Abstract
The human 56-kD U2AF65-associated protein (hUAP56), a member of the DExD/H box protein family of RNA-dependent ATPases, is required for the stable binding of U2 snRNP to the pre-mRNA branchpoint. Here we identify a highly conserved Saccharomyces cerevisiae homolog of hUAP56, yUAP/Sub2p. yUAP/Sub2p can be functionally substituted for by hUAP56 and, like its human counterpart, is an essential pre-mRNA splicing factor. yUAP/Sub2p is required for formation of the commitment complex, the precursor for U2 snRNP addition. In conjunction with previous studies, we conclude that at least two DExD/H box proteins, Prp5p and yUAP/Sub2p, mediate the U2 snRNP-branchpoint interaction.
Keywords: pre-mRNA splicing, hUAp56, SUB2, U2AF, DEAD box protein, U2 snRNP binding, commitment complex
Pre-mRNA splicing occurs in a large and structurally dynamic complex, the spliceosome, within which the two-step transesterification reactions occur. To assemble the spliceosome, four ribonucleoprotein particles (U1, U2, U5, and U4/U6), together with a large number of non-snRNP proteins, interact with the pre-mRNA in a highly ordered and stepwise pathway (Will and Luhrmann 1997; Burge et al. 1999). The earliest event that specifically targets the pre-mRNA to the splicing pathway is formation of the U1 snRNP/pre-mRNA commitment complex both in yeast (Seraphin and Rosbash 1991) and in mammals (Michaud and Reed 1991; Jamison et al. 1992). In yeast, two distinct commitment complexes, CC1 and CC2, have been identified: CC1 requires only the 5′ splice site, whereas formation of CC2 requires both the 5′ splice site and the branchpoint (Seraphin and Rosbash 1989). The likely mammalian counterpart of CC2 is the E complex, whose formation requires both the 5′ splice site and the polypyrimidine (Py) tract adjacent to the branchpoint.
Although commitment complex assembly is ATP independent, many of the subsequent steps in spliceosome assembly and splicing require ATP-hydrolysis, the first of which is the stable binding of U2 snRNP to the pre-mRNA branchpoint. These ATP-dependent events are mediated by a series of splicing factors that are members of the DExD/H box protein family, the founding member of which is eIF-4A, a known RNA helicase (Kramer 1996; Staley and Guthrie 1998). Several of these DExD/H box splicing factors have been shown to possess an ATP-dependent RNA unwinding activity (Laggerbauer et al. 1998; Raghunathan and Guthrie 1998; Wagner et al. 1998; Wang et al. 1998) and are thought to use ATP hydrolysis as a driving force to modulate specific RNA structural rearrangements during spliceosome assembly. Among yeast protein splicing factors, seven belong to the DExD/H box protein family, and four of these have known human orthologs (for review, see Staley and Guthrie 1998).
To date, Prp5p is the only yeast DExD/H box protein shown to be required for the stable binding of U2 snRNP to the branchpoint (Dalbadie-McFarland and Abelson 1990; Ruby et al. 1993). In mammals, hUAP56 is the only known DExD/H box protein required for stable U2 snRNP binding (Fleckner et al. 1997). hUAP56 is an essential splicing factor, recruited to the pre-mRNA dependent on the Py-tract and U2AF65. hUAP56 interacts with U2AF65 directly, which presumably directs hUAP56 to the vicinity of the branchpoint. A variety of metazoan homologs of hUAP56 have been identified, from C. elegans through man, all of which contain a DECD motif as opposed to the canonical DEAD/H motif.
In this article, we identify yUAP/Sub2p, the S. cerevisiae homolog of hUAP56, and analyze its role in pre-mRNA splicing and spliceosome assembly. Our results indicate that hUAP56 is both structurally and functionally conserved from yeast to man and have mechanistic implications regarding early events of spliceosome assembly.
