Abstract
The UAP56 gene has been shown to be required for prespliceosome assembly in mammals. We report here the isolation of the Schizosaccharomyces pombe ortholog of this gene by heterologous complementation of a combined PRP40HA3/nam8Δ defect in budding yeast. The Saccharomyces cerevisiae ortholog, YDL084w/SUB2, is also able to suppress this defect. We show that SUB2 is involved in splicing in vivo as well as in vitro. Sub2 defective extracts form a stalled intermediate that contains U2snRNP and can be chased into functional spliceosomes. Our experiments also suggest a role for this protein in events that precede prespliceosome formation. Data reported here as well as in the accompanying papers strongly implicate Sub2p in multiple steps of the spliceosome assembly process.
Keywords: splicing, hUAP56, SUB2, SPAC17G6.14c, helicase, U2snRNP, prespliceosome
Splicing takes place in a highly dynamic and transient ribonucleoprotein particle called the spliceosome. Central to the process of spliceosome assembly and catalysis is the role played by the small nuclear ribonucleoprotein particles (U1, U2, U4, U5, and U6 snRNP), which are composed of one snRNA and several proteins. A large number of nonspliceosomal proteins are also involved in the process (Moore et al. 1993; Staley and Guthrie 1998).
The interaction of U1snRNP with the pre-mRNA defines the first ATP-independent stable complex that can be detected by gel shift analysis in yeast, which is called the commitment complex 1 (CC1; Seraphin and Rosbash 1989). This interaction is stabilized by U1snRNP-associated proteins, among which U1Cp, Prp40p, and Nam8p are believed to play a direct role (Kao and Siliciano 1996; Tang et al. 1997; Puig et al. 1999). A second ATP-independent complex, CC2, contains two additional proteins, Mud2p and BBPp, which recognize the branch-point sequence on the pre-mRNA.
The entry of the U2snRNP particle, which leads to the formation of the prespliceosome, defines the first ATP-requiring step. Then the preformed tri-snRNP, U4/U6-U5 interacts with the prespliceosome and marks the first spliceosomal complex.
ATP is not needed for the two transesterification steps that ultimately lead to exon joining. However, the multiple rearrangements that take place in the spliceosome during assembly and after splicing require ATP. A class of proteins belonging to a superfamily of RNA-dependent ATPases (RNA unwindases or helicases) plays a pivotal role as “spliceosome chaperones” (Staley and Guthrie 1998; de la Cruz et al. 1999). The strong sequence and structural similarities to the ATP-dependent DNA helicases has led to the hypothesis that a major role for these proteins is to unwind RNA–RNA duplexes. However, it is still a major challenge to define the precise function and mechanism of action of these proteins and, to date, only two of the splicing RNA helicases have been assigned a somewhat precise role: Prp28p and Brr2p (Raghunathan and Guthrie 1998; Staley and Guthrie 1998, 1999). On the basis of the conserved motif II, the RNA helicases can be assigned to different subfamilies, such as DEAD, DEAH, or DExH.
In yeast, the DEAD box unwindase, Prp5p, has been shown to be required for the interaction of U2 with the branch point (Ruby et al. 1993; O'Day et al. 1996; Wiest et al. 1996). In mammals, another putative RNA-dependent ATPase, UAP56, has been shown to be required for the formation of the prespliceosome (Fleckner et al. 1997). This protein is tethered to the branchpoint complex through interactions with U2AF65, the putative ortholog of Mud2p.
In this article, we describe the molecular characterization of the S. cerevisiae SUB2 gene, the yeast ortholog of human UAP56. SUB2 plays a role in splicing in vivo and is absolutely required for spliceosome assembly and splicing in vitro. Data presented here and in the accompanying reports implicate Sub2p in at least two steps of spliceosome formation that precede and follow U2snRNP addition.
Results and Discussion
Isolation of the S. pombe ortholog of SUB2/yUAP56
The yeast Prp40p and Nam8p proteins have both been implicated in the stabilization of the U1snRNP-pre-mRNA interaction (Kao and Siliciano 1996; Puig et al. 1999). We noticed that a cold-sensitive phenotype is associated with the deletion of the nonessential NAM8 gene when a tagged version of Prp40p (Prp40HA3) is present in the cell as the only source of this protein. This defect is splicing related, as the double mutant (nam8Δ, PRP40HA3 ) strain displays less efficient splicing in vivo and assembles commitment complex poorly in vitro (data not shown). This phenotype is not linked to mutations in the PRP40 orf, as occasional revertants lack most or all of the HA tag. Transformation of this strain with a high–copy number S. cerevisiae genomic library allowed the isolation of wild-type PRP40 (11 times), NAM8 (six times), and SAR1 (three times), which confirms the origin of the defect. Note that SAR1 was previously isolated as a multicopy suppressor of PRP40 mutants (Kao and Siliciano 1996) and other splicing defects (Chen et al. 1998).
