Abstract
Epigenetic changes in tumours are associated not only with cancer development and progression, but also with resistance to chemotherapy. Aberrant DNA methylation at CpG islands and associated epigenetic silencing are observed during the acquisition of drug resistance. However, it remains unclear whether all of the observed changes are drivers of drug resistance, causally associated with response of tumours to chemotherapy, or are passenger events representing chance DNA methylation changes. Systematic approaches that link DNA methylation and expression with chemosensitivity will be required to identify key drivers. Such drivers will be important prognostic or predicitive biomarkers, both to existing chemotherapies, but also to epigenetic therapies used to modulate drug resistance.
Keywords: CpG islands, DNA methylation, drug resistance, epigenetics, histones, ovarian cancer, therapies
Introduction
Although there have been substantial advances in current chemotherapeutic strategies, clinical drug resistance remains a major obstacle to successful cancer treatment and is still a limiting factor in patient survival [Broxterman et al. 2009]. This problem is particularly obvious in the treatment of ovarian cancer [Agarwal and Kaye, 2003]. Although around 80% of ovarian cancer patients initially respond to primary chemotherapy, the majority will relapse and eventually develop resistance to currently available treatment options. Conventionally, standard treatment consists of platinum-based drugs (carboplatin/cisplatin) which are either given as a single agent or in combination with the taxane, paclitaxel [Greenlee et al. 2001]. The chemotherapeutic activity of platinum drugs is mainly based on their ability to form DNA adducts and the 1,2-d (GpG) intrastrand crosslink has been particularly implicated. Here, the platinum coordinates the N7 atoms of adjacent guanosines in the DNA strand, which is believed to be poorly repaired in cells, with persistence of the lesion ultimately interfering with replication and transcription leading to cell death [Wang and Lippard, 2005; Kartalou and Essigmann, 2001]. Ovarian cancer is defined clinically as being ‘platinum resistant’ if the tumour recurrence occurs less than 6 months after completion of platinum-based first-line chemotherapy. An improved response to second-line treatment is seen if the tumour recurs later than 6 months following the end of first-line treatment and chances for a ‘platinum-sensitive’ response are greatly enhanced if the relapse occurs after 12 months.
CpG island methylation and drug resistance
A number of genetic alterations have been suggested to underlie the phenomenon of drug resistance, such as alterations in genes involved in DNA repair, drug uptake, apoptosis and cell cycle checkpoints [Broxterman et al. 2009; Luqmani, 2005]. However, in recent years it has become increasingly apparent that aberrant epigenetic mechanisms may also play a crucial role in drug resistance (for a summary of epigenetic mechanisms, see Figure 1). DNA methylation is one of the major epigenetic mechanisms controlling gene expression and cell differentiation [Bird, 1996]. Regions of high CpG dinucleotide density, called CpG islands, are frequently located in the promoters of house-keeping genes and are usually free of methylation in normal cells [Bird, 2002]. In cancer, CpG islands can become hypermethylated, contributing for example to silencing of tumour suppressor genes. This has been demonstrated for cancer susceptibility genes such as BRCA1 [Press et al. 2008]. Furthermore, methylation of the CpG island linked to BRCA1 is associated with good response to platinum-based chemotherapy in ovarian cancer [Teodoridis et al. 2005]. Mutation of the BRCA1/BRCA2 genes is also associated with good response to platinum-based chemotherapy [Cass et al. 2003]. Conversely, reversion mutations at BRCA2 have been shown to be associated with platinum resistance [Edwards et al. 2008]. It remains to be established whether reversal of BRCA methylation is a mechanism of acquired resistance. In ovarian cancer cell lines, methylation of the FANCF gene has been observed to be associated with increased sensitivity to cisplatin [Taniguchi et al. 2003]. FANCF is crucial for the activation of a DNA repair complex containing BRCA1 and BRCA2. Treatment with the demethylating agent 2′-deoxy-5-azacytidine led to demethylation of the FANCF gene and reduced sensitivity towards cisplatin in these cell line models [Taniguchi et al. 2003]; however, again, the relevance of FANCF methylation to clinical outcome following chemotherapy is still to be established.
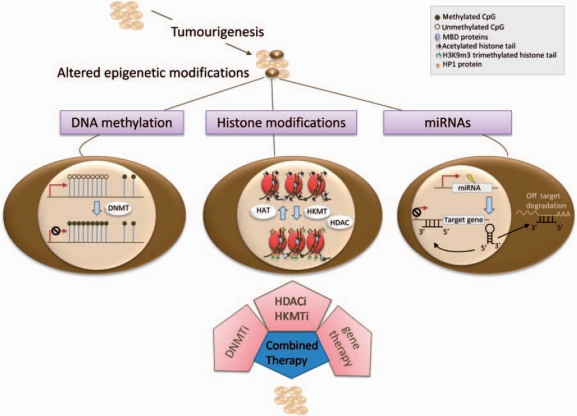
Figure 1.
Possible targets for epigenetic therapy approaches in chemoresistant cells. Aberrant DNA hypermethylation (dark circles) at CpG islands can lead to transcriptional inactivation of genes and is frequently observed in tumours. Inhibition of the enzymes catalysing DNA methylation (DNMTs) leads to a genome-wide decrease of DNA methylation levels, thereby potentially re-activating vital anticancer genes. Each histone modification is established via specific enzymes (HDACs and HKMTs) catalysing the addition or removal of certain marks. Targeting aberrant hypoacetylation via HDACi can result in the re-expression of former transcriptionally incompetent chromatin. Similarly, the inhibition of certain aberrantly active histone methyltransferases (HKMTs) prevents methyl marks which may lead to repression of genes, as seen following the binding of HP1 (orange circles) to H3K9me3. Next to the two major epigenetic mechanisms of DNA methylation and histone modification, aberrant expression of miRNAs (small non-protein-coding RNAs of 21–23 nucleotides) has been correlated with tumourigenesis. Here, miRNAs potentially act on two pathways, the transcriptional silencing mechanism and the translational silencing mechanism. If in either case the miRNAs promiscuously bind sequences, for instance via mutation, the former specific gene regulation is out of control, possibly contributing to tumourigenesis and chemoresistance. In ovarian cancer, for example, downregulation of the miRNA let-7i increases resistance to cisplatin and is associated with shorter progression-free survival time of patients with late-stage ovarian cancer [Yang et al. 2008]. Although not yet available, gene therapy may be a tool to either re-establish lost endogenous miRNA expression or silence aberrant miRNA expression thereby complementing existing epigenetic therapies.