Results
Identification of a highly conserved yeast homolog of hUAP56
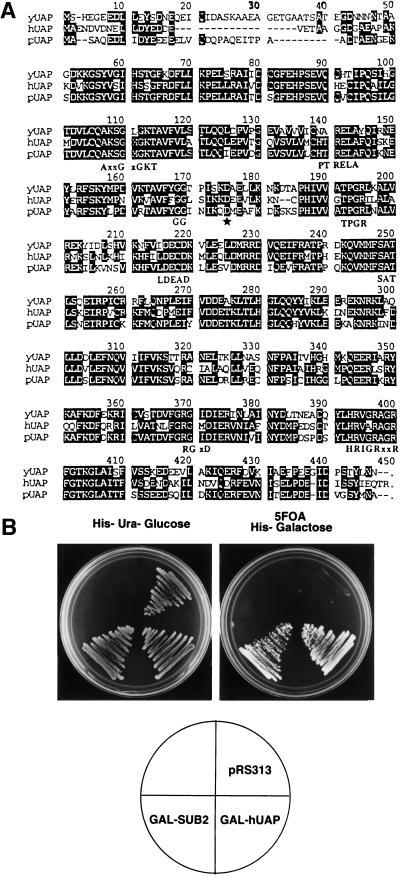
hUAP56 is required for the U2 snRNP-branchpoint interaction (Fleckner et al. 1997). In yeast, Prp5p is the only DExD/H box protein known to be required for this step of spliceosome assembly, raising the possibility that Prp5p may be the hUAP56 homolog. However, a search of the complete S. cerevisiae genome revealed an uncharacterized open reading frame whose protein product is far more similar to hUAP56 than Prp5p. Figure 1A shows an alignment between hUAP56, the S. cerevisiae protein and a putative Schizosaccharomyces pombe homolog. The S. cerevisiae protein is 65% identical and 83% similar to hUAP56, whereas Prp5p is only 30% identical and 55% similar to hUAP56. Moreover, the similarity between Prp5p and hUAP56 is confined to the seven signature motifs of DExD/H box proteins, whereas the similarity between the yeast protein and human UAP56 occurs throughout the entire protein. Finally, hUAP56 possesses a DECD sequence, as does the yeast protein, whereas Prp5p contains a DEAD sequence. On the basis of this unusually high homology and the experiments presented below, we conclude that this S. cerevisiae protein is the true homolog of hUAP56. Significantly, the gene encoding yUAP has been implicated previously in splicing as a high-copy suppressor of an snRNP biogenesis mutant, brr1-1, and referred to as SUB2 (Noble and Guthrie 1996; Kistler and Guthrie 2001). We therefore will refer to the gene as SUB2 and to its protein product as yUAP/Sub2p.
Figure 1.
A highly conserved Saccharomyces cerevisiae homolog of hUAP56. (A) Amino acid alignment. The S. cerevisiae yUAP/Sub2p amino acid sequence was aligned to human UAP56 (hUAP) and the hypothetical Schizosaccharomyces pombe UAP (pUAP) found in database searches. Identical amino acids in all three homologs are shaded. Conserved motifs characteristic of DExD/H box protein family are indicated at the bottom of the alignment. Star indicates the mutated aspartic acid residue in sub2-100, a temperature-sensitive allele of SUB2. (B) hUAP56 can functionally substitute for yUAP/Sub2p. Growth of cells on 5FOA, galactose plate is shown.
Using standard gene disruption procedures (Guthrie and Fink 1991; see Materials and Methods), we found that SUB2 was essential for viability (data not shown). The significant amino acid similarity between hUAP56 and yUAP/Sub2p (Fig. 1A) prompted us to ask whether hUAP56 can functionally substitute for yUAP/Sub2p. Plasmids in which hUAP56 or yUAP/Sub2p were expressed from the GAL1 promoter were transformed, in parallel, into a SUB2 deletion strain. Figure 1B shows that both hUAP56 and yUAP/Sub2p transformants grew on 5FOA plates, which selects against retention of the URA3-marked wild-type yUAP/Sub2p expression plasmid. Thus, hUAP56 can functionally substitute for yUAP/Sub2p, indicating that yUAP/Sub2p and hUAP56 are structurally and functionally conserved.