We decided to exploit this phenotype to screen for heterologous proteins from S. pombe, Arabidopsis thaliana, and HeLa cells that would be able to suppress the 22°C slow growth defect. We will discuss here results obtained from the S. pombe experiment. Four clones survived the entire selection process. Three of them encoded the same gene, the S. pombe SAR1. The fourth one was identical to the S. pombe SPAC17G6.14c gene, a putative ortholog of the human UAP56 gene, which has been shown previously to be involved in splicing (Fleckner et al. 1997). The putative S. cerevisiae ortholog, SUB2 (ORF YDL084w) was previously described (Staley and Guthrie 1998; Kistler and Guthrie 2001) as a suppressor of the snRNP biogenesis mutant brr1-1 (Noble and Guthrie 1996a,b) even though its role in splicing had not been determined. These three well-conserved proteins (Fig. 1A) belong to the DEAD box family of RNA-dependent ATPases (DEcD subfamily; de la Cruz et al. 1999).
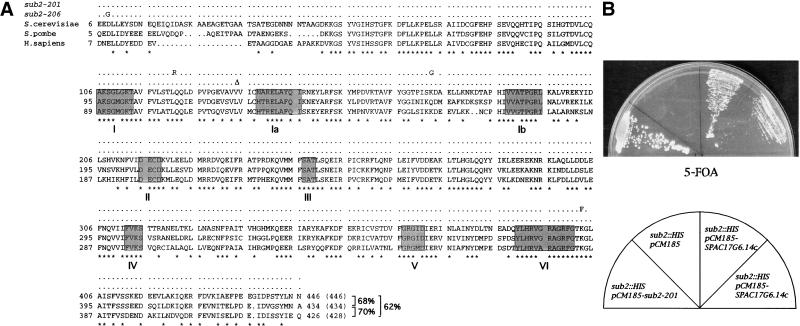
Figure 1.
(A) Alignement of SPAC17G6.14c (Schizosaccharomyces pombe), SUB2 (Saccharomyces cerevisiae) and hUAP56 (Homo sapiens). The mutations in sub2-201 and sub2-206 Ts strains are shown on top of the alignment. *: identical residues in the three proteins. The eight helicase conserved motifs are boxed. Percentage identity is indicated. (B) Complementation of the sub2 null phenotype by the S. pombe SPAC17G6.14c gene (two independent clones). As a positive control, the sub2 null phenotype was complemented by the thermosensitive mutant sub2-201 on a TRP1 marked plasmid (pCM185). Note that sub2 null cells (sector sub2::HIS, pCM185) are barely visible in this experiment because of the very slow growth rate.
Moderate overexpression of S. cerevisiae Sub2p was able to suppress the combined PRP40HA3/nam8Δ defect, as with the S. pombe SPAC17G6.14c gene. We deleted SUB2 in a diploid strain and showed by tetrad analysis that the gene is not essential in this genetic background, although sub2 null haploids are severely impaired in growth and are thermosensitive at 34°C (data not shown). Note, however, that SUB2 requirement for growth depends on strain background (Lopez et al. 1998; Kistler and Guthrie, this issue; Zhang and Green, this issue).
We constructed a sub2 null haploid strain containing a URA3 marked, plasmid-borne copy of the gene under the control of a tetracycline repressible promoter (pCM188-SUB2). The resulting strain, DLY23, grew normally in the absence of the repressor (doxycycline).
The S. pombe SPAC17G6.14c gene cloned on a TRP1-marked centromeric plasmid was able to complement the loss of S. cerevisiae SUB2, as shown by growth of 5-FOA selected Δsub2, SPAC17G6.14c cells, although the complementation was not complete (Fig. 1B). We conclude that the S. pombe SPAC17G6.14c and the S. cerevisiae SUB2 genes are true orthologs.
Mature mRNA levels of intron-containing genes are affected in vivo by SUB2 depletion or inactivation
We decided to pursue this work by studying the potential role of the yeast S. cerevisiae protein, Sub2p, in splicing. In the presence of doxycycline, the growth rate of strain DLY23 started to decline between 10 and 12 h and attained a constant doubling time of 5.5–6 h by 16 h. Using a myc-tagged construct, we verified that Sub2p levels are reduced to <2.5% of the initial amount by 14.5 h of growth in repression conditions (data not shown). However, because the growth rate of the genetically depleted strain is significantly higher than the growth rate of sub2 null cells (8.5 h), it is likely that the depletion is not complete.
