Abstract
Mammalian U2AF65 and UAP56 are required for prespliceosome (PS) formation. We tested the predictions that the yeast UAP56 homolog, SUB2, is required for the same step and functions collaboratively with MUD2, the yeast homolog of U2AF65. Unexpectedly, sub2-1 extracts accumulate PS-like complexes. Moreover, deletion of MUD2 exacerbates the cs phenotype of sub2 alleles yet suppresses both the ts sub2-1 and the lethal Δsub2 phenotypes. We propose that Sub2 functionally interacts with Mud2 both before and after PS formation. In the absence of Mud2, Sub2 function becomes dispensable.
Keywords: Pre-mRNA splicing, Sub2, Mud2, DEAD-box protein
The assembly, function, and disassembly of the spliceosome is a multistep, ATP-dependent pathway that requires the participation of at least seven members of the “DEAD-box” protein family (Staley and Guthrie 1998). These proteins exhibit RNA-stimulated ATP binding/hydrolysis activity and, in some cases, ATP-dependent RNA unwinding (Wagner et al. 1998; Wang et al. 1998). In several cases, these ATPases appear to catalyze conformational rearrangements that result in mutually exclusive RNA switches (Staley and Guthrie 1998), providing a plausible strategy for the precise coordination of this multistep pathway. However, the molecular mechanisms by which these factors function remain unclear.
The earliest ATP-dependent step in spliceosome assembly in both yeast and mammals is the basepairing of the U2 snRNA with the branchpoint region of the intron to form the prespliceosome (PS) (Konarska and Sharp 1986, 1987; Cheng and Abelson 1987; Liao et al. 1992; Das et al. 2000). In mammals, this event requires prior binding of the branchpoint region by branchpoint binding protein, mBBP/SF1, and of the pyrimidine tract by U2AF65 in an ATP-independent, early (E) complex (Ruskin et al. 1988; Kramer and Utans 1991; Michaud and Reed 1991). Although the U2 snRNP has been detected in the mammalian E complex, mutations in the branchpoint sequence (BPS) that affect base pairing do not affect this association, suggesting that the U2 present in E complex is not base paired with the BPS (Das et al. 2000). It is likely that the sequence-specific binding of mBBP is mutually exclusive with the subsequent base pairing of the U2 snRNA with the BPS (Berglund et al. 1997, 1998). An additional factor, the U2AF65-associated protein of 56kD, UAP56, is a DEAD-box protein that is also required in mammals for U2 snRNP addition in vitro (Fleckner et al. 1997). The mechanism by which UAP56 promotes PS formation in mammals is unknown. It has been proposed to facilitate the association of the U2 snRNP with the pre-mRNA substrate through either affecting the availability of the branchpoint base-pairing region of the U2 snRNA or by modulating the accessibility of the branchpoint region of the pre-mRNA substrate (Fleckner et al. 1997).
Yeast contain homologs of both BBP, yBBP/ScSF1 (Abovich and Rosbash 1997), and U2AF65, Mud2 (Abovich et al. 1994). These factors bind the BPS in the absence of ATP to form the commitment complex (CC2) before PS formation (Abovich et al. 1994; Abovich and Rosbash 1997). The DEAD-box protein Prp5 is required for stable, ATP-dependent association of the U2 snRNP with the BPS to form the PS (Ruby et al. 1993). Available data suggest that Prp5 promotes a conformational rearrangement of the U2 snRNP (O'Day et al. 1996; Wiest et al. 1996). It was originally proposed that UAP56 was the mammalian counterpart of Prp5 (Fleckner et al. 1997). Subsequently, as we describe here, SUB2 has been identified as the UAP56 homolog. Moreover, a human homolog of Prp5 has recently been identified (C. Newnham and C. Query, pers. comm.). Thus, two ATPases may be required to promote efficient PS formation during splicing in both yeast and mammals.
Here, we report biochemical and genetic analyses that suggest that Sub2 may play multiple roles in the splicing pathway, both before and after PS formation. Remarkably, the essential function(s) of Sub2 can be partially bypassed by deletion of the nonessential MUD2 gene, suggesting that in the absence of this likely protein target, the essential function of Sub2 becomes dispensable.
Results and Discussion
SUB2 encodes an essential homolog of human UAP56
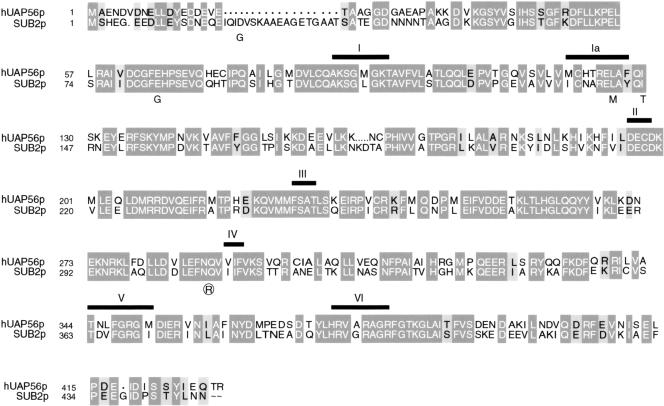
We initially identified the suppressor of brr1-1 (SUB2) in a selection for high copy suppressors of the cold-sensitive growth defect of this snRNP biogenesis mutant (Noble 1995; Noble and Guthrie 1996). SUB2 encodes a DEAD-box protein with 63% identity and 78% similarity to human UAP56 (Fig. 1). This homology extends beyond the conserved helicase domain of the two proteins (Fig. 1). Moreover, this level of homology is comparable to that previously reported for yeast and human spliceosomal DEXH-box proteins with demonstrated similarity in function (Ono et al. 1994; Gee et al. 1997; Zhou and Reed 1998).
