at the Institute of Molecular Agrobiology, National University of Singapore. We thank Qi-Wen Niu for excellent technical assistance, Yan Wu for help with confocal microscopy, and Jean Wenger (Novartis) for flumioxazin. We thank members of Chua laboratory for critical reading of the manuscript and Cordelia Bolle and Peter D. Hare for fruitful discussions. S.G.M. is a recipient of a NATO Science Fellowship administered through the Norwegian Research Council. T.K. is a recipient of a Human Frontier Science Fellowship. Work done in our laboratory was supported in part by a grant (GM 44640) from The National Institute of Health to N.-H.C.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Chua@rockvax.rockefeller.edu; FAX (212) 327-8327.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.850101.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Arnould S, Camadro JM. The domain structure of protoporphyrinogen oxidase, the molecular target of diphenyl ether-type herbicides. Proc Natl Acad Sci. 1998;95:10553–10558. doi: 10.1073/pnas.95.18.10553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer J, Chen K, Hiltbunner A, Wehrli E, Eugster M, Schnell D, Kessler F. The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature. 2000;13:203–207. doi: 10.1038/35003214. [DOI] [PubMed] [Google Scholar]
- Bolle C, Koncz C, Chua N-H. PAT1, a new member of the GRAS family, is involved in phytochrome A signal transduction. Genes & Dev. 2000;14:1269–1278. [PMC free article] [PubMed] [Google Scholar]
- Bowler C, Neuhaus G, Yamagata H, Chua N-H. Cyclic GMP and calcium mediate phytochrome phototransduction. Cell. 1994;77:73–81. doi: 10.1016/0092-8674(94)90236-4. [DOI] [PubMed] [Google Scholar]
- Buche C, Poppe C, Schafer E, Kretsch T. eid1: A new Arabidopsismutant hypersensitive in phytochrome A-dependent high-irradiance responses. Plant Cell. 2000;12:547–558. [PMC free article] [PubMed] [Google Scholar]
- Chen X, Schnell DJ. Protein import into chloroplasts. Trends Cell Biol. 1999;9:222–227. doi: 10.1016/s0962-8924(99)01554-8. [DOI] [PubMed] [Google Scholar]
- Choi G, Yi H, Lee J, Kwon YK, Soh MS, Shin B, Luka Z, Hahn TR, Song PS. Phytochrome signalling is mediated through nucleoside diphosphate kinase 2. Nature. 1999;401:610–613. doi: 10.1038/44176. [DOI] [PubMed] [Google Scholar]
- Clough SJ, Bent AF. Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Chory J. Light control of plant development. Annu Rev Cell Dev Biol. 1998;13:203–229. doi: 10.1146/annurev.cellbio.13.1.203. [DOI] [PubMed] [Google Scholar]
- Fankhauser C, Yeh KC, Lagarias J C, Zhang H, Elich TD, Chory J. PKS1, a substrate phosphorylated by phytochrome that modulates light signaling in Arabidopsis. Science. 1999;284:1539–1541. doi: 10.1126/science.284.5419.1539. [DOI] [PubMed] [Google Scholar]
- Furuya M. Molecular properties and biogenesis of phytochrome I and II. Adv Biophys. 1989;25:133–167. doi: 10.1016/0065-227x(89)90006-3. [DOI] [PubMed] [Google Scholar]
- ————— Phytochromes: Their molecular species, gene families, and functions. Annu Rev Plant Physiol Plant Mol Biol. 1993;44:617–645. [Google Scholar]
- Gegenheimer P. Preparation of extracts from plants. Methods Enzymol. 1990;182:174–193. doi: 10.1016/0076-6879(90)82016-u. [DOI] [PubMed] [Google Scholar]
- Hoecker U, Xu Y, Quail PH. SPA1: A new genetic locus involved in phytochrome A–specific signal transduction. Plant Cell. 1998;10:19–33. doi: 10.1105/tpc.10.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoecker U, Tepperman JM, Quail PH. SPA1, a WD-repeat protein specific to phytochrome A signal transduction. Science. 1999;284:496–499. doi: 10.1126/science.284.5413.496. [DOI] [PubMed] [Google Scholar]
- Holland IB, Blight MA. ABC-ATPases, adaptable energy generators fuelling transmembrane movement of a variety of molecules in organisms from bacteria to humans. J Mol Biol. 1999;293:381–399. doi: 10.1006/jmbi.1999.2993. [DOI] [PubMed] [Google Scholar]