Splicing of endogenous U3 snRNAs or RP51A genes (data not shown) was not affected in these conditions, up to 16 h in the presence of doxycyclin. On the other side, levels of mature mRNA from endogenous RPL32 or RP51b genes, which are both inefficiently spliced, were already affected after 8 h in the presence of doxycycline and strongly reduced after 16 h (Fig. 2; data not shown). Levels of U1 and U6 snRNAs remained unchanged during the whole time course (Fig. 2A; data not shown).
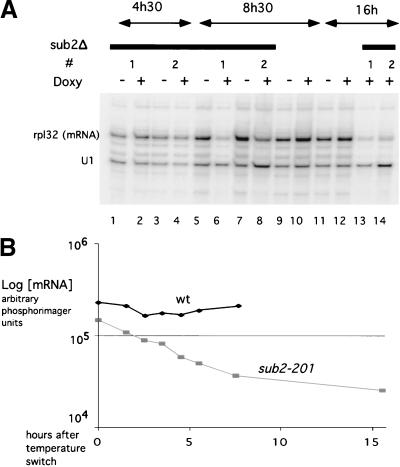
Figure 2.
In vivo analysis of RPL32 mature transcripts by primer extension. (A) Strain DLY23 (lanes 1–8 and 13–14) or an isogenic wild-type strain expressing SUB2 on a centromeric plasmid (lanes 9–12), treated (+ doxy) or untreated (− doxy) with doxycycline during the times indicated on top. RNAs from two different clones (1 and 2) were primer extended with radioactive oligonucleotides against the U1snRNA and the RPL32 gene (mature products indicated on the right). The decrease in mature transcript for RPL32 at 8h30 and 16h (+ doxy/− doxy, normalized to U1 levels) is respectively 3.08 ± 0.6 (n = 2) and 7.35 ± 0.86 (n = 2). (B) Quantitation of a primer extension experiment done on RPL32 and U1snRNA genes. RNAs extracted from a sub2-201 Ts strain after switch to the nonpermissive temperature for the times indicated. A non-Ts strain was analyzed in parallel. Values are normalized to the average levels of endogenous U1snRNA.
We screened a library of SUB2 variants for thermosensitive (ts) mutants. Two of the five ts mutants obtained, sub2-201 and sub2-206 (Fig. 1 for sequence), displayed normal growth at the permissive temperature and were used for further studies. In both cases, growth started to decline 3.5–4 h after the switch at the nonpermissive temperature (37°C) and arrested completely after 7 h. Sub2p protein levels were not significantly affected during growth at the nonpermissive temperature (data not shown). Analysis by primer extension of endogenous RPL32 transcripts during the heat-inactivation period confirmed—in both ts mutants (Fig. 2B; data not shown)—a strong and fast reduction in the ratio between the mature mRNA and the pre-mRNA.
These experiments suggest that Sub2p is involved in splicing in vivo. However, it behaves as an efficiency rather than an essential factor, because expression of some intron-containing genes is not strongly affected by its depletion or inactivation.
Sub2p is required for splicing in vitro
We then examined the effects of Sub2p depletion and inactivation on in vitro splicing and spliceosome assembly. In spite of multiple efforts, we could not efficiently immunodeplete extracts prepared from Sub2p myc-tagged strains. Extracts prepared from a Sub2p genetically depleted strain were inactive for splicing a RP51A derived intron (Δ2; Seraphin and Rosbash 1991) and a natural U3 snRNA intron (Fig. 3A), which are both efficiently spliced in a wild-type extract.
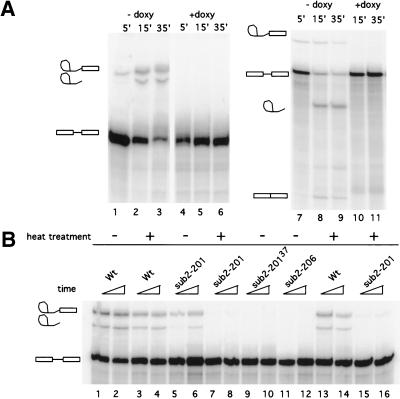
Figure 3.