Figure 1.
Sub2 is highly homologous to UAP56. Alignment of UAP56 and Sub2. Sequences were aligned with CLUSTALW and MacBoxShade. Dark gray boxes highlight identical amino acids, light gray boxes highlight similar residues. Thick black bars above the aligned sequences highlight the seven conserved motifs (I, Ia, II–VI) of helicase superfamily 2. Mutations present in sub2-1 (plain letters) and sub2-5 (circled letters) are indicated below the altered residue.
To test the prediction that SUB2 encodes a functional homolog of UAP56, we first examined the impact of a loss of Sub2 function. Previous systematic deletion studies in yeast have shown that deletion of the SUB2 ORF has variable effects on viability depending on strain background (Lopez et al. 1998). In our hands, deletion of SUB2 is lethal in two different strain backgrounds (Materials and Methods). To examine the impact on splicing, we generated a strain in which SUB2 expression was under control of the glucose-repressible GAL1 promoter. A shift to glucose media caused growth to slow after 12 h, then halt after 28 h (data not shown). In vivo genetic depletion of SUB2 causes U3 pre-mRNA levels to increase up to sixfold, as monitored by primer extension of RNA derived from this strain (data not shown). Taken together, these data indicate Sub2 plays an early, essential role in pre-mRNA splicing.
Isolation of two classes of conditional sub2 mutants
We next used mutagenic PCR (Leung et al. 1989) to generate conditional sub2 alleles. We obtained six alleles, four of which (sub2-1 through sub2-4) are both cold sensitive and temperature sensitive (cs/ts), while two (sub2-5 and sub2-6) are cold sensitive (cs). We focused on the alleles from each class with the strongest growth defect, sub2-1 and sub2-5. The minimal number of mutations required to confer the growth defects for each allele has not been determined; nonetheless, we note an interesting correlation in the location of the mutations and the different growth phenotypes of sub2-1 and sub2-5.
The cs sub2-5 allele contains a single mutation (Q308R) near helicase motif IV (Fig. 1). Available crystal structures of superfamily 2 (SF2) helicases and related superfamily 1 (SF1) helicases place this motif in a structural domain implicated in RNA binding/helicase activity (Pause et al. 1993; Korolev et al. 1997, 1998; Yao et al. 1997; Bird et al. 1998; Velankar et al. 1999). In contrast, the sub2-1 allele contains multiple mutations (D22G, E83G, L142M, and I146T) in the vicinity of the SF2 helicase motifs I and Ia (Fig. 1). These motifs have been shown to be required for ATP binding/hydrolysis and thus the coordination of conformational changes in the ATP binding domain with the function of the other motifs of SF2 helicases (Pause and Sonenberg 1992; Hotz and Schwer 1998; Edwalds-Gilbert et al. 2000). Recent crystal structures of SF1 and SF2 helicases place motifs I and Ia in a structural domain distinct from that containing motif IV (Bird et al. 1998; Korolev et al. 1998; Johnson and McKay 1999). Thus, the differences in growth phenotypes of sub2-1 and sub2-5 probably reflect inactivation of different aspects of wild-type Sub2 function.
Evidence for Sub2 function after U2 addition
We analyzed the in vitro splicing activity of temperature-inactivated extracts derived from sub2-1 and sub2-5 cells grown at permissive temperature. We have been unable to detect any defects in splicing activity or spliceosome assembly in sub2-5 extracts (data not shown). Possibly the sub2-5 allele is defective for an in vivo splicing function that is not rate-limiting in vitro.
In contrast, after 37°C preincubation, sub2-1 extracts accumulate uncleaved pre-mRNA, with a concomitant decrease in lariat intermediate, excised lariat, and mRNA (Fig. 2a, lanes 1–4). This early splicing defect is probably caused by a block in activity, rather than reduced kinetics, because incubation up to 120 min did not increase the amount of splicing intermediates and products (data not shown). This phenotype is specifically caused by heat inactivation of sub2-1, as sub2-1 extracts preincubated on ice (lanes 5–8) and SUB2 extracts preincubated at 37°C (lanes 9–12) or on ice (lanes 13–16) show similar levels of splicing activity. Thus, consistent with our expectation for a functional homolog of UAP56, Sub2 function is required early in splicing.