In contrast to BRCA and FANCF where epigenetic or genetic inactivation of the gene is associated with drug sensitivity, inactivation of genes involved in engaging an apoptotic response would lead to drug resistance [Teodoridis et al. 2005]. For instance, methylation of the DNA mismatch repair gene MLH1 and transcriptional silencing occurs in cisplatin-resistant ovarian cell line models. MLH1 has been shown to be necessary for engagement of a variety of downstream cellular responses to alkylating agents and cisplatin-induced DNA damage [Papouli et al. 2004; Stojic et al. 2004]. It has been argued that, since mismatch repair (MMR) proteins can recognize and bind to certain types of damage in DNA, this is necessary for MMR-dependent engagement of DNA damage responses such as activation of p53, p73 and other downstream apoptosis signalling pathways [Stojic et al. 2004; Shimodaira et al. 2003; Duckett et al. 1999]. Hence, loss of MLH1 expression may lead to reduced engagement of apoptosis either due to reduced cycles of futile repair [Karran and Hampson, 1996], reduced stalling (or increased bypass) of lesions in DNA during DNA replication [Moreland et al. 1999] or direct signalling of cell death pathways [Yoshioka et al. 2006]. Acquired methylation at the MLH1 locus has been observed following platinum-based chemotherapy in ovarian cancer, which has been shown to be associated with poor patient survival [Gifford et al. 2004].
Increased methylation of DAPK might also have an implication in chemoresistance. Hypermethylation of DAPK occurs frequently in tumours such as colon and breast tumours [Yamaguchi et al. 2003; Lehmann et al. 2002] and is an indicator of poor clinical outcome in lung cancer patients [Tang et al. 2000]. The calcium/calmodulin-regulated serine/threonin kinase DAPK has a pro-apoptotic role which is mediated via interferon (IFN)-γ, Fas and tumour necrosis factor (TNF)-related apoptosis-inducing ligand (TRAIL) [Tang et al. 2004; Cohen et al. 1999; Inbal et al. 1997]. Highlighting the potential role of DAPK in chemoresistance, it has been shown that TRAIL-resistant lung cancer cell lines can be resensitized by using DNMT inhibitors leading to demethylation and re-expression of DAPK [Tang et al. 2004]. Following onto this, in gastric cancer patients the promoter methylation of DAPK was also shown to correlate with shorter progression-free survival and lower response rates to chemotherapeutic treatment with 5-fluorouracil [Kato et al. 2008]. Consequently, the methylation of members of the apoptotic and anti-apoptotic machinery such as DAPK have the potential to influence apoptosis and hence chemosensitivity.
Epigenetic drug resistance drivers
It has been proposed that drug resistance is a polygenic phenotype caused by the cumulative effect of multiple drug resistance or sensitivity associated genes, rather than phenotypic effects on drug sensitivity of a few loci [Glasspool et al. 2006]. Indeed large numbers of gene expression changes are observed in tumours and during acquired drug resistance [Konstantinopoulos et al. 2008], as well as extensive changes in DNA methylation, which then may lead to the drug-resistant phenotype. One of the current challenges is to identify the key changes which drive a cell towards a drug-resistant phenotype among the vast majority of epigenetic changes caused, for instance, by global demethylation or localized hypermethylation of CpG islands [Ehrlich, 2006]. Many of these global changes could have occurred by chance either as part of a methylator phenotype or simply as random silencing events [Issa, 2004]. In other words: how do we discriminate a ‘driver’ which, in analogy to tumourigenesis, has a functional effect that provides the cell with a selective advantage from a ‘passenger’ that has no functional impact but was incidentally acquired [Greenman et al. 2007]?
For driver mutations of cancer it has been estimated that only 8–16% of all reported mutations within protein kinase genes represent true drivers [Carter et al. 2009; Greenman et al. 2007]. Moreover, a substantial number of these changes are only present at a low frequency [Wood et al. 2007]. By analogy, it might be only a very specific subset of epigenetic changes transforming a sensitive tumour cell into a resistant one. Few large-scale studies have addressed the issue of acquired drug resistance in ovarian cancer on an epigenome-wide basis. Dai et al. [2008] analysed methylation changes associated with acquired cisplatin resistance in isogenic ovarian cancer cell lines and showed that hypermethylation of CpG islands is prevalent during the acquisition of drug resistance. To take this further, Li and colleagues [2009] combined methylation with expression data and also identified extensive hypermethylation associated with epigenetic repression of loci implicated in cell adhesion as well as hypomethylation associated with activation of genes involved in the PI3K/AKT, transforming growth factor (TGF)-beta and cell cycle progression pathways. However, to pinpoint the epigenetic changes driving chemoresistance more precisely, it will be necessary to systematically investigate combined methylation, gene expression and resensitization in order to identify key drivers of the resistance phenotype. As an example, MLH1 epigenetic silencing is selected for during platinum treatment of ovarian tumour cell lines and re-expression of MLH1 either by demethylation or gene re-introduction resensitizes ovarian tumour cells to subsequent chemotherapeutic treatment [Plumb et al. 2000] and might, therefore, represent one of the key genes driving chemoresistance in the cell line models examined. However, this gives no indication of how many other genes may be key drivers of resistance, either in this model or in tumours in general. The actual proportion of epigenetically altered genes driving chemoresistance is still elusive. For future studies, systematic approaches will be required confirming a substantial effect of these methylation changes in terms of their ability to reverse drug resistance. If a small subset of genes drives drug resistance, this would be a valuable set of biomarkers for stratifying patients who may benefit from epigenetic approaches to resensitize tumours to chemotherapy.