yUAP/Sub2p is an essential pre-mRNA splicing factor
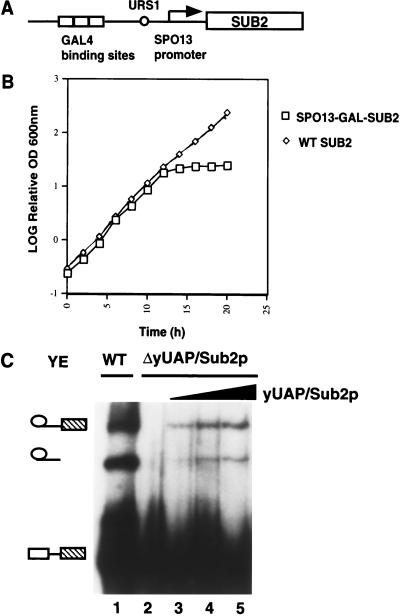
To investigate the role of yUAP/Sub2p in splicing, we constructed a yeast strain conditionally expressing yUAP/Sub2p from the GAL1 promoter. The strain grew on both glucose- and galactose-containing media (data not shown), indicating that the low level of uninduced expression from the GAL1 promoter was sufficient to support cell growth. To circumvent this problem, we expressed yUAP/Sub2p from a more tightly controlled promoter, SPO13–GAL (Fig. 2A). The SPO13 promoter is normally repressed by a cis-acting upstream element (URS1; Wang et al. 1987). Repression can be overridden by a strong transcriptional activator, such as GAL4 (Vidal et al. 1996). Figure 2B shows that the growth of this strain halted ∼12 h after shift from galactose- to glucose-containing media.
Figure 2.
yUAP/Sub2p is an essential pre-mRNA splicing factor. (A) Schematic diagram of a conditional yUAP/Sub2p expression construct. (B) Growth curve of strain MZ105, which conditionally expresses yUAP/Sub2p. Strain MZ102a, which expresses yUAP/Sub2p constitutively, was used as a control. Cultures of MZ105 and MZ102a in 4% galactose containing medium were centrifuged and the cells resuspended in rich medium containing 4% glucose at time zero. Every 2 h, the OD600 of the cultures was measured and the concentration of cells adjusted to remain between OD600 1.0 and 3.0. (C) Splicing analysis. Splicing reactions were carried out at 25°C for 30 min. In lane 1, splicing extract was prepared from strain MZ105 grown in galactose-containing media. In lanes 2–5, splicing extract was prepared from strain MZ105 16 h following shift from galactose- to glucose-containing media. In lanes 3–5, 0.1 μL, 0.5 μ, and 1 μL of GST-yUAP/Sub2p (0.1 mg/mL) was added to the reaction mixture.
To assess the role of yUAP/Sub2p in splicing, extracts were prepared from the SPO13–GAL–SUB2-containing strain after growth in glucose-containing media, which represses expression of yUAP/Sub2p, or galactose-containing media, which induces expression of yUAP/Sub2p. Figure 2C shows that splicing extracts prepared from cells expressing yUAP/Sub2p were active (lane 1), whereas the yUAP/Sub2p-depleted extract failed to support splicing (lane 2). Addition of yeast-derived GST-yUAP/Sub2p to the yUAP/Sub2p-depleted extract restored splicing (lanes 3–5). Thus, like its human counterpart, yUAP/Sub2p is an essential splicing factor.
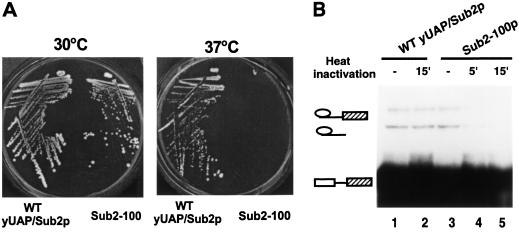
The direct role of yUAP/Sub2p in pre-mRNA splicing was confirmed by isolation and characterization of a conditional (temperature-sensitive) allele of yUAP, sub2-100. A yeast strain carrying sub2-100 grew normally at 30°C, whereas at 37°C, growth of this strain was severely inhibited (Fig. 3A). Sequence analysis revealed that the temperature-sensitive phenotype of sub2-100 was conferred by a single point mutation (adenine to guanine) resulting in the replacement of the aspartic acid at position 175 with glycine (see Fig. 1A). We note that D175 is not in any of the DExD/H box protein consensus motifs but is highly conserved from yeast to man.
Figure 3.
A temperature-sensitive mutant of yUAP/Sub2p. (A) Growth phenotype of sub2-100 containing strain, MZ306. (WT yUAP/Sub2), yeast strain MZ102b carrying SUB2. (Sub2-100), yeast strain MZ306 carrying a mutant allele of SUB2, sub2–100. (B) Splicing analysis. Splicing extracts were derived from strain MZ102b (lanes 1,2) and MZ306 grown at 30°C (lanes 3,4,5), respectively. Splicing reactions were carried out at 25°C for 30 min. In lane 4, the splicing extract was preincubated at 37°C for 5 min. In lanes 2 and 5, splicing extracts were preincubated at 37°C for 15 min.