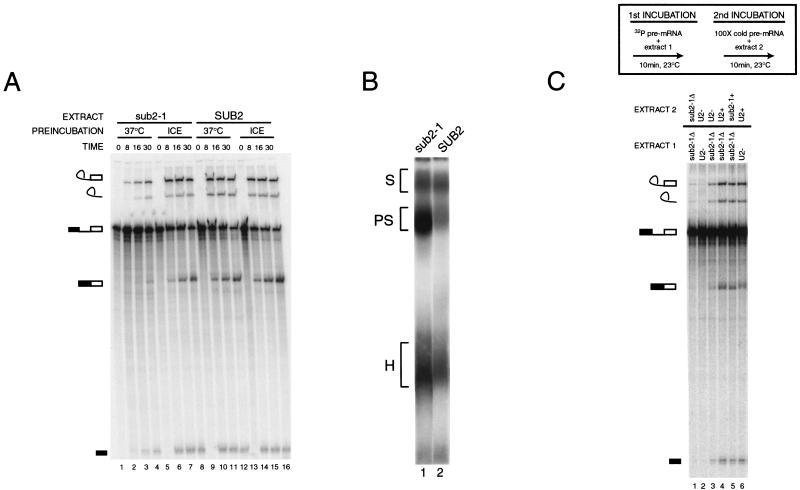
Figure 2.
Biochemical phenotype of heat-inactivated sub2-1 mutant. (A) Time course of in vitro splicing activity of sub2-1 extracts. sub2-1 (lanes 1–8) or SUB2 (lanes 9–16) extracts were preincubated at 37°C (lanes 1–4 and 9–12) or on ice (lanes 5–8 and 13–16) for 15 min and then assayed for splicing activity with radiolabeled pre-actin RNA at 23°C for 0, 8, 16, and 30 min. (B) Spliceosome assembly phenotype of sub2-1 extract. Spliceosome assembly of 37°C preincubated samples from the 16-min timepoint shown in A (sub2-1, lane 3 and SUB2, lane 11) was assessed by native gel analysis (Cheng and Abelson 1986). Spliceosome complexes corresponding to H (heterogenous), PS (prespliceosome), and S (spliceosome) are indicated at left; lane 1, sub2-1; lane 2, SUB2. (C) Chase of splicing defect in sub2-1 extract. Heat-preincubated sub2-1 (lanes 1, 3–5) or U2-inactivated extracts (lanes 2,6) were incubated with radiolabeled preactin RNA in a two-step incubation schematized in the box above the gel. Extracts used in each stage of the incubation experiment are indicated above each lane. sub2-1Δ, 37°C preincubated extract; U2-, U2 oligo-inactivated extract; sub2-1+, mock- (ice) preincubated extract; U2+, mock oligo-inactivated extract.
Native gel analysis (Cheng and Abelson 1987) was performed to test the prediction that heat-inactivated sub2-1 extracts would be blocked before formation of the PS. Surprisingly, a complex that comigrates with the PS in SUB2 extracts preincubated at 37°C (Fig. 2b, lane 2) accumulates in the sub2-1 heat-inactivated extracts (Fig. 2b, lane 1). Depletion of ATP prior to addition of labeled pre-actin substrate abolishes formation of the sub2-1 PS (data not shown). Northern analysis of the native gel shown in Figure 2B confirmed the presence of U2 snRNA in the sub2-1 complex (data not shown). Taken together, these data indicate that Sub2 may function after, rather than before, PS formation in yeast.
To determine if the sub2-1 complex is a bona fide splicing intermediate, we tested if the splicing activity defect of heat-inactivated sub2-1 extracts could be chased. We first incubated radiolabeled pre-mRNA in heat-inactivated sub2-1 extracts and then performed a second (chase) incubation with different extracts in the presence of cold competitor pre-mRNA (Fig. 2C; Materials and Methods). As expected, heat-inactivated sub2-1 extracts were inactive in the chase (lane 1). In contrast, U2-inactivated extracts (Fig. 2C, lane 3), mock-U2-inactivated extracts (Fig. 2C, lane 4), and mock-preincubated sub2-1 extracts (Fig. 2C, lane 5) were all competent to chase the labeled preactin substrate into splicing intermediates and products. These data suggest that the PS complex that accumulates in the sub2-1 extract is a competent splicing intermediate, rather than an aberrant, “dead-end” complex.
Our efforts to complement the sub2-1 splicing defect with Sub2 purified from Escherichia coli or yeast have been unsuccessful to date, despite the fact that both of these versions of Sub2 fully complement a Δsub2 allele when expressed in yeast (data not shown). Furthermore, efficient immunoprecipitation of epitope tagged Sub2 from splicing extract requires high salt (data not shown), suggesting that Sub2 may be part of a complex. It may be significant in this regard that a U2-inactivated extract (Fig. 2C, lane 2) is only partially competent to chase the precursor that accumulates in the sub2-1 heat-inactivated extract (Fig. 2C, lane 3). This same U2-inactivated extract can be fully chased by a mock-U2-inactivated extract (Fig. 2C, lane 6). While we cannot rule out a nonspecific impact of combining two defective extracts, these data raise the possibility that heat inactivation of the sub2-1 extract may also affect Sub2-interacting factors in the U2 snRNP. Consistent with this possibility, Northern analysis of U2 snRNA levels suggests a slight decrease (less than twofold, data not shown) in levels of U2 in the sub2-1 extracts regardless of preincubation treatment. Whether this is a direct or indirect effect of the presence of the sub2-1 allele is unclear, but further indicates an interaction between sub2-1 and U2 snRNP function.
Evidence for functional interactions between Sub2 and Mud2
Because UAP56 interacts with U2AF65, we tested for genetic interactions between the sub2 alleles and MUD2 (Table 1). Deletion of the nonessential MUD2 gene exacerbates the cold-sensitive (cs) growth defects of both sub2-1 and sub2-5. In contrast, the temperature-sensitive (ts) growth defect of sub2-1 is partially suppressed by the Δmud2 allele. Exacerbation of the cs growth defect is consistent with a defect in a collaborative function between Sub2 and Mud2, while suppression of the ts growth defect of sub2-1 suggests an antagonistic interaction between Sub2 and Mud2.