In vitro splicing experiments. (A) Left panel, RP51AΔ2 RNA substrate, incubated with DLY23 extracts prepared in the absence (lanes 1–3) or presence (lanes 4–6) of doxycycline, for the times indicated. Right panel: U3 snRNA substrate. (B) In vitro splicing with Ts mutant extracts. Heat treatment is at 37°C for 20 (lanes 3–4 and 7–8) or 15 min (lanes 13–14 and 15–16). Lanes 9–10: extract prepared from sub2-201 mutant after a switch to 37°C for 3 h. Splicing intermediates and products are indicated.
We prepared splicing extracts from sub2-201 and sub2-206 at the permissive temperature of 24°C and, after a 3- and 6-h shift, to the nonpermissive temperature (37°C), that is, before and after the decline in the growth rate. Extracts from sub2-206 were inactive even when prepared at the permissive temperature of 24°C (Fig. 3B, lanes 11,12). Extracts from sub2-201 were active when prepared at the permissive temperature (lanes 5,6) but were inactive when the growth temperature was shifted to 37°C for 3 h (lanes 9,10). Most important, the active extracts were inactivated in vitro by a 20-min incubation at 37°C (lanes 7,8). Thus, in contrast to the effects observed in vivo, Sub2p appears to be absolutely required for splicing in vitro.
yUAP56/SUB2 plays a dual role in spliceosome assembly
Spliceosome assembly can be followed by analyzing the formation of complexes on native gels. Commitment complexes 1 (CC1) and 2 (CC2) can be resolved from the prespliceosome and the spliceosome that comigrate in these gels.
Labeled pre-mRNA was incubated in splicing conditions with Sub2p genetically depleted or mock-depleted extracts, and the complexes formed were resolved on a nondenaturing gel (Fig. 4). A complex of slower mobility than the commitment complex accumulates in a Sub2p genetically depleted extract, which migrates roughly as a (pre-)spliceosome (complex X; cf. Fig. 4, lanes 1–4 with 5–8). Complex X formation was dependent on the presence of ATP (Fig. 4, lane 16) and a functional U2snRNA (Fig. 4, lanes 13–15). This indicates that in the presence of limiting amounts of Sub2p, spliceosome assembly can proceed through the first ATP-requiring step. However, CC2 also reproducibly accumulates in Sub2p-depleted but not in mock-depleted extracts (cf. lanes 2–4 with lanes 6–8; see below), suggesting a slow rate of (pre-)spliceosome formation.
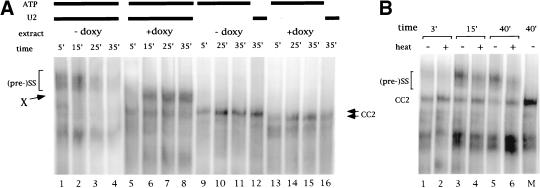
Figure 4.
In vitro spliceosomes and commitment complex formation. (A) Extracts from DLY23 strain prepared in the absence (lanes 1–4 and 9–12) or presence (lanes 5–8 and 13–16) of doxycycline. For commitment complex formation, the extract was U2snRNA inactivated (lanes 9–11 and 13–15) or ATP depleted before incubation with the RNA (lanes 12,16). (B) Complex gel analysis of spliceosome assembly reactions from sub2-201 extracts. Heat treatment (even lanes) was for 20 min at 37°C. Mock treatment (odd lanes) was on ice. M: CC2 marker lane (U2snRNA inactivated extract). The ratios CC2/(pre-)spliceosome are as follows: lane 1: 0.87; lane 2: 1.42; lane 3: 0.33; lane 4: 0.69; lane 5: 0.27; lane 6: 0.90. The increase in this ratio on heat treatment is as follow: 3 min: 1.62; 15 min: 2.11; 40 min: 3.33.
To examine commitment complex formation, we either inactivated U2snRNP by oligonucleotide-directed RNAseH cleavage or ATP-depleted the extracts before incubation with the RNA (lanes 9–16). CC2 migrates reproducibly faster and forms with a reduced rate in Sub2p-depleted extracts (Fig. 4A, lanes 13–16; data not shown) compared to the wild type (lanes 9–12).
Migration of CC1 in the Sub2p genetically depleted extract was also altered when complex gel analysis was performed with an RNA bearing a mutation in the branchpoint (Δ2BPGG; data not shown). However, because we have been unable to reveal the presence of Sub2p in both complexes by immunoprecipitation and affinity selection of biotinylated RNA (D. Libri, unpubl.), the basis for the faster migration of commitment complexes in Sub2p depleted extract is not clear at this time.