Epigenetic modulators of drug resistance
It is widely recognized that a wide variety of epigenetic changes are prevalent in cancer [Jones and Baylin, 2007]. Importantly, unlike genetic mutations, epigenetic alterations are reversible and, therefore, the executing enzymes provide attractive drug targets for new therapies. Growing understanding of the mechanisms and enzymes governing epigenetic regulation has resulted in a variety of new drugs and possible drug targets for cancer treatment. These epigenetic therapies have the potential to re-activate or silence aberrantly regulated genes, thereby reversing many aspects of cancer phenotypes, including drug resistance. Examples of epigenetic therapies undergoing preclinical and clinical evaluation or registered for use in certain cancers are shown in Table 1.
Table 1.
Classes and names of DNA methylation inhibitors and histone deacetylase inhibitors, their targets and clinical status.
| Type of epigenetic therapy | Class of compound | Compound | Target | Development stage |
|---|---|---|---|---|
| DNA methylation inhibitor | Nucleoside analogue | 5′-azacytidine (Vidaza) | DNMTs | Approved for MDS, CMML and AML |
| 5-aza-2′-deoxycytidine (Decitabine/ Dacogen) | DNMTs | Approved for MDS | ||
| Zebularine | DNMTs | Preclinical | ||
| Non-nucleoside analogue | RG 108 | DNMT1 | Preclinical | |
| Procaine | DNMT1 | Preclinical | ||
| Procainamide | DNMT1 | Preclinical | ||
| Hydralazine | DNMT1 | Phase II | ||
| Antisense | MG98 | DNMT1 | Phase II | |
| Histone deacetylase inhibitor | Hydroxamate | Suberoylanilide hydroxamic acid (SAHA, Vorinostat) | Class I, II | Approved for advanced CTCL |
| PXD101 | Class I, II | Phase II | ||
| LAQ824, LBH589 | Class I, II | Phase I | ||
| Trichostatin A | Class I, II | Preclinical | ||
| Oxamflatin, Scriptaid, SBHA | N/A | Preclinical | ||
| Pyroxamide | Class I, unknown effect on class II | Phase I | ||
| SK-7041, SK-7068 | HDAC1, 2 | Preclinical | ||
| Tubacin | HDAC6 | Preclinical | ||
| Aliphatic acid | Valproic acid (VPA) | Class I, II | Phase II, III | |
| Phenylbutyrate | Class I, II | Phase I, II | ||
| Savicol | N/A | Phase I, II | ||
| AN-9 (prodrug) | N/A | Phase I, II | ||
| Baceca | Class I | Phase I, II | ||
| Benzamide | MS-275 (SNDX-275) | HDAC1, 2, 3 and slightly 8 | Phase I, II | |
| MGCD0103 | HDAC1, 2, 3, 11 | Phase I, II | ||
| Cyclic tetrapeptide | Depsipeptide (FK228) | Class I | Phase I, II | |
| Trapoxin A | Class I, II | Preclinical | ||
| Apicidin | HDAC1, 3 | Preclinical | ||
| CHAPs | Class I | Preclinical |
AML, acute myeloid leukaemia; CHAP, cyclic hydroxamic-acid-containing peptide; CMML, chronic myelomonocytic leukaemia; CTCL, cutaneous T-cell lymphoma; DNMT, DNA methyltransferase; DNMTi, DNMT inhibitor; HDAC, histone deacetylase; HDACi, HDAC inhibitor; MDS, myelodysplastic syndrome; N/A, not available; SBHA, suberic bishydroxamic acid. Adapted from Bolden et al. [2006], Yoo and Jones [2006] and Xu et al. [2007].
DNA methylation inhibitors
So far, DNA methylation inhibitors (DNMTi) represent the most widely studied class of epigenetic therapeutic agents. The US Food and Drug Administration (FDA) approved 5-azacytidine (Vidaza) and Decitabine (5-aza-2′-deoxycytidine, Dacogen), cytidine ribose and deoxyribose nucleoside analogues, respectively [Issa, 2007]. Both drugs exert their effect by becoming incorporated into DNA, and inhibiting the DNA methyltransferase (DNMT) by forming covalent adducts with the enzyme, leading to its sequestration and cellular depletion. As a result of reduced DNMT activity during subsequent cell division DNA methylation is increasingly diminished. Consequently, cytosine methylation is reduced in newly replicated DNA, but not in the DNA of resting or non-dividing cells. In addition, Decitabine has been shown to induce depletion of DNMT1 through proteosomal degradation [Ghoshal et al. 2005]. In cell line models, these nucleoside DNA methylation inhibitors proved to effect methylation and to re-activate epigenetically silenced tumour suppressor genes [Mund et al. 2006]. Importantly, Decitabine induced re-expression of genes known to mediate drug response, for example MLH1, resulting in enhanced chemosensitivity to cytotoxic drugs [Plumb et al. 2000].
Vidaza and Decitabine have been clinically tested and have shown substantial therapeutic potential in haematological cancers such as leukaemias [Fenaux et al. 2009; Stewart et al. 2009; Issa et al. 2005; Issa et al. 2004]. A randomized phase III trial with myelodysplastic syndrome (MDS) patients supported the potency of 5-azacytidine to improve overall survival time of patients when compared with other therapies [Fenaux et al. 2009]. However, the effectiveness of DNMTi in solid tumours has been limited so far [Graham et al. 2009]. Several reasons may account for this, including their short half-life in plasma combined with the relatively low proliferation rate of solid tumours cells, limiting the amount of aza-nucleosides becoming incorporated into DNA. To overcome this problem, treatments with prolonged exposures to demethylating agents at lower doses may be necessary, in order to reverse methylation and restore gene activity rather than triggering cytotoxicity as seen with higher doses of DNMTi [Issa et al. 2004].