Table 1.
Genetic interactions between Δmud2 and sub2 mutants
| Relevant genotype SUB2, MUD2
|
Growth phenotype at different temperatures
|
||
|---|---|---|---|
| 16°Ca
|
25°Cb
|
37°Cc
|
|
| SUB2, MUD2 | +++ | +++ | +++ |
| SUB2, Δmud2 | +++ | +++ | +++ |
| sub2-1, MUD2 | + | ++ | +/− |
| sub2-1, Δmud2 | +/− | ++ | + |
| sub2-5, MUD2 | +/− | ++ | ++ |
| sub2-5, Δmud2 | − | ++ | ++ |
| Δsub2, MUD2 | − | − | − |
| Δsub2, Δmud2 | − | ++ | − |
Note. Growth on YEPD plates by streaking isogenic strains bearing the designated sub2 allele covering a Δsub2 at different temperatures in the presence or absence of a chromosomal MUD2 (wild-type growth, +++; mild defect, ++; poor growth, +; severe defects, +/−; no growth −).
Growth at 16°C after 14 ds.
Growth at 25°C after 5 ds.
Growth at 37°C after 5 ds.
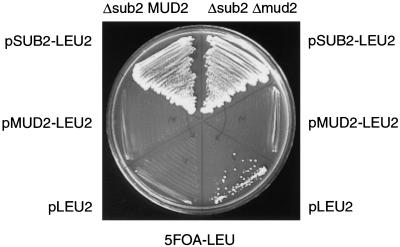
Because the sub2-1 mutant contains mutations in ATPase motifs, the absence of Mud2 would be predicted to suppress a defect in an ATP-dependent function of Sub2. An extreme extension of this hypothesis is that the sole function of Sub2 is to remove Mud2 to promote the formation of a productive PS. To test this hypothesis, we asked if deletion of MUD2 could suppress a SUB2 deletion. Strikingly, deletion of MUD2 restores viability to the otherwise lethal deletion of SUB2 at 25°C (Table 1). In the absence of both MUD2 and SUB2 plasmids, the Δsub2Δmud2 strain is capable of growth (Fig. 3, lower segment). The presence of MUD2 abrogates the growth of the Δsub2Δmud2 strain (Fig. 3, middle segment), while a SUB2 plasmid confers wild-type growth to the Δsub2Δmud2 strain (Fig. 3, top segment). These genetic interactions indicate that Sub2 may function in an ATP-dependent step to promote removal of Mud2. Thus, in the absence of Mud2, Sub2 function becomes dispensable.
Figure 3.
Impact of Δmud2 on the lethality of Δsub2. Growth on 5-FOA-LEU media of Δsub2MUD2/pSUB-URA3 CEN strain (left side of plate) compared with growth of Δsub2Δmud2/pSUB2-URA3 CEN strain (right side of plate) transformed with either pSUB2-LEU2 CEN (upper segment), pMUD2-LEU2 CEN (middle segment), or pLEU2 CEN (lower segment) plasmids.
A Sub2–Mud2-dependent splicing pathway
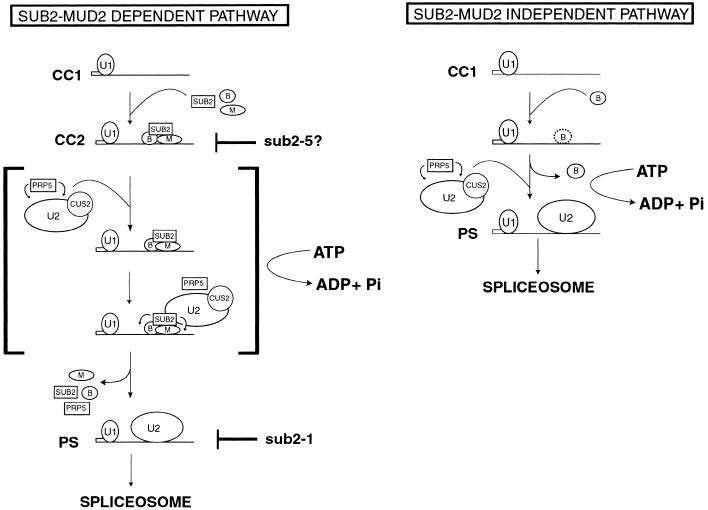
Taken together, our data are consistent with a two-step model for Sub2 function (Fig. 4, left panel). The exacerbation of the cs growth phenotype of sub2-5 by deletion of MUD2 suggests that Sub2 first functions in collaboration with Mud2. Sub2 may interact in an ATP-independent fashion with the pre-mRNA substrate either via Mud2 or via Mud2 and other components of the CC2 complex. Indeed, such a function is predicted for Sub2 based on previous studies of the human homolog, UAP56 (Fleckner et al. 1997). Although we have been unable to detect a biochemical defect in CC formation or stability in sub2-5 extracts (data not shown), this hypothesis for Sub2 function is supported by results in the accompanying communications.
Figure 4.