Figure 3B shows that extracts prepared from the sub2-100 expression strain, grown at the permissive temperature, exhibited splicing activity comparable to that of the wild-type extract (cf. lanes 1 and 3). However, this extract was unable to support splicing following preincubation at 37°C for 15 min (lane 5). In contrast, the splicing activity of the wild-type extract was unaffected by the 37°C preincubation (cf. lanes 1 and 2).
yUAP/Sub2p is required for formation of CC2
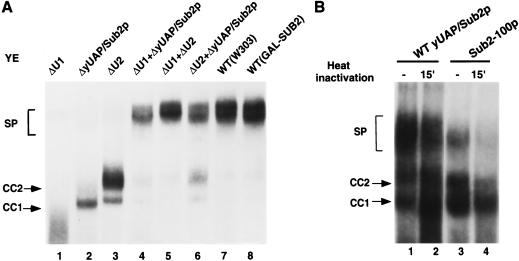
To assess the role of yUAP/Sub2p in complex assembly, we first used a native gel assay to determine the splicing complexes formed in a yUAP/Sub2p-depleted extract. As controls, U1- and U2-depleted extracts were analyzed in parallel. Figure 4A shows, as reported previously, that U1 snRNA depletion abolished formation of all splicing complexes (lane 1), U2 snRNA depletion resulted in arrest at the CC2 complex (lane 3), and a combination of U1- and U2-depleted extracts fully supported spliceosome assembly (lane 5). The yUAP/Sub2p-depleted extract (lane 2) was unable to form any ATP-dependent spliceosome, analogous to results in HeLa cell extracts (Fleckner et al. 1997). Unexpectedly, the branchpoint-dependent commitment complex, CC2, was also absent, whereas formation of the CC1 complex, which requires only the 5′ splice site, was not significantly affected. The yUAP/Sub2p-depleted extract efficiently complemented both U1 snRNA- and U2 snRNA-depleted extracts (lanes 4 and 6).
Figure 4.
yUAP/Sub2p is required for commitment complex formation. (A) Analysis of splicing complex formation using U1 snRNA-, U2 snRNA-, or yUAP/Sub2p-depleted extracts. yUAP/Sub2p-, U1 snRNA-, or U2 snRNA-depleted splicing extract was derived from strain MZ105, BS-Y82, or BS-Y88, respectively, 16 h after shift from galactose- to glucose-containing media. Splicing reactions (40% extract) were carried out and analyzed on a native gel. In lanes 4–6, 20% of the indicated extracts were used. In lanes 7 and 8, extracts were prepared from strain W303 and MZ105 grown in galactose-containing media. Commitment (CC1 and CC2) and spliceosomal (SP) complexes are indicated. (B) Analysis of splicing complex formation using sub2-100 containing extracts. Splicing extracts were derived from strain MZ102b (lanes 1,2), and MZ306 grown at 30°C (lanes 3,4), respectively. Splicing reactions (40% extract) were carried out for 20 min and analyzed on a native gel. In lanes 2 and 4, splicing extracts were preincubated at 37°C for 15 min before assembling the splicing reaction mixture.
The direct role of yUAP/Sub2p in splicing complex assembly was further verified by analyzing the sub2-100 extract. Figure 4B shows that extract prepared from the sub2-100 expression strain, grown at the permissive temperature, supported spliceosome assembly (lane 3). However, following preincubation at 37°C for 15 min, the sub2100 extract was unable to support spliceosome assembly (lane 4). By contrast, in the wild-type extract, spliceosome assembly was unaffected by the 37°C preincubation (cf. lanes 1 and 2). Significantly, in the heat-inactivated sub2-100 extract, the level of CC2 was also substantially reduced (cf. lanes 2 and 4), confirming a role for yUAP/Sub2p in CC2 formation.