- Hsieh H-L, Okamoto H, Wang M, Ang L-H, Matsui M, Goodman H, Deng XW. FIN219, an auxin-regulated gene, defines a link between phytochrome A and the downstream regulator COP1 in light control of Arabidopsisdevelopment. Genes & Dev. 2000;14:1958–1970. [PMC free article] [PubMed] [Google Scholar]
- Hudson M, Ringli C, Boylan MT, Quail PH. The FAR1 locus encodes a novel nuclear protein specific to phytochrome A signaling. Genes & Dev. 1999;13:2017–2027. doi: 10.1101/gad.13.15.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs JM, Jacobs NJ. Porphyrin accumulation and export by isolated barely (Hordeum vulgareL.) plastids: Effect of diphenyl ether herbicides. Plant Physiol. 1993;101:1181–1188. doi: 10.1104/pp.101.4.1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johanningmeier U. Possible control of transcript levels by chlorophyll precursors in Chlamydomonas. Eur J Biochem. 1988;177:417–424. doi: 10.1111/j.1432-1033.1988.tb14391.x. [DOI] [PubMed] [Google Scholar]
- Kendrick RE, Kronenberg GHM. Photomorphogenesis in Plants. 2nd ed. Dordrecht: Kluwer; 1994. [Google Scholar]
- Kost B, Spielhofer P, Chua N-H. A GFP-mouse talin fusion protein labels plant actin filaments in vivo and visualises the actin cytoskeleton in growing pollen tubes. Plant J. 1998;16:393–401. doi: 10.1046/j.1365-313x.1998.00304.x. [DOI] [PubMed] [Google Scholar]
- Kunkel T, Tomizawa KI, Kern R, Furuya M, Chua N-H, Schafer E. In vitro formation of a photoreversible adduct of phycocyanobilin and tobacco apophytochrome B. Eur J Biochem. 1993;215:587–594. doi: 10.1111/j.1432-1033.1993.tb18069.x. [DOI] [PubMed] [Google Scholar]
- Lee HJ, Duke MV, Duke SO. Cellular localization of protoporphyrin-oxidizing activities of etiolated barely (Hordeum vulgareL.) leaves. Plant Physiol. 1993;102:881–889. doi: 10.1104/pp.102.3.881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lermontova I, Grimm B. Overexpression of plastidic protoporphyrinogen IX oxidase leads to resistance to the diphenyl-ether herbicide acifluorfen. Plant Physiol. 2000;122:75–84. doi: 10.1104/pp.122.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Juez E, Paul Jarvis R, Takeuchi A, Page AM, Chory J. New Arabidopsiscue mutants suggest a close connection between plastid- and phytochrome regulation of nuclear gene expression. Plant Physiol. 1998;118:803–815. doi: 10.1104/pp.118.3.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YP, Li ZS, Rea PA. AtMRP1 gene of Arabidopsisencodes a glutathione S-conjugate pump: Isolation and functional definition of a plant ATP-binding cassette transporter gene. Proc Natl Acad Sci. 1997;94:8243–8248. doi: 10.1073/pnas.94.15.8243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu YP, Li Z-S, Drozdowicz YM, Hortensteiner S, Martinoia E, Rea PA. AtMRP2, an ArabidopsisATP binding cassette transporter able to transport glutathione S-conjugates and chlorophyll catabolites: Functional comparisons with Atmrp1. Plant Cell. 1998;10:267–282. doi: 10.1105/tpc.10.2.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matringe M, Camadro JM, Block MA, Joyard J, Scalla R, Labbe P, Douce R. Localisation within chloroplasts of protoporphyrinogen oxidase, the target enzyme for diphenylether-like herbicides. J Biol Chem. 1992;267:4646–4651. [PubMed] [Google Scholar]
- Mayfield SP. Chloroplast gene regulation: Interaction of the nuclear and chloroplast genomes in the expression of photosynthetic proteins. Curr Opin Cell Biol. 1990;2:509–513. doi: 10.1016/0955-0674(90)90135-2. [DOI] [PubMed] [Google Scholar]
- McFadden GI. Endosymbiosis and evolution of the plant cell. Curr Opin Plant Biol. 1999;2:513–519. doi: 10.1016/s1369-5266(99)00025-4. [DOI] [PubMed] [Google Scholar]