Furthermore, recent clinical trials have aimed to optimize currently available treatments by combining those with DNMTis to enhance susceptibility to conventional chemotherapeutic agents. For example, Decitabine has been studied as a modulator of resistance in combination with carboplatin in solid tumours in a phase I clinical trial [Appleton et al. 2007]. Encouragingly, demethylation rates were reported in peripheral blood cells as indicated by MAGE1A promoter methylation levels. Following on from the initial success, a randomized phase II trial was started in patients with recurrent ovarian cancer that had progressed within 6–12 months following the first round of platinum therapy [Glasspool et al. 2009]. However, the study was closed after a planned interim analysis due to poor feasibility and lack of efficacy of the combination. One possibility for the lack of efficacy seen in the phase II trial may be due to reduced dose intensity of the carboplatin and/or due to the additional myelosuppression associated with the Decitabine treatment. In order for these agents to progress within the solid tumour setting, it will be important to develop agents that are less myelosuppressive in combination with an altered schedule. For example, studies in haematological malignancies have used a 1 h infusion of Decitabine over 5–10 days. Regarding ovarian cancer, studies are currently underway combining platinum chemotherapy with similar schedules [Matei and Nephew, 2010]. Another feasible explanation may be that the demethylation of certain genes leads to an adverse effect counteracting the carboplatin sensitizing effect of demethylation. Again highlighting the importance of patient selection based on their tumour methylation profile for treatments and future studies.
More recently, much attention has been focused on developing small molecule inhibitors of DNMTs. RG 108 is one example of such a nonnucleoside compound which inhibits DNA methylation by directly blocking the active site of the enzyme [Brueckner et al. 2005]. RG 108 has been shown to restore the activity of epigenetically silenced tumour suppressor genes such as p16Ink4 in human colon cancer cell lines, notably without the drug-induced toxicity usually observed with traditional DNMTi [Stresemann et al. 2006; Brueckner et al. 2005]. However, further improvement of the drug is highly desirable in terms of its cell uptake and efficacy [Brueckner et al. 2007]. The potential of non-nucleoside inhibitors to more specifically inhibit DNMTs, without causing non-specific DNA damage associated with nucleoside DNMT inhibitors, suggests that this class of compounds may have potential to be less-toxic therapies. However, clinical studies are needed to reveal more about their clinical applicability.
Histone modulators
Histone deacetylase inhibitors
The second intensively studied class of drugs targeting epigenetic silencing mechanisms are the histone deacetylase inhibitors (HDACis). Histone deacetylases (HDACs) can regulate the chromatin conformation through the removal of acetyl groups from the lysine residue of histone tails. Inhibiting these enzymes promotes accumulation of the acetylated form of histone proteins, ultimately leading to less-condensed packaging of genes in chromatin which may lead to the re-expression of silenced tumour suppressor genes. However, HDACis also target various other proteins, including key molecules regulating tumour cell growth, which may be responsible for the observed induction of different phenotypes in transformed cells such as proliferation arrest, differentiation and apoptosis [Egger et al. 2004]. Although the actual mechanism of growth inhibition might not necessarily be due to acetylation of histones per se, the fact that normal cells are usually not affected by HDACi-induced cell death at low concentrations supports their suitability as specific anticancer agents [Warrener et al. 2003]. Currently, a variety of up to 15 different HDACis are under investigation in phase I–III clinical trials [Marks and Xu, 2009].
The only HDACi approved by the FDA is Vorinostat (SAHA), a hydroxide acid derivative, registered for treatment of cutaneous T-cell lymphoma (since 2006). Vorinostat has shown proven efficacy in the treatment of haematologic malignancies [Garcia-Manero et al. 2008; Duvic et al. 2007; Olsen et al. 2007]. It has also been tested as a single agent in clinical phase I/II trials in various solid tumours including head and neck, breast and thyroid cancer. However, response rates suggested that the activity of this HDACi is low in solid tumour types [Batty et al. 2009]. Recently, Vorinostat has been shown to enhance the efficacy of the cytotoxic agents carboplatin and paclitaxel in patients with advanced non-small cell lung cancer in the setting of a clinical phase II trial [Ramalingam et al. 2010]. However, due to increased toxicity optimization of the schedule seems to be required for this combination. The synergy of HDACi and conventional chemotherapeutic agents might be a promising route for the treatment of solid tumours.
Despite the initial encouraging results, monotherapy with DNMTis is limited due to the observed toxicity and the eventual re-methylation of genes [Plumb et al. 2004]. Now, there is growing interest in combined epigenetic therapies involving DNMTis and HDACis. For example, the DNMTi treatment can act synergistically with an HDACi in restoring gene expression [Cameron et al. 1999]. It has been proposed that DNA hypermethylation can lead to compact nucleosomes resistant to acetylation, thereby dominating silencing. A sequential administration of DNMTi prior to the HDACi administration appears to be important for sufficient efficacy in the treatment of solid tumours. Preclinical studies and early clinical trials are exploring the combination of epigenetic remodelling agents [Griffiths and Gore, 2008]. For example, the sequential treatment with Decitabine and the hydroxamate HDACi, PXD101 (Belinostat), has been tested in a cisplatin-resistant human ovarian tumour xenograft model [Steele et al. 2009; Plumb et al. 2004]. Combined treatment resulted in a marked increase in expression of epigenetically silenced MLH1 and MAGE1A compared with treatment with Decitabine alone supporting the idea that combinatorial epigenetic therapy might improve sensitivity to chemotherapeutic agents. A recent clinical phase I/II trial combining the short fatty acid Valproic acid (VPA) (HDACi) with Decitabine was done in patients with leukaemia [Garcia-Manero et al. 2006]. Here, it emerged that this combination led to a transient reversal of epigenetic marks such as methylation of the p15 promoter and deacetylation of histone H3 and H4. However, neither the level of DNA demethylation nor the level of acetylation could be correlated to clinical response.