Role of the yUAP/Sub2p DExD/H box consensus motifs
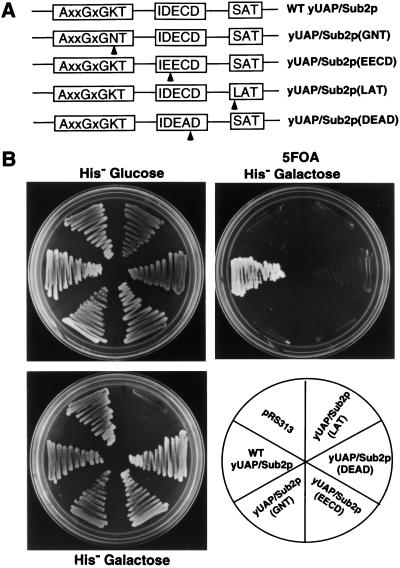
The fact that yUAP/Sub2p is essential for the formation of the CC2 complex, which is ATP independent, prompted us to examine the role of the DExD/H box family consensus motifs, several of which are required for ATP utilization. We designed a series of point mutations in yUAP/Sub2p based on the known role of these motifs in other DExD/H box proteins, in particular, of eIF-4A (Schmid and Linder 1991; Pause and Sonenberg 1992; Pause et al. 1993). The GKT motif was mutated to GNT to eliminate ATP binding; the DECD motif was changed to EECD or the SAT motif was mutated to LAT to eliminate putative RNA helicase activity (see Fig. 5A). Mutant or wild-type yUAP/Sub2p derivatives were placed under control of the GAL1 promoter and introduced into a SUB2 deletion strain. To test the ability of these yUAP/Sub2p mutants to support cell growth, transformants were restreaked onto 5-FOA, galactose-containing plates to shuffle out the wild-type SUB2-containing, URA3-marked plasmid. Figure 5B shows that cells expressing wild-type yUAP/Sub2p grew on 5-FOA, galactose plates, whereas cells transformed with mutant yUAP/Sub2p expression vector, or vector alone, did not. Thus, the GKT, DECD, and SAT motifs are essential for yUAP/Sub2p function, indicating that in addition to the ATP-independent role in commitment complex formation, yUAP/Sub2p has an ATP-dependent function (see Kistler and Guthrie 2001; Libri et al. 2001).
Figure 5.
Role of DExD/H box protein family consensus motifs in yUAP/Sub2p function. (A) Amino acid sequences of the mutated motifs. Arrows indicate the mutated residues. (B) Effects of yUAP/Sub2p mutations on cell growth. Strains used in top panel were derived from strain MZ102b and contain plasmid pMZ11 (SUB2, URA3, CEN/ARS) and an integrated copy of wild-type yUAP/Sub2p (MZ104) or various yUAP/Sub2p mutants (MZ106, MZ107, MZ108, and MZ109) expressed from the GAL1 promoter. Growth of cells carrying the indicated genes on His− glucose or 5FOA His− galactose is shown. In the bottom panel, W303 was used as the parental strain. Growth of cells on a His− galactose plate is shown.
To monitor the effects of overexpression of mutant yUAP/Sub2p on cell growth, mutant yUAP/Sub2p expression vectors were introduced into a wild-type yeast strain and the resulting transformants restreaked onto galactose-containing plates to induce mutant yUAP/Sub2p expression. Figure 5B shows that yUAP/Sub2p(LAT) transformants failed to grow; in contrast, yUAP/Sub2p(GNT) and yUAP/Sub2p(EECD) had little impact on cell growth. These results indicate that the SAT to LAT mutation in yUAP/Sub2p conferred a dominant-negative phenotype, analogous to the results with Prp2p (Plumpton et al. 1994).
A characteristic feature of all known hUAP56 homologs is the presence of a DECD motif (Fleckner et al. 1997; see Fig. 1A). This high conservation suggested that the cysteine residue may be required for UAP function. We mutated the DECD motif in yUAP/Sub2p to DEAD, and Figure 5B shows that yeast harboring yUAP/Sub2p (DEAD) were inviable. We conclude that complete yUAP/Sub2p function requires the cysteine residue of the DECD motif.