Models for a Sub2-Mud2-dependent splicing pathway and a Sub2-Mud2-independent splicing pathway. Spliceosomal complexes formed during the early ATP-independent and ATP-dependent stages of splicing are diagrammed. CC1, commitment complex 1; CC2, commitment complex 2; PS, prespliceosome. Circled B, BBP; Circled M, Mud2. The U1 snRNP, U2 snRNP, and Sub2, Prp5, and Cus2 are denoted by labeled shapes. Hypothesized defects in Sub2 functions by different sub2 alleles are indicated at right in the Sub2–Mud2 dependent pathway model. Small arrows from Prp5 and Sub2 indicate activity on potential substrates in the presence of ATP. A possible scenario for the functions of both Prp5 and Sub2 during the ATP-dependent conversion of CC2 to PS complex is shown surrounded by brackets in the Sub2–Mud2 dependent pathway model. Potentially less stable binding of BBP with BPS in the absence of Mud2 is denoted by a dotted outline for the BBP symbol in the Sub2–Mud2 independent pathway.
In sub2-100 extracts, the formation of CC2 complexes is blocked (Zhang and Green 2001), while in sub2-201 extracts, CC2 accumulates with a concomitant decrease in PS (Libri et al. 2001). We think it is likely that the variability in the severity of the biochemical defects detected before PS formation reflects differences in the mutations present in sub2-5 (Q308R), sub2-100 (D175G), and sub2-201 (D175G, L405F). The fact that all three groups have obtained independent genetic or biochemical evidence implicating defects in Sub2 function before PS formation strongly supports the hypothesis that wild-type Sub2 may first function early in splicing, before U2 addition.
In contrast, Δmud2 suppression of the ts phenotype of the sub2-1 allele indicates a second function of Sub2, antagonistic to Mud2. This interpretation is strongly supported by the Δmud2 bypass suppression of Δsub2 lethality (Fig. 3). Because Mud2 has been detected in CC2 complexes but not in PS complexes (Abovich et al. 1994; Rutz and Seraphin 1999), the ATP-dependent function of Sub2 may be to promote the removal of Mud2 (and perhaps BBP) from the branchpoint region to facilitate the stable/productive interaction of the U2 snRNP with the pre-mRNA substrate. An important future goal is to test whether Mud2 is present in the PS complex that accumulates in the sub2-1 extracts (Fig. 4, left panel, bracketed intermediate).
A Sub2–Mud2 independent splicing pathway
The fact that MUD2 is inessential has been thought to reflect the diminished role of the pyrimidine tract in yeast versus mammalian introns (Abovich et al. 1994; Abovich and Rosbash 1997). Indeed, although Mud2 enhances the efficiency with which yeast BBP binds to introns, this reaction can occur in the complete absence of Mud2 (Berglund et al. 1997, 1998). In the Sub2-Mud2 independent pathway proposed in Figure 4 (right panel), BBP binds in the absence of Mud2, albeit more weakly, and can be displaced from the BPS without the requirement for Sub2.
It remains to be seen whether the removal of BBP is also affected in sub2 mutants. We have been unable to detect genetic interactions between our sub2 alleles and the original BBP mutant, msl5 (Abovich and Rosbash 1997). Newly reported mutant alleles may be informative in this regard (Rutz and Seraphin 2000). However, the finding that several conditional msl5 mutants show no impairment of splicing in vitro (Rutz and Seraphin 2000) suggests that BBP may also be dispensable in the Sub2-Mud2 independent pathway.
Implications
While our finding that the essential requirement for the SUB2 ATPase can be bypassed by deletion of the nonessential splicing factor MUD2 was unanticipated, we believe that these results have wider significance with regard to the role of ATP in splicing. Prp28 is an essential DEAD-box protein required to promote the exchange of U1 for U6 at the 5′ splice site; reduced function prp28 alleles become lethal in the presence of a hyperstabilized base-pairing interaction between U1 and the 5′ss (Staley and Guthrie 1999). As reported by Chang and colleagues (Chen et al. 2001), mutations in the RNA-binding domain of the U1C snRNP protein render Prp28 dispensable and suppress the lethal phenotype conferred by combining prp28-1 with a hyperstabilized U1-5′ss interaction. Thus, the ATP-dependent role of Prp28 appears to be essential only for counteracting the stabilizing effect of U1C on the U1-5′ss interaction. Similarly, the ATP-dependent function of Sub2 appears to be essential only for counteracting the interaction of Mud2 (and, thus, perhaps BBP) at the BPS. Taken together, these results strengthen the notion that the ATP-dependent steps of splicing are not an essential mechanistic feature but, rather have evolved for the refinement of the splicing pathway, allowing multiple opportunities for regulation and fidelity maintenance.
It will be extremely interesting to further investigate these and other scenarios in which energy-requiring transitions in splicing can be bypassed. For example, deletion of the nonessential splicing factor CUS2 allows formation of the PRP5-dependent PS in the absence of ATP in vitro (Perriman and Ares 2000). Such studies provide a promising inroad to a deeper mechanistic understanding of how DEAD-box proteins utilize ATP to catalyze conformational rearrangements during the course of splicing, and how such events contribute to the accuracy and activity of the essential function of the spliceosome.
Materials and methods
Yeast methods
All methods for manipulation of yeast, including media preparation, growth conditions, transformation, plasmid recovery, and 5-fluoro-orotic acid (5-FOA) selection, were performed according to standard methods (Guthrie and Fink 1991). Additional techniques used in this study are noted where applied. The strains utilized in this work are derived from W303, unless otherwise indicated.