We next examined spliceosomes formed in extracts prepared from the ts strain sub2-201 (Fig. 4B). In vitro heat-inactivated extracts behaved essentially as the genetically depleted extracts, and accumulation of both CC2 and an ATP-dependent complex was consistently observed. CC2 accumulation was already detected in splicing reactions performed with mock-inactivated sub2-201 extracts. On heat inactivation, CC2 accumulation increased with a concomitant decrease in the (pre-)spliceosomal complexes (cf. lanes 1,3,5 with lanes 2,4,6; the CC2/(pre-)spliceosome ratio increased up to threefold on heat treatment in this experiment). Note that increase in CC2 and (pre-)spliceosome decrease were not observed when wild-type extracts were used (data not shown).
Data presented here and in the accompanying reports indicate that Sub2p is required for steps that follow the addition of U2snRNP; they also strongly suggest a role in the formation of the prespliceosome.
The complexes formed in Sub2p-depleted or -inactivated extracts are functional intermediates
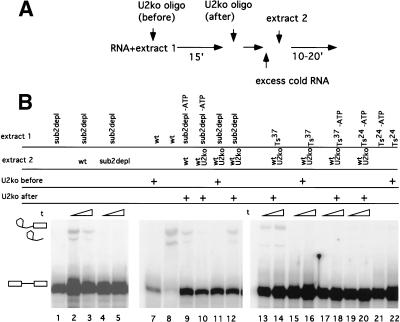
The complex that accumulates in the Sub2p depleted splicing reaction, complex X, might be a functional stalled intermediate or a dead-end complex. We performed chase experiments to address this question. In these experiments, radioactive RNA is incubated in the mutant extract for a time sufficient to accumulate the stalled/dead-end complex. An excess of cold competitor RNA is then added to prevent ex novo formation of radioactive complex, and fresh wild-type extract is added to chase the complexes.
Splicing reactions assembled with a Sub2p-depleted extract can be chased by a wild-type extract but not by a depleted extract (Fig. 5, lanes 1–5). As commitment complex is present in the stalled Sub2p-depleted reactions, it is possible that splicing products evolve from CC rather than from complex X. To rule out this possibility, radioactive RNA was preincubated with Sub2p-depleted extracts in the presence of ATP, and free U2snRNP was inactivated by oligonucleotide-directed RNAseH digestion. These splicing reactions were then chased by the addition of excess cold competitor RNA and U2snRNP-inactivated wild-type extract. As the only source of active U2snRNP in these reactions is complex X (in the complex, U2snRNA is protected from degradation), the appearance of splicing products (Fig. 5, lane 12) unambiguously indicates that complex X is a stalled functional intermediate in the splicing process. When the preincubation step was performed in the absence of ATP and ATP was added after the U2snRNP inactivation, we did not observe the appearance of splicing products (Fig. 5, lane 10), which verifies the effectiveness of this U2 inactivation “on the fly.” Finally, when the chase in the latter conditions (preincubation in the absence of ATP) was performed with a wild-type (non–U2 inactivated) extract, splicing products were observed (Fig. 5, lane 9). Extracts prepared from the heat-inactivated sub2-201 strain responded identically to the same chase experiments as described for the genetically depleted extracts (Fig. 5, lanes 13–22).
Figure 5.
Chase experiments. (A) Scheme of the experiment. (B) (subdepl) Sub2p genetically depleted extracts. (wt) DLY23 extracts prepared in the absence of doxycycline; (Ts24) extracts from sub2-201 prepared at the permissive temperature non–heat inactivated; (Ts37) extracts heat inactivated in vitro for 20 min at 37°C. (wtU2ko) extracts in which U2snRNA has been inactivated by oligonucleotide directed RNAseH cleavage; (oligo before) U2snRNA was inactivated before the first incubation step. (Oligo after) U2snRNA was inactivated after the first incubation step; (−ATP) the extract was ATP depleted before the first incubation step.
From this set of experiments, we conclude that the complexes formed in Sub2p-depleted and heat-inactivated extracts are stalled intermediates in the spliceosome assembly pathway.
The entry of the tri-particle U4/U6-U5 snRNP follows U2snRNP addition in spliceosome assembly. How far do Sub2p-defective extracts go in the spliceosome assembly process? To answer this question, we assembled (pre-)spliceosomes on biotinylated RNAs and analyzed the pattern of associated snRNAs after selection with streptavidine-agarose beads. Sub2p-defective extracts allowed the selection of U2 but not of U4, U5, and U6 snRNAs (data not shown). In accordance with these results, Kistler and Guthrie (2001) report the accumulation of U2snRNA-containing prespliceosomes in heat-inactivated sub2-1 extracts.