Histone methyltransferase inhibitors
The discovery of histone methyltransferases (HKMT) has opened up another promising avenue to target aberrant epigenetic mechanisms [Lachner et al. 2003]. HKMTs are chromatin-modifying enzymes which establish methyl marks on the lysine of histone tail proteins. Depending on the position of the methyl mark the histone code can be read from binding proteins which then lead to transcriptional activation or repression of genes. Deregulation of HKMT activity has been linked to tumour development, especially high levels of the H3K27 methyltransferase EZH2 (polycomb group protein, enhancer of zeste homologue) have recently been observed in a variety of tumours including prostate, breast and melanoma and are associated with poor prognosis [Yu et al. 2007; Bachmann et al. 2006; Varambally et al. 2002]. Subsequent efforts to find appropriate inhibitors for EZH2 led to the discovery of 3-deazaneplanocin A (DZNep). DZNep is an S-adenosylhomocysteine hydrolase inhibitor capable of depleting EZH2 and other components of the Polycomb Repressive Complex 2 (PRC2) from cells in vitro and induces apoptosis in breast cancer cells [Tan et al. 2007]. However, subsequent studies showed that DZNep is not specific to EZH2 but is generally inhibiting methyltransferase activity thereby affecting global histone methylation levels [Miranda et al. 2009]. This is most likely due to its indirect mechanism of inhibition which blocks S-adenosyl methionine (SAM)-dependent methyltransferases through byproduct inhibition.
Nevertheless, DZNep is being explored to improve the epigenetic effect of HDACi at an early preclinical stage. Results seem encouraging, for example, treatment of an acute myeloid leukaemia cell line with combined DZNep and the HDACi panobinostat highly increased rates of apoptotic cell death in comparison with treatment with either drug alone [Fiskus et al. 2009]. However, there is a need for the development of novel, more selective inhibitors of HKMT which is supported by studies performed in glioblastoma multiform, an aggressive form of malignant glioma. Here, it was demonstrated that pharmacologic disruption of EZH2 by DZNep strongly impaired glioblastoma multiforme cancer stem cell self-renewal in vitro and tumour-initiating capacity in vivo due to repression of c-myc, a proto-oncogene mediating cell growth [Suva et al. 2009]. Therefore, the development of novel inhibitors with greater biological specificity is highly sought and may provide valuable tools for more targeted therapies.
Also in light of combined epigenetic therapy the current efforts to develop compounds targeting more distinct classes of epigenetic enzymes should greatly enhance their efficacy. The pleiotropic effects observed with anticancer agents already in the clinic can often be a drawback and limit their value in therapy. More selectively acting compounds should aid in overcoming toxic effects currently seen. Combination studies indicate that sequential administration of drugs against different classes of enzymes might also increase efficacy of treatment. Although still at an early stage, future inhibitors of HKMT may represent a new class of compounds which could offer more selectivity compared with the currently available options.
Conclusions
It is becoming increasingly clear that epigenetic changes occurring during the acquisition of drug resistance are substantial and complex and might even outnumber genetic alterations. There are clear examples of the potential of epigenetic alterations at specific loci affecting drug resistance. However, in order to successfully target abnormal epigenetic changes in the clinic, it will be necessary to comprehensively identify the key events driving this process. Consequently, a therapy specifically targeting epigenetically deregulated genes could help in overcoming resistance. Longitudinal studies monitoring tumours acquiring resistance during the course of chemotherapy could provide an opportunity to map genome-wide epigenetic changes which may allow the correlation to acquisition of drug resistance. However, the magnitude of changes and heterogeneity among tumours will make such analyses challenging. Here, it will be crucial to perform epigenetic profiling of an appropriately sized panel of tumours pre- and postchemotherapy in order to achieve sufficient power for statistical analysis. In addition, those large-scale studies will also greatly expand the repertoire of available biomarkers which are vital for stratification of patients and for monitoring efficacy of existing epigenetic therapies.
Current efforts to isolate and characterize so-called cancer sustaining (stem) cells may yield new insights into genetic and epigenetic mechanisms governing chemoresistance [Ferrandina et al. 2008; Zhang et al. 2008; Agarwal and Kaye, 2003]. One of the features of such tumour sustaining cells is their inherent resistance to a number of chemotherapeutics, making them a suitable model for drug-resistance studies. Cancer sustaining cells have stem-cell-like properties in the way that they have the capacity to recapitulate the original tumour and promote recurrence.
Although successfully used in haematological malignancies DNMTi and HDACi work on a multitude of targets. The concern is that the genome-wide re-expression of aberrantly and normally regulated genes could lead to conflicting effects concerning response to chemotherapy. Therefore, the development of more targeted (epigenetic) therapies might be a prerequisite of successfully preventing drug resistance. The idea of reversing the malignant epigenetic marks of a small subset of genes driving drug resistance in order to sensitize tumours to chemotherapy will require novel strategies based on our increased understanding of the complexities of epigenetic regulation.
Funding
This research received funding from Cancer Research UK.
Conflict of interest statement
The authors declare they have no conflicts of interest.