SUB2 plasmid shuffle strain
To obtain a haploid SUB2 shuffle strain, the SUB2 ORF was replaced by the HIS3 coding sequences through one-step gene replacement into the wild-type diploid strain yCG465. Disruption of the chromosomal SUB2 in the resulting HIS+ strain, yCG466, was confirmed by whole-cell PCR. yCG466 was transformed with pCG788 (SUB2 in pRS316 [Sikorski and Hieter 1989]) and sporulated. Dissection of tetrads yielded four viable spores in which the HIS+ phenotype segregated 2 : 2. All viable HIS+ haploids were also URA+ and inviable on 5-FOA media. yCG470 (Δsub2::HIS3/pCG788) was used as the SUB2 shuffle strain. Disruption of the chromosomal SUB2 locus with the kan-MX6 marker (Longtine et al. 1998) was also lethal in a haploid in the SS330 strain background (Ruby et al. 1993).
Strains for analysis of genetic interactions between sub2 and Δ mud2
Strain yCG472 (Δsub2Δmud2) was generated from yCG470 by replacement of the chromosomal MUD2 with the kan-MX6 marker through one-step gene replacement (Longtine et al. 1998). Disruption of MUD2 in yCG472 was confirmed by whole-cell PCR. yCG472 was transformed with wild-type SUB2 (pCG817), sub2-1 (pCG838), and sub2-5 (pCG840) in the LEU2 CEN vector pRS315 (Sikorski and Hieter 1989). The growth of these and an isogenic MUD2 (yCG470) strain was assessed on YEPD after loss of pCG788 on 5-FOA (Table 1). In addition, yCG472 and its isogenic parent, yCG470, were transformed with pRS315, MUD2-pRS315 (Abovich et al. 1994), or SUB2-pRS315 (pCG817). Growth of these strains in the absence of pCG788 was assessed on 5-FOA-LEU plates. Growth of viable strains was tested on YEPD (Table 1).
Plasmids
The original SUB2 plasmid isolate, pSN120, was identified in a selection for high copy suppressors of the cold-sensitive growth defect of the brr1-1 mutant (Noble 1995) using a GAL-cDNA library (Liu et al. 1992). A plasmid bearing the genomic version of SUB2 (ORF YDL084W) was cloned in two steps. The 1939 bp SalI-XbaI SUB2 fragment from the Saccharomyces cerevisiae λ cosmids 70700 and 70751 (American Type Culture Collection) was cloned into a SalI-XbaI-cleaved pBS (KS+) vector (Stratagene) to generate pCG782, and the remaining 181 bp of upstream 5′UTR sequences was PCR amplified from the same λ cosmid DNA and subcloned into pBS (KS+) to make pCG783. pCG788 (YDL084W in pRS316) was generated by three-way ligation of KpnI-XbaI-digested pRS316, KpnI-SalI-digested pCG783, and SalI-XbaI-digested pCG782. A SUB2 LEU2-CEN plasmid (pCG817) was generated by subcloning the ApaI-NotI SUB2 fragment from pCG788 into pRS315.
Generation of conditional alleles of SUB2
Mutagenic PCR conditions (Leung et al. 1989) were used to amplify the SUB2 ORF (final concentrations of Mn2+ used in PCR reactions ranged from 0 mM MnCl2 to 0.01 mM MnCl2), using pCG817 as a template and primers oCG252 (5′-CCTTGT TTTATATATTCATTGTTCA-3′) and oCG253 (5′-GAAAAT TTCAAGATTTTTTCCAAAT-3′). The resulting PCR products were cotransformed into yCG470 with the purified 7171-bp PstI-NdeI vector fragment of pCG817 (Muhlrad et al. 1992). Transformants were selected on SD-LEU media at 25°C and then replica plated to 5-FOA media at 25°C, 16°C, and 37°C. The efficiency of mutagenesis ranged from 1.7% to 24.6%, depending on [Mn2+]. A total of 25,000 transformants were screened. Six sub2 alleles (sub2-1 though sub2-6; pCG838 through pCG843) were growth impaired at either 16°C or both 37°C and 16°C and were sequenced by the Biomolecular Resource Center DNA sequencing facility at the University of California, San Francisco. The software program EditView was used in conjunction with the NCBI BLAST alignment server (http://www.ncbi.nlm.nih.gov/BLAST) to identify mutations in these recovered sub2 alleles.
In vitro splicing assays and native gel analysis
Splicing extracts derived from yCG484 (sub2-1 strain), yCG485 (sub2-5 strain), and yCG483 (SUB2 strain) were prepared (Umen and Guthrie 1996). Spliceosome assembly analysis was performed by native gel analysis (Cheng and Abelson 1987). Preincubation conditions required to inactivate sub2-1 extracts but not SUB2 extracts varied depending on protein concentration of each extract prep and the presence or absence of splicing reaction buffer. Effective conditions remained consistent with each extract preparation and ranged from 15-min preincubation at 37°C or 40°C in the presence of splicing buffer components (2 mM ATP, 2.5 mM MgCl2, 0.06 M Kphoshate at pH 7, 3% PEG 8000), to 45-min preincubation at 37°C in buffer D (20 mM Hepes at pH 7.9, 2 mM EDTA, 1.5 mM MgCl2, 50 mM KCl, 20% glycerol, 0.5 mM DTT). Mock preincubations were performed on ice.