- Neff MM, Fankhauser C, Chory J. Light: An indicator of time and place. Genes & Dev. 2000;14:257–271. [PubMed] [Google Scholar]
- Neuhaus G, Bowler C, Kern R, Chua N-H. Calcium/calmodulin-dependent and -independent phytochrome signal transduction pathways. Cell. 1993;73:937–952. doi: 10.1016/0092-8674(93)90272-r. [DOI] [PubMed] [Google Scholar]
- Ni M, Tepperman JM, Quail PH. PIF3, a phytochrome-interacting factor necessary for normal photoinduced signal transduction, is a novel basic helix–loop–helix protein. Cell. 1998;95:657–667. doi: 10.1016/s0092-8674(00)81636-0. [DOI] [PubMed] [Google Scholar]
- Oelmuller R. Photooxidative destruction of chloroplasts and its effect on nuclear gene expression and extraplastidic enzyme levels. Photochem Photobiol. 1989;49:229–239. [Google Scholar]
- Oelmuller R, Levitan I, Bergfeld R, Rajasekhar VK, Mohr H. Expression of nuclear genes are affected by treatments acting on the plastids. Planta. 1986;168:482–492. doi: 10.1007/BF00392267. [DOI] [PubMed] [Google Scholar]
- Parinov S, Sevugan M, De Ye, Yang WC, Kumaran M, Sundaresan V. Analysis of flanking sequences from dissociation insertion lines: A database for reverse genetics in Arabidopsis. Plant Cell. 1999;11:2263–2270. doi: 10.1105/tpc.11.12.2263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parks BM, Quail PH. Phytochrome-deficient hy1 and hy2 long hypocotyl mutants of Arabidopsisare defective in phytochrome chromophore biosynthesis. Plant Cell. 1991;3:1177–1186. doi: 10.1105/tpc.3.11.1177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt LH. Distribution and localization of phytochrome within the plant. In: Kendrick RE, Kronenberg GHM, editors. Photomorphogenesis in plants. 2nd ed. Dordrecht: Kluwer; 1994. [Google Scholar]
- Quail PH, Boylan MT, Parks BM, Short TW, Xu Y, Wagner D. Phytochromes: Photosensory perception and signal transduction. Science. 1995;268:675–680. doi: 10.1126/science.7732376. [DOI] [PubMed] [Google Scholar]
- Reinbothe S, Reinbothe C. Regulation of chlorophyll biogenesis in angiosperm. Plant Physiol. 1996;111:1–7. doi: 10.1104/pp.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider E, Hunke S. ATP-binding-cassette (ABC) transport systems: Functional and structural aspects of the ATP-hydrolyzing subunits/domains. FEMS Microbiol Rev. 1998;22:1–20. doi: 10.1111/j.1574-6976.1998.tb00358.x. [DOI] [PubMed] [Google Scholar]
- Soh MS, Hong SH, Hanzawa H, Furuya M, Nam HG. Genetic identification of FIN2, a far red light specific signaling component of Arabidopsis thaliana. Plant J. 1998;16:411–419. doi: 10.1046/j.1365-313x.1998.00307.x. [DOI] [PubMed] [Google Scholar]
- Streatfield SJ, Weber A, Kinsman EA, Hausler RE, Li J, Post-Beittenmiller D, Kaiser WM, Pyke KA, Flugge UI, Chory J. The phosphoenolpyruvate/phosphate translocator is required for phenolic metabolism, palisade cell development, and plastid-dependent nuclear gene expression. Plant Cell. 1999;11:1609–1622. doi: 10.1105/tpc.11.9.1609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaresan V, Springer P, Volpe T, Haward S, Jones JD, Dean C, Ma H, Martienssen R. Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes & Dev. 1995;9:1797–1810. doi: 10.1101/gad.9.14.1797. [DOI] [PubMed] [Google Scholar]
- Susek R, Ausubel FM, Chory J. Signal transduction mutants of Arabidopsisuncouple nuclear CAB and RBSC gene expression from chloroplast development. Cell. 1993;74:787–799. doi: 10.1016/0092-8674(93)90459-4. [DOI] [PubMed] [Google Scholar]
- Whitelam GC, Devlin PF. Roles of different phytochromes in Arabidopsisphotomorphogenesis. Plant Cell Environ. 1997;20:752–758. [Google Scholar]
- Whitelam GC, Johnson E, Peng J, Carol P, Anderson ML, Cowl JS, Harberd NP. Phytochrome A null mutants of Arabidopsisdisplay a wild-type phenotype in white light. Plant Cell. 1993;5:757–768. doi: 10.1105/tpc.5.7.757. [DOI] [PMC free article] [PubMed] [Google Scholar]