References
- Agarwal R., Kaye S.B. (2003) Ovarian cancer: strategies for overcoming resistance to chemotherapy. Nat Rev Cancer 3: 502–516 [DOI] [PubMed] [Google Scholar]
- Appleton K., Mackay H.J., Judson I., Plumb J.A., McCormick C., Strathdee G., et al. (2007) Phase I and pharmacodynamic trial of the DNA methyltransferase inhibitor decitabine and carboplatin in solid tumors. J Clin Oncol 25: 4603–4609 [DOI] [PubMed] [Google Scholar]
- Bachmann I.M., Halvorsen O.J., Collett K., Stefansson I.M., Straume O., Haukaas S.A., et al. (2006) EZH2 expression is associated with high proliferation rate and aggressive tumor subgroups in cutaneous melanoma and cancers of the endometrium, prostate, and breast. J Clin Oncol 24: 268–273 [DOI] [PubMed] [Google Scholar]
- Batty N., Malouf G.G., Issa J.P. (2009) Histone deacetylase inhibitors as anti-neoplastic agents. Cancer Lett 280: 192–200 [DOI] [PubMed] [Google Scholar]
- Bird A. (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16: 6–21 [DOI] [PubMed] [Google Scholar]
- Bird A.P. (1996) The relationship of DNA methylation to cancer. Cancer Surv 28: 87–101 [PubMed] [Google Scholar]
- Bolden J.E., Peart M.J., Johnstone R.W. (2006) Anticancer activities of histone deacetylase inhibitors. Nat Rev Drug Discov 5: 769–784 [DOI] [PubMed] [Google Scholar]
- Broxterman H.J., Gotink K.J., Verheul H.M. (2009) Understanding the causes of multidrug resistance in cancer: a comparison of doxorubicin and sunitinib. Drug Resist Updat 12: 114–126 [DOI] [PubMed] [Google Scholar]
- Brueckner B., Garcia Boy R., Siedlecki P., Musch T., Kliem H.C., Zielenkiewicz P., et al. (2005) Epigenetic reactivation of tumor suppressor genes by a novel small-molecule inhibitor of human DNA methyltransferases. Cancer Res 65: 6305–6311 [DOI] [PubMed] [Google Scholar]
- Brueckner B., Kuck D., Lyko F. (2007) DNA methyltransferase inhibitors for cancer therapy. Cancer J 13: 17–22 [DOI] [PubMed] [Google Scholar]
- Cameron E.E., Bachman K.E., Myohanen S., Herman J.G., Baylin S.B. (1999) Synergy of demethylation and histone deacetylase inhibition in the re-expression of genes silenced in cancer. Nat Genet 21: 103–107 [DOI] [PubMed] [Google Scholar]
- Carter H., Chen S., Isik L., Tyekucheva S., Velculescu V.E., Kinzler K.W., et al. (2009) Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations. Cancer Res 69: 6660–6667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cass I., Baldwin R.L., Varkey T., Moslehi R., Narod S.A., Karlan B.Y. (2003) Improved survival in women with BRCA-associated ovarian carcinoma. Cancer 97: 2187–2195 [DOI] [PubMed] [Google Scholar]
- Cohen O., Inbal B., Kissil J.L., Raveh T., Berissi H., Spivak-Kroizaman T., et al. (1999) DAP-kinase participates in TNF-alpha- and Fas-induced apoptosis and its function requires the death domain. J Cell Biol 146: 141–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai W., Teodoridis J.M., Graham J., Zeller C., Huang T.H., Yan P., et al. (2008) Methylation Linear Discriminant Analysis (MLDA) for identifying differentially methylated CpG islands. BMC Bioinformatics 9: 337–337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duckett D.R., Bronstein S.M., Taya Y., Modrich P. (1999) hMutSalpha and MutLalpha dependent phosphorylation of p53 in response to DNA methylator damage. Proc Natl Acad Sci USA 96: 12384–12388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duvic M., Talpur R., Ni X., Zhang C., Hazarika P., Kelly C., et al. (2007) Phase 2 trial of oral vorinostat (suberoylanilide hydroxamic acid, SAHA) for refractory cutaneous T-cell lymphoma (CTCL). Blood 109: 31–39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edwards S.L., Brough R., Lord C.J., Natrajan R., Vatcheva R., Levine D.A., et al. (2008) Resistance to therapy caused by intragenic deletion in BRCA2. Nature 451: 1111–1115 [DOI] [PubMed] [Google Scholar]
- Egger G., Liang G., Aparicio A., Jones P.A. (2004) Epigenetics in human disease and prospects for epigenetic therapy. Nature 429: 457–463 [DOI] [PubMed] [Google Scholar]
- Ehrlich M. (2006) Cancer-linked DNA hypomethylation and its relationship to hypermethylation. Curr Top Microbiol Immunol 310: 251–274 [DOI] [PubMed] [Google Scholar]
- Fenaux P., Mufti G.J., Hellstrom-Lindberg E., Santini V., Finelli C., Giagounidis A., et al. (2009) Efficacy of azacitidine compared with that of conventional care regimens in the treatment of higher-risk myelodysplastic syndromes: a randomised, open-label, phase III study. Lancet Oncol 10: 223–232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrandina G., Bonanno G., Pierelli L., Perillo A., Procoli A., Mariotti A., et al. (2008) Expression of CD133-1 and CD133-2 in ovarian cancer. Int J Gynecol Cancer 18: 506–514 [DOI] [PubMed] [Google Scholar]
- Fiskus W., Wang Y., Sreekumar A., Buckley K.M., Shi H., Jillella A., et al. (2009) Combined epigenetic therapy with the histone methyltransferase EZH2 inhibitor 3-deazaneplanocin A and the histone deacetylase inhibitor panobinostat against human AML cells. Blood 114: 2733–2743 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manero G., Kantarjian H.M., Sanchez-Gonzalez B., Yang H., Rosner G., Verstovsek S., et al. (2006) Phase 1/2 study of the combination of 5-aza-2′-deoxycytidine with valproic acid in patients with leukemia. Blood 108: 3271–3279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Manero G., Yang H., Bueso-Ramos C., Ferrajoli A., Cortes J., Wierda W.G., et al. (2008) Phase 1 study of the histone deacetylase inhibitor vorinostat (suberoylanilide hydroxamic acid [SAHA]) in patients with advanced leukemias and myelodysplastic syndromes. Blood 111: 1060–1066 [DOI] [PubMed] [Google Scholar]