In vitro chase experiments
Heat-inactivated sub2-1 extracts or U2-inactivated extracts were first incubated under standard splicing conditions (40% extract, 2 mM ATP, 2.5 mM MgCl2, 0.6 M Kphosphate buffer at pH 7, 3% PEG 8000) in a volume of 5 μL with 0.4 nM of uniformly labeled α-[32P] pre-ACT1 substrate for 10 min at 23°C. A second chase incubation was then carried out for another 10 min at 23°C. This chase incubation was initiated in the presence of 100-fold molar excess cold pre-ACT1, added in addition to 5 μL of splicing mix containing heat-preincubated or mock-preincubated sub2-1 extracts or U2-depleted or U2 mock-depleted splicing extract.
In vitro U2 snRNA depletions
U2 snRNA depletions were performed as previously described, using a final concentration of 1 μM U2 oligo (5′-CAGATACT ACACTTG-3′) (McPheeters et al. 1989), and incubating for 15 min at 25°C as recently described (Perriman and Ares 2000). In mock-depleted extracts, water was substituted for the U2 oligo during the preincubation at 25°C.
Acknowledgments
We thank Meng Zhang, Michael Green, Domenico Libri, T.-H. Chang, Jon Staley and Jie Pan for communicating unpublished results and extensive collaborative discussions; Cathy Collins, Kent Duncan, Maki Inada, Karen Kim, Pascal Preker, Stephen Rader, Tommaso Villa, Manny Ares, Liz Blackburn, and Alan Frankel for critical comments on this manuscript; and Lucita Esperas and Carol Pudlow for excellent technical assistance. A.K. was an HHMI predoctoral fellow in the Program in Biological Sciences Graduate Program at the University of California, San Francisco. C.G. is an American Cancer Research Professor of Molecular Genetics. This work was supported by National Institutes of Health grant GM21119.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL guthrie@cgl.ucsf.edu; FAX (415) 502-5306.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.851601.
References
- Abovich N, Rosbash M. Cross-intron bridging interactions in the yeast commitment complex are conserved in mammals. Cell. 1997;89:403–412. doi: 10.1016/s0092-8674(00)80221-4. [DOI] [PubMed] [Google Scholar]
- Abovich N, Liao XC, Rosbash M. The yeast MUD2 protein: An interaction with PRP11 defines a bridge between commitment complexes and U2 snRNP addition. Genes & Dev. 1994;8:843–854. doi: 10.1101/gad.8.7.843. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Chua K, Abovich N, Reed R, Rosbash M. The splicing factor BBP interacts specifically with the pre-mRNA branchpoint sequence UACUAAC. Cell. 1997;89:781–787. doi: 10.1016/s0092-8674(00)80261-5. [DOI] [PubMed] [Google Scholar]
- Berglund JA, Abovich N, Rosbash M. A cooperative interaction between U2AF65 and mBBP/SF1 facilitates branchpoint region recognition. Genes & Dev. 1998;12:858–867. doi: 10.1101/gad.12.6.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird LE, Subramanya HS, Wigley DB. Helicases: A unifying structural theme? Curr Opin Struct Biol. 1998;8:14–18. doi: 10.1016/s0959-440x(98)80004-3. [DOI] [PubMed] [Google Scholar]
- Chen, J.Y.-F., Stands, L., Staley, J.P., Jackups, Jr., R., Latus, L.J., and Chang, T.-H. 2001. Specific alterations of U1-C protein or U1 small nuclear RNA can eliminate the requirement of Prp28p, an essential DEAD-box splicing factor. Mol. Cell (in press). [DOI] [PubMed]
- Cheng S-C, Abelson J. Spliceosome assembly in yeast. Genes & Dev. 1987;1:1014–1027. doi: 10.1101/gad.1.9.1014. [DOI] [PubMed] [Google Scholar]
- Das R, Zhou Z, Reed R. Functional association of U2 snRNP with the ATP-independent spliceosomal complex E. Mol Cell. 2000;5:779–787. doi: 10.1016/s1097-2765(00)80318-4. [DOI] [PubMed] [Google Scholar]
- Edwalds-Gilbert G, Kim DH, Kim SH, Tseng YH, Yu Y, Lin RJ. Dominant negative mutants of the yeast splicing factor Prp2 map to a putative cleft region in the helicase domain of DExD/H-box proteins. Rna. 2000;6:1106–1119. doi: 10.1017/s1355838200992483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcárcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Gee S, Krauss SW, Miller E, Aoyagi K, Arenas J, Conboy JG. Cloning of mDEAH9, a putative RNA helicase and mammalian homologue of Saccharomyces cerevisiae splicing factor Prp43. Proc Natl Acad Sci. 1997;94:11803–11807. doi: 10.1073/pnas.94.22.11803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. In: Guthrie C, Fink GR, editors. Methods in Enzymology. San Diego, CA: Academic Press; 1991. [PubMed] [Google Scholar]
- Hotz HR, Schwer B. Mutational analysis of the yeast DEAH-box splicing factor Prp16. Genetics. 1998;149:807–815. doi: 10.1093/genetics/149.2.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson ER, McKay DB. Crystallographic structure of the amino terminal domain of yeast initiation factor 4A, a representative DEAD-box RNA helicase. Rna. 1999;5:1526–1534. doi: 10.1017/s1355838299991410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konarska MM, Sharp PA. Electrophoretic separation of complexes involved in the splicing of precursors to mRNAs. Cell. 1986;46:845–855. doi: 10.1016/0092-8674(86)90066-8. [DOI] [PubMed] [Google Scholar]