The role of Sub2p in splicing
What is the role of Sub2p in splicing? Results presented in this report as well as in Kistler and Guthrie (2001) clearly indicate a role for SUB2 after the addition of U2snRNP and, most likely, before the entry of the tri-snRNP U4/U6-U5. This implication was not previously documented in the mammalian system (Fleckner et al. 1997), most likely because spliceosome assembly was blocked before U2snRNP addition. The U2snRNP-containing complex that we observe in Sub2p-defective cells (complex X) behaves as a bona fideprespliceosome in our chase experiments. Also, we could not detect the presence of Mud2p in complex X (D. Libri, unpubl.), which is consistent with the absence of this protein in the normal prespliceosome (Rutz and Seraphin 1999). However, complex X cannot be converted to a functional spliceosome in the absence of Sub2p, which might be indicative of more subtle differences with a true prespliceosome, as, for instance, with an improperly positioned U2snRNP.
An alternative possibility is that U2 and/or the U4/U6-U5 particles are somehow defective and unable to associate efficiently in the mature spliceosome. This possibility is supported by the isolation of SUB2 as a suppressor of the snRNP biogenesis mutant brr1-1 (Noble and Guthrie 1996a,b; Staley and Guthrie 1998). We are currently performing experiments in this direction.
Several lines of evidence also strongly implicate a function for Sub2p at or before U2snRNP addition, which is consistent with the proposed role for hUAP56 in the mammalian system (Fleckner et al. 1997). The most direct evidence for a role in U2snRNP addition is provided by the accumulation of CC2 and decrease of prespliceosomes in Sub2p-defective extracts, although assembly was not completely arrested at this stage (Fig. 4). We also observed a slower rate of formation for the CC2 complex (Fig. 4A), which might be related to the existence of an ATP-independent role for Sub2p (Zhang and Green, this issue). Zhang and Green observe an accumulation of CC1 and a failure in CC2 formation with Sub2p-defective extracts (Zhang and Green, this issue). We do not fully understand the reason why in these conditions CC1 accumulates instead of CC2. It is likely that subtle differences in experimental conditions and/or allelic differences between sub2-100 and sub2-201 underlie such apparent discrepancy. However, it is possible that in both cases the block in spliceosome assembly reflects a failure in U2snRNP addition and not, in the case of sub2-100, a defect in the transition from CC1 to CC2. This would be supported by work from B. Seraphin and colleagues suggesting that accumulation of CC2 might not be limiting (and/or not absolutely required) for (pre-)spliceosome formation and splicing (Rutz and Seraphin 1999, 2000). In these experiments, BBP-defective extracts that do not accumulate CC2 still support (pre-)spliceosome formation and splicing.
The suppression by Sub2p overexpression of the combined PRP40(HA)3/nam8Δ defect is compatible with a defect in U2snRNP addition. The effect of this synergistic double U1snRNP modification on the formation/stability of CC1 does not exclude a subsequent defect that might be preferentially suppressed by Sub2p overexpression. More precisely, recent studies have revealed genetic and biochemical interactions between Prp40p and proteins involved in the entry of U2snRNP, such as Prp5p, ySAP155/Hsh155p, and USA3p/Ybv2p (P. Siliciano, pers. comm.), which might implicate Prp40p in prespliceosome formation. It is possible that a defective Prp40(HA)3p, possibly destabilized from its interaction with the U1snRNP by the absence of Nam8p (Gottschalk et al. 1998), cannot promote the interaction between U1 and U2snRNPs and subsequent prespliceosome formation.
Finally, we show that the S. pombe ortholog of SUB2 can functionally replace the S. cerevisiae gene, which testifies to a conservation of functions over a relatively large evolutionary distance. Differences are likely to exist, however, in the mode of action of the helicase in the different systems. The very poor conservation of Mud2p with both SpU2AF and hU2AF, for instance, as well as our negative immunoprecipitation experiments (data not shown), argue against a Mud2p-dependent recruitment of Sub2p to the spliceosome and raises the question of what directs Sub2p to its site(s) of action. The isolation and characterization of the three ortholog genes coding for this DEcD helicase provides an important tool for parallel work in the three systems, which will certainly contribute to the understanding of the precise role and mechanism of action of this factor.
Material and methods
Strains
Strain PRP40HA3/nam8Δ was constructed by deleting the NAM8 gene with the Kanr gene disruption module (Wach et al. 1994) in a PRP40HA3-tagged strain (Kao and Siliciano 1996; kindly provided by P. Siliciano). The genotype of this strain (DLY9) is: MATa, ura3-52, trp1-289, his3-52, lys2-801, ade2-101, prp40::HIS3, (pSE358-PRP40HA3), nam8::Kanr.