- Ghoshal K., Datta J., Majumder S., Bai S., Kutay H., Motiwala T., et al. (2005) 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol 25: 4727–4741 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Gifford G., Paul J., Vasey P.A., Kaye S.B., Brown R. (2004) The acquisition of hMLH1 methylation in plasma DNA after chemotherapy predicts poor survival for ovarian cancer patients. Clin Cancer Res 10: 4420–4426 [DOI] [PubMed] [Google Scholar]
- Glasspool R.M., Gore M., Rustin G., McNeish I., Wilson R., Pledge S., et al. (2009) Randomized phase II study of decitabine in combination with carboplatin compared with carboplatin alone in patients with recurrent advanced ovarian cancer. J Clin Oncol 27: 15s–15s [Google Scholar]
- Glasspool R.M., Teodoridis J.M., Brown R. (2006) Epigenetics as a mechanism driving polygenic clinical drug resistance. Br J Cancer 94: 1087–1092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graham J.S., Kaye S.B., Brown R. (2009) The promises and pitfalls of epigenetic therapies in solid tumours. Eur J Cancer 45: 1129–1136 [DOI] [PubMed] [Google Scholar]
- Greenlee R.T., Hill-Harmon M.B., Murray T., Thun M. (2001) Cancer statistics, 2001. CA Cancer J Clin 51: 15–36 [DOI] [PubMed] [Google Scholar]
- Greenman C., Stephens P., Smith R., Dalgliesh G.L., Hunter C., Bignell G., et al. (2007) Patterns of somatic mutation in human cancer genomes. Nature 446: 153–158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths E.A., Gore S.D. (2008) DNA methyltransferase and histone deacetylase inhibitors in the treatment of myelodysplastic syndromes. Semin Hematol 45: 23–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inbal B., Cohen O., Polak-Charcon S., Kopolovic J., Vadai E., Eisenbach L., et al. (1997) DAP kinase links the control of apoptosis to metastasis. Nature 390: 180–184 [DOI] [PubMed] [Google Scholar]
- Issa J.P. (2004) CpG island methylator phenotype in cancer. Nat Rev Cancer 4: 988–993 [DOI] [PubMed] [Google Scholar]
- Issa J.P. (2007) DNA methylation as a therapeutic target in cancer. Clin Cancer Res 13: 1634–1637 [DOI] [PubMed] [Google Scholar]
- Issa J.P., Garcia-Manero G., Giles F.J., Mannari R., Thomas D., Faderl S., et al. (2004) Phase 1 study of low-dose prolonged exposure schedules of the hypomethylating agent 5-aza-2′-deoxycytidine (decitabine) in hematopoietic malignancies. Blood 103: 1635–1640 [DOI] [PubMed] [Google Scholar]
- Issa J.P., Gharibyan V., Cortes J., Jelinek J., Morris G., Verstovsek S., et al. (2005) Phase II study of low-dose decitabine in patients with chronic myelogenous leukemia resistant to imatinib mesylate. J Clin Oncol 23: 3948–3956 [DOI] [PubMed] [Google Scholar]
- Jones P.A., Baylin S.B. (2007) The epigenomics of cancer. Cell 128: 683–692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karran P., Hampson R. (1996) Genomic instability and tolerance to alkylating agents. Cancer Surveys 28: 69–85 [PubMed] [Google Scholar]
- Kartalou M., Essigmann J.M. (2001) Recognition of cisplatin adducts by cellular proteins. Mutat Res 478: 1–21 [DOI] [PubMed] [Google Scholar]
- Kato K., Iida S., Uetake H., Takagi Y., Yamashita T., Inokuchi M., et al. (2008) Methylated TMS1 and DAPK genes predict prognosis and response to chemotherapy in gastric cancer. Int J Cancer 122: 603–608 [DOI] [PubMed] [Google Scholar]
- Konstantinopoulos P.A., Spentzos D., Cannistra S.A. (2008) Gene-expression profiling in epithelial ovarian cancer. Nat Clin Pract Oncol 5: 577–587 [DOI] [PubMed] [Google Scholar]
- Lachner M., O'Sullivan R.J., Jenuwein T. (2003) An epigenetic road map for histone lysine methylation. J Cell Sci 116: 2117–2124 [DOI] [PubMed] [Google Scholar]
- Lehmann U., Celikkaya G., Hasemeier B., Langer F., Kreipe H. (2002) Promoter hypermethylation of the death-associated protein kinase gene in breast cancer is associated with the invasive lobular subtype. Cancer Res 62: 6634–6638 [PubMed] [Google Scholar]
- Li M., Balch C., Montgomery J.S., Jeong M., Chung J.H., Yan P., et al. (2009) Integrated analysis of DNA methylation and gene expression reveals specific signaling pathways associated with platinum resistance in ovarian cancer. BMC Med Genomics 2: 34–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luqmani Y.A. (2005) Mechanisms of drug resistance in cancer chemotherapy. Med Princ Pract 14(Suppl 1): 35–48 [DOI] [PubMed] [Google Scholar]
- Marks P.A., Xu W.S. (2009) Histone deacetylase inhibitors: Potential in cancer therapy. J Cell Biochem 107: 600–608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matei D.E., Nephew K.P. (2010) Epigenetic therapies for chemoresensitization of epithelial ovarian cancer. Gynecol Oncol 116: 195–201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miranda T.B., Cortez C.C., Yoo C.B., Liang G., Abe M., Kelly T.K., et al. (2009) DZNep is a global histone methylation inhibitor that reactivates developmental genes not silenced by DNA methylation. Mol Cancer Ther 8: 1579–1588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreland N.J., Illand M., Kim Y.T., Paul J., Brown R. (1999) Modulation of drug resistance mediated by loss of mismatch repair by the DNA polymerase inhibitor aphidicolin. Cancer Res 59: 2102–2106 [PubMed] [Google Scholar]
- Mund C., Brueckner B., Lyko F. (2006) Reactivation of epigenetically silenced genes by DNA methyltransferase inhibitors: basic concepts and clinical applications. Epigenetics 1: 7–13 [DOI] [PubMed] [Google Scholar]
- Olsen E.A., Kim Y.H., Kuzel T.M., Pacheco T.R., Foss F.M., Parker S., et al. (2007) Phase IIb multicenter trial of vorinostat in patients with persistent, progressive, or treatment refractory cutaneous T-cell lymphoma. J Clin Oncol 25: 3109–3115 [DOI] [PubMed] [Google Scholar]
- Papouli E., Cejka P., Jiricny J. (2004) Dependence of the cytotoxicity of DNA-damaging agents on the mismatch repair status of human cells. Cancer Res 64: 3391–3394 [DOI] [PubMed] [Google Scholar]
- Plumb J.A., Steele N., Finn P.W., Brown R. (2004) Epigenetic approaches to cancer therapy. Biochem Soc Trans 32: 1095–1097 [DOI] [PubMed] [Google Scholar]