- ————— Interactions between small nuclear ribonucleoprotein particles in formation of spliceosomes. Cell. 1987;49:763–774. doi: 10.1016/0092-8674(87)90614-3. [DOI] [PubMed] [Google Scholar]
- Korolev S, Hsieh J, Gauss GH, Lohman TM, Waksman G. Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP. Cell. 1997;90:635–647. doi: 10.1016/s0092-8674(00)80525-5. [DOI] [PubMed] [Google Scholar]
- Korolev S, Yao N, Lohman TM, Weber PC, Waksman G. Comparisons between the structures of HCV and Rep helicases reveal structural similarities between SF1 and SF2 super-families of helicases. Protein Sci. 1998;7:605–610. doi: 10.1002/pro.5560070309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A, Utans U. Three protein factors (SF1, SF3 and U2AF) function in pre-splicing complex formation in addition to snRNPs. EMBO J. 1991;10:1503–1509. doi: 10.1002/j.1460-2075.1991.tb07670.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Chen E, Goeddel DV. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- Liao XC, Colot HV, Wang Y, Rosbash M. Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res. 1992;20:4237–4245. doi: 10.1093/nar/20.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes & Dev. 2001;15:36–41. doi: 10.1101/gad.852101. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H, Krizek J, Bretscher A. Construction of a GAL1-regulated yeast cDNA expression library and its application to the identification of genes whose overexpression causes lethality in yeast. Genetics. 1992;132:665–673. doi: 10.1093/genetics/132.3.665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopez MC, Sanchez M, Ferminan E, Dominguez A. Disruption of six Saccharomyces cerevisiae genes from chromosome IV and basic phenotypic analysis of deletion mutants. Yeast. 1998;14:1199–1208. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1199::AID-YEA309>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- McPheeters DS, Fabrizio P, Abelson J. In vitro reconstitution of functional yeast U2 snRNPs. Genes & Dev. 1989;3:2124–2136. doi: 10.1101/gad.3.12b.2124. [DOI] [PubMed] [Google Scholar]
- Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes & Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Muhlrad D, Hunter R, Parker R. A rapid method for localized mutagenesis of yeast genes. Yeast. 1992;8:79–82. doi: 10.1002/yea.320080202. [DOI] [PubMed] [Google Scholar]
- Noble SM. The BRR genes encode novel factors involved in RNA processing. PhD thesis. San Francisco: University of California; 1995. [Google Scholar]
- Noble SM, Guthrie C. Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996;15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- O'Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- Ono Y, Ohno M, Shimura Y. Identification of a putative RNA helicase (HRH1), a human homolog of yeast Prp22. Mol Cell Biol. 1994;14:7611–7620. doi: 10.1128/mcb.14.11.7611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perriman R, Ares M., Jr ATP can be dispensable for prespliceosome formation in yeast. Genes & Dev. 2000;14:97–107. [PMC free article] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes & Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Ruskin B, Zamore PD, Green MR. A factor, U2AF, is required for U2 snRNP binding and splicing complex assembly. Cell. 1988;52:207–219. doi: 10.1016/0092-8674(88)90509-0. [DOI] [PubMed] [Google Scholar]
- Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. Rna. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sikorski RS, Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- ————— An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Umen JG, Guthrie C. Mutagenesis of the yeast gene PRP8 reveals domains governing the specificity and fidelity of 3′ splice site selection. Genetics. 1996;143:723–739. doi: 10.1093/genetics/143.2.723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Velankar SS, Soultanas P, Dillingham MS, Subramanya HS, Wigley DB. Crystal structures of complexes of PcrA DNA helicase with a DNA substrate indicate an inchworm mechanism. Cell. 1999;97:75–84. doi: 10.1016/s0092-8674(00)80716-3. [DOI] [PubMed] [Google Scholar]
- Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein Prp22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wagner JD, Guthrie C. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr Biol. 1998;8:441–451. doi: 10.1016/s0960-9822(98)70178-2. [DOI] [PubMed] [Google Scholar]
- Wiest DK, O′Day CL, Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- Yao N, Hesson T, Cable M, Hong Z, Kwong AD, Le HV, Weber PC. Structure of the hepatitis C virus RNA helicase domain. Nat Struct Biol. 1997;4:463–467. doi: 10.1038/nsb0697-463. [DOI] [PubMed] [Google Scholar]
- Zhang M, Green MR. Identification and characterization of yUAP/Sub2p, a yeast homolog of the essential human pre-mRNA splicing factor hUAP56. Genes & Dev. 2001;15:30–35. doi: 10.1101/gad.851701. (this issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Reed R. Human homologs of yeast Prp16 and Prp17 reveal conservation of the mechanism for catalytic step II of pre-mRNA splicing. EMBO J. 1998;17:2095–2106. doi: 10.1093/emboj/17.7.2095. [DOI] [PMC free article] [PubMed] [Google Scholar]