The SUB2 gene was deleted in a diploid strain (W303, kind gift of D. Thomas) with the S. pombe HIS5 module (Longtine et al. 1998). Sporulation and all subsequent steps of tetrads dissection were performed at 26°C. Strain DLY23 was constructed by first expressing pCM188-SUB2 in strain W303 (haploid) and deleting the endogenous SUB2 gene with the S. pombe HIS5 module (Longtine et al. 1998). Disruptions were verified by PCR with internal and external primers. The growth rate of a haploid sub2 null strain (obtained either by 5-FOA selection of strain DLY23 or from one of the slow growing tetrads) was estimated to be 8.5 h. Ts strains have been obtained by cloning a PCR fragment containing the SUB2 coding region into the TRP1 marked vector pCM185 in strain DLY23. Transformants have been cured of the pCM188-SUB2 plasmid and assayed for thermosensitivity at 37°C. Five clones have been isolated and two, sub2-201 and sub2-206, were analyzed further. Plasmid recovery and retransformation confirmed that the Ts phenotype was linked to mutations on the SUB2 gene.
Plasmid constructions
Cloning of the SUB2 gene was performed by homologous recombination directly into yeast strains. Briefly, a BamHI–NotI linearized and dephosphorylated vector was cotransformed with a PCR fragment containing the SUB2 coding region and short homology regions with the vector at the extremities. pCM188 (marker URA3) and pCM185 (marker TRP1), centromeric vectors which differ for the number of Tet operators (respectively 2 and 7; Gari et al. 1997), were used for most experiments. The S. pombe orthologue gene was cloned in vector pCM185 with the same method.
Growth conditions
Expression of the SUB2 gene in strain DLY23 was switched off by the addition of 5 μg/mL of doxycyclin. We verified that this dose was not toxic for growth. Ts strains were grown at 24°C for extracts and RNA preparations and switched to 37°C as required.
Library screening
Strain DLY9 was transformed with a cDNA S. pombe library in vector pFL61 (kind gift of F. Lacroute and M. Minet). Transformants were directly plated at 22°C and fast-appearing colonies were selected. Clones that restored a slow growth phenotype after plasmid loss were used for plasmid recovery and retesting.
Oligonucleotides
SUB2 was cloned in pCM188 and pCM185 using oligonucleotides (5′-CTAAATTACCGGATCAATTCGGGGGATCCATGTCACACGAAGG TGAAGA-3′) and (5′-TGCGGCCCTCCTGCAGGGCCCTAGCGGCCG CTTAATTATTCAAATAAGTGG-3′) for PCR. The S. pombe SPAC17G6.14c was cloned in pCM185 using oligonucleotides (5′-CTAAATTACCGGA TCAATTCGGGGGATCCATGGCATCTGCTCAGGAGGAT-3′) and (5′-TGCGGCCCTCCTGCAGGGCCCTAGCGGCCGCCTTATGCAT TCATGTATGAGCCA-3′). Primers for RPL32 and U3snRNA primer extension were, respectively, GTGGACTTGTAACCTAAGGTG and GAA GAGTCAAAGAGTGAC.
Splicing extracts, in vitro splicing, and spliceosome assembly
Splicing extracts were prepared essentially as described by Umen and Guthrie (1995). Native gels for commitment complex and spliceosome analysis and in vitro splicing reactions were as described (Seraphin and Rosbash 1989). U2snRNA was inactivated by the addition of oligonucleotide CAGATACTACACTTG to the final concentration of 1 μM 5′ before incubation with the RNA. Splicing extracts were ATP depleted by incubation at 25°C with 2 mM glucose for 10 min. Δ2BPGG RNA contains a double A to G mutation of the branch-point sequence (UACUAAC to UACUggC).
Extracts from sub2-201 Ts strain were inactivated in vitro with a 20-min incubation at 37°C. Chase experiments were performed as follows: extract 1 was incubated in the conditions described for 15 min; when required, U2snRNA was inactivated with a 5-min incubation before the chase; a 200-fold molar excess of cold RNA was then added before the addition of extract 2.
Acknowledgments
We would like to thank J. Banroques, J. Hamm, M. Green, J. Marie, M. Rosbash, L. Sperling, B. Seraphin, and K. Tanner for critical reading of the manuscript. Thanks also to M. Green, C. Guthrie, A. Kistler, and M. Zhang for communicating results before publication and for fruitful discussions. We are also indebted to F. Lacroute and M. Minet for the gift of the S. pombe cDNA library, E. Herrero for gift of pCM plasmids, Y. Kerjan for help with tetrad dissection, and P. Siliciano for gift of the original PRP40(HA)3 strain and for communicating results before publication. Finally, we thank A. Adoutte for constant support. This work was supported by the CNRS and the Fondation pour la Recherche Medicale.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL Libri@cgm.cnrs-gif.fr; FAX 33-1-69823877.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.852101.