- Plumb J.A., Strathdee G., Sludden J., Kaye S.B., Brown R. (2000) Reversal of drug resistance in human tumor xenografts by 2′-deoxy-5-azacytidine-induced demethylation of the hMLH1 gene promoter. Cancer Res 60: 6039–6044 [PubMed] [Google Scholar]
- Press J.Z., De Luca A., Boyd N., Young S., Troussard A., Ridge Y., et al. (2008) Ovarian carcinomas with genetic and epigenetic BRCA1 loss have distinct molecular abnormalities. BMC Cancer 8: 17–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramalingam S.S., Maitland M.L., Frankel P., Argiris A.E., Koczywas M., Gitlitz B., et al. (2010) Carboplatin and Paclitaxel in combination with either vorinostat or placebo for first-line therapy of advanced non-small-cell lung cancer. J Clin Oncol 28: 56–62 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimodaira H., Yoshioka-Yamashita A., Kolodner R.D., Wang J.Y. (2003) Interaction of mismatch repair protein PMS2 and the p53-related transcription factor p73 in apoptosis response to cisplatin. Proc Natl Acad Sci U S A 100: 2420–2425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steele N., Finn P., Brown R., Plumb J.A. (2009) Combined inhibition of DNA methylation and histone acetylation enhances gene re-expression and drug sensitivity in vivo. Br J Cancer 100: 758–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart D.J., Issa J.P., Kurzrock R., Nunez M.I., Jelinek J., Hong D., et al. (2009) Decitabine effect on tumor global DNA methylation and other parameters in a phase I trial in refractory solid tumors and lymphomas. Clin Cancer Res 15: 3881–3888 [DOI] [PubMed] [Google Scholar]
- Stojic L., Mojas N., Cejka P., Di Pietro M., Ferrari S., Marra G., et al. (2004) Mismatch repair-dependent G2 checkpoint induced by low doses of SN1 type methylating agents requires the ATR kinase. Genes Dev 18: 1331–1344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stresemann C., Brueckner B., Musch T., Stopper H., Lyko F. (2006) Functional diversity of DNA methyltransferase inhibitors in human cancer cell lines. Cancer Res 66: 2794–2800 [DOI] [PubMed] [Google Scholar]
- Suva M.L., Riggi N., Janiszewska M., Radovanovic I., Provero P., Stehle J.C., et al. (2009) EZH2 is essential for glioblastoma cancer stem cell maintenance. Cancer Res 69: 9211–9218 [DOI] [PubMed] [Google Scholar]
- Tan J., Yang X., Zhuang L., Jiang X., Chen W., Lee P.L., et al. (2007) Pharmacologic disruption of Polycomb-repressive complex 2-mediated gene repression selectively induces apoptosis in cancer cells. Genes Dev 21: 1050–1063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang X., Khuri F.R., Lee J.J., Kemp B.L., Liu D., Hong W.K., et al. (2000) Hypermethylation of the death-associated protein (DAP) kinase promoter and aggressiveness in stage I non-small-cell lung cancer. J Natl Cancer Inst 92: 1511–1516 [DOI] [PubMed] [Google Scholar]
- Tang X., Wu W., Sun S.Y., Wistuba I.I., Hong W.K., Mao L. (2004) Hypermethylation of the death-associated protein kinase promoter attenuates the sensitivity to TRAIL-induced apoptosis in human non-small cell lung cancer cells. Mol Cancer Res 2: 685–691 [PubMed] [Google Scholar]
- Taniguchi T., Tischkowitz M., Ameziane N., Hodgson S.V., Mathew C.G., Joenje H., et al. (2003) Disruption of the Fanconi anemia-BRCA pathway in cisplatin-sensitive ovarian tumors. Nat Med 9: 568–574 [DOI] [PubMed] [Google Scholar]
- Teodoridis J.M., Hall J., Marsh S., Kannall H.D., Smyth C., Curto J., et al. (2005) CpG island methylation of DNA damage response genes in advanced ovarian cancer. Cancer Res 65: 8961–8967 [DOI] [PubMed] [Google Scholar]
- Varambally S., Dhanasekaran S.M., Zhou M., Barrette T.R., Kumar-Sinha C., Sanda M.G., et al. (2002) The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature 419: 624–629 [DOI] [PubMed] [Google Scholar]
- Wang D., Lippard S.J. (2005) Cellular processing of platinum anticancer drugs. Nat Rev Drug Discov 4: 307–320 [DOI] [PubMed] [Google Scholar]
- Warrener R., Beamish H., Burgess A., Waterhouse N.J., Giles N., Fairlie D., et al. (2003) Tumor cell-selective cytotoxicity by targeting cell cycle checkpoints. FASEB J 17: 1550–1552 [DOI] [PubMed] [Google Scholar]
- Wood L.D., Parsons D.W., Jones S., Lin J., Sjoblom T., Leary R.J., et al. (2007) The genomic landscapes of human breast and colorectal cancers. Science 318: 1108–1113 [DOI] [PubMed] [Google Scholar]
- Xu W.S., Parmigiani R.B., Marks P.A. (2007) Histone deacetylase inhibitors: molecular mechanisms of action. Oncogene 26: 5541–5552 [DOI] [PubMed] [Google Scholar]
- Yamaguchi S., Asao T., Nakamura J., Ide M., Kuwano H. (2003) High frequency of DAP-kinase gene promoter methylation in colorectal cancer specimens and its identification in serum. Cancer Lett 194: 99–105 [DOI] [PubMed] [Google Scholar]
- Yang N., Kaur S., Volinia S., Greshock J., Lassus H., Hasegawa K., et al. (2008) MicroRNA microarray identifies Let-7i as a novel biomarker and therapeutic target in human epithelial ovarian cancer. Cancer Res 68: 10307–10314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo C.B., Jones P.A. (2006) Epigenetic therapy of cancer: past, present and future. Nat Rev Drug Discov 5: 37–50 [DOI] [PubMed] [Google Scholar]
- Yoshioka K., Yoshioka Y., Hsieh P. (2006) ATR kinase activation mediated by MutSalpha and MutLalpha in response to cytotoxic O6-methylguanine adducts. Mol Cell 22: 501–510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Rhodes D.R., Tomlins S.A., Cao X., Chen G., Mehra R., et al. (2007) A polycomb repression signature in metastatic prostate cancer predicts cancer outcome. Cancer Res 67: 10657–10663 [DOI] [PubMed] [Google Scholar]
- Zhang S., Balch C., Chan M.W., Lai H.C., Matei D., Schilder J.M., et al. (2008) Identification and characterization of ovarian cancer-initiating cells from primary human tumors. Cancer Res 68: 4311–4320 [DOI] [PMC free article] [PubMed] [Google Scholar]