References
- Chen EJ, Frand AR, Chitouras E, Kaiser CA. A link between secretion and pre-mRNA processing defects in Saccharomyces cerevisiae and the identification of a novel splicing gene, RSE1. Mol Cell Biol. 1998;18:7139–7146. doi: 10.1128/mcb.18.12.7139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de la Cruz J, Kressler D, Linder P. Unwinding RNA in Saccharomyces cerevisiae: DEAD-box proteins and related families. Trends Biochem Sci. 1999;24:192–198. doi: 10.1016/s0968-0004(99)01376-6. [DOI] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Gari E, Piedrafita L, Aldea M, Herrero E. A set of vectors with a tetracycline-regulatable promoter system for modulated gene expression in Saccharomyces cerevisiae. Yeast. 1997;13:837–848. doi: 10.1002/(SICI)1097-0061(199707)13:9<837::AID-YEA145>3.0.CO;2-T. [DOI] [PubMed] [Google Scholar]
- Gottschalk A, Tang J, Puig O, Salgado J, Neubauer G, Colot HV, Mann M, Seraphin B, Rosbash M, Luhrmann R, et al. A comprehensive biochemical and genetic analysis of the yeast U1 snRNP reveals five novel proteins. Rna. 1998;4:374–393. [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Siliciano PG. Identification of Prp40, a novel essential yeast splicing factor associated with the U1 small nuclear ribonucleoprotein particle. Mol Cell Biol. 1996;16:960–967. doi: 10.1128/mcb.16.3.960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Longtine MS, McKenzie A, III, Demarini DJ, Shah NG, Wach A, Brachat A, Philippsen P, Pringle JR. Additional modules for versatile and economical PCR-based gene deletion and modification in Saccharomyces cerevisiae. Yeast. 1998;14:953–961. doi: 10.1002/(SICI)1097-0061(199807)14:10<953::AID-YEA293>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- Lopez MC, Sanchez M, Ferminan E, Dominguez A. Disruption of six Saccharomyces cerevisiae genes from chromosome IV and basic phenotypic analysis of deletion mutants. Yeast. 1998;14:1199–1208. doi: 10.1002/(SICI)1097-0061(19980930)14:13<1199::AID-YEA309>3.0.CO;2-V. [DOI] [PubMed] [Google Scholar]
- Moore MJ, Query CC, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, Atkins JF, editors. The RNA world. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1993. pp. 303–357. [Google Scholar]
- Noble SM, Guthrie C. Identification of novel genes required for yeast pre-mRNA splicing by means of cold-sensitive mutations. Genetics. 1996;143:67–80. doi: 10.1093/genetics/143.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996;15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- O'Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- Puig O, Gottschalk A, Fabrizio P, Seraphin B. Interaction of the U1 snRNP with nonconserved intronic sequences affects 5′ splice site selection. Genes & Dev. 1999;13:569–580. doi: 10.1101/gad.13.5.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes & Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Rutz B, Seraphin B. Transient interaction of BBP/ScSF1 and Mud2 with the splicing machinery affects the kinetics of spliceosome assembly. Rna. 1999;5:819–831. doi: 10.1017/s1355838299982286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— A dual role for BBP/ScSF1 in nuclear pre-mRNA retention and splicing. EMBO J. 2000;19:1873–1886. doi: 10.1093/emboj/19.8.1873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Rosbash M. Identification of functional U1 snRNA-pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- ————— The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA-pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991;10:1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- ————— An RNA switch at the 5′ splice site requires ATP and the DEAD box protein Prp28p. Mol Cell. 1999;3:55–64. doi: 10.1016/s1097-2765(00)80174-4. [DOI] [PubMed] [Google Scholar]
- Tang J, Abovich N, Fleming ML, Seraphin B, Rosbash M. Identification and characterization of a yeast homolog of U1 snRNP-specific protein C. EMBO J. 1997;16:4082–4091. doi: 10.1093/emboj/16.13.4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umen JG, Guthrie C. A novel role for a U5 snRNP protein in 3′ splice site selection. Genes & Dev. 1995;9:855–868. doi: 10.1101/gad.9.7.855. [DOI] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- Wiest DK, O'Day CL, Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]