Discussion
In this article, we have identified and characterized yUAP/Sub2p, a yeast homolog of hUAP56. yUAP/Sub2p is essential for viability and can be replaced by hUAP56. Consistent with our previous work on hUAP56 (Fleckner et al. 1997), yUAP/Sub2p is an essential splicing factor required for stable binding of U2 snRNP to the pre-mRNA branchpoint. Surprisingly, we found that yUAP/Sub2p was required for the formation of the branchpoint-dependent CC2 commitment complex, whereas formation of branchpoint-independent CC1 commitment complex was unaffected by yUAP/Sub2p depletion or inactivation. This result suggests a role for yUAP/Sub2p in early 3′ splice site/branchpoint recognition or in 5′–3′ splice sites communication. Consistent with this idea, in the accompanying manuscripts SUB2 is shown to genetically interact with MUD2 (Kistler and Guthrie 2001) and PRP40/Namp8 (Libri et al. 2001). MUD2, PRP40, and Namp8 have all been implicated in commitment complex formation. Because CC2 complex formation occurs in the absence of ATP, the role of yUAP/Sub2p in this step must be unrelated to its putative ATP-dependent RNA helicase or ATPase activity. However, the DExD/H box motifs required for ATP utilization are essential for yUAP/Sub2p function, indicating that yUAP/Sub2p must also be required for the subsequent ATP-consuming steps. Accordingly, the accompanying manuscripts (Kistler and Guthrie 2001; Libri et al. 2001) demonstrate a role for yUAP/Sub2p subsequent to the ATP-dependent U2 snRNP-branchpoint interaction. The different complex assembly defects observed in the three studies are most likely explained by the fact that distinct yUAP/Sub2p mutants were analyzed. Taken together, the results of the three studies indicate that yUAP/Sub2p acts at multiple steps and in both an ATP-dependent and ATP-independent manner during spliceosome assembly.
Prp5p is another yeast DExD/H box protein required for entry of U2 snRNP into the spliceosome (Dalbadie-McFarland and Abelson 1990). Both genetic and biochemical studies have suggested that U2 snRNA is the target of Prp5p (Ruby et al. 1993; Wells and Ares 1994; O'Day et al. 1996; Wells et al. 1996; Wiest et al. 1996) and that Prp5p may facilitate U2 snRNP binding by exposing the U2 snRNA branchpoint recognition region (O'Day et al. 1996). It will be important to determine how the two DExD/H box proteins, yUAP/Sub2p and Prp5p, act in concert to promote the U2 snRNP-branchpoint interaction.
Materials and methods
Plasmids
The yeast SUB2 gene was cloned by PCR amplification of yeast genomic DNA and inserted into plasmid pRS416 (URA3, CEN/ARS) and pRS313 (HIS, CEN/ARS) at the BamHI site to generate plasmid pMZ11 and pMZ12. For hUAP56 or yUAP/Sub2p expression, pMZ13 (GAL–hUAP HIS3) or pMZ14 (GAL–yUAP/Sub2p HIS3) were constructed by inserting the human UAP56 or yUAP/Sub2p coding sequence into pSW107 (Walker et al. 1997) under control of the GAL1 promoter. Plasmid pMZ19 (SPO13–GAL–yUAP/Sub2p HIS3), used for conditional yUAP/Sub2p expression, contains the SPO13–GAL promoter derived from pMV253-13 (Vidal et al. 1996) and the yUAP/Sub2p coding sequence in the pRS313 backbone.
Synthetic oligonucleotides were designed to introduce point mutations in the SUB2 gene by a two-round PCR strategy (Higuchi et al. 1988). Mutated yUAP/Sub2p fragments were subcloned into the wild-type yUAP/Sub2p backbone in plasmid pMZ14, generating plasmids pMZ15 (GAL–yUAP/Sub2p–GNT), pMZ16 (GAL–yUAP/Subp2–EECD), pMZ17 (GAL–yUAP/Sub2p–LAT), and pMZ18 (GAL–yUAP/Sub2p–DEAD), respectively. All mutations were confirmed by sequencing.
Yeast strains
Stains for conditional U1 or U2 snRNA expression, BS-Y82 (GAL-U1) and BS-Y88 (GAL-U2), were obtained from M. Rosbash (Seraphin and Rosbash 1989). All strains constructed in this study were derived from W303 (ade2-1 his3-11 his3-15 leu2-3 leu2-112 trp1-1 ura3-1 can-100). Growth of yeast cells in rich or synthetic medium was performed by standard procedures (Guthrie and Fink 1991).
To disrupt the SUB2 gene, a 2073-bp HpaI–StuI fragment containing the entire yUAP/Sub2p coding sequence was replaced with TRP1 gene. This null SUB2 allele was introduced into diploid W303 by one-step gene replacement (Guthrie and Fink 1991). Correct integration in the resulting strain MZ101 (SUB2/sub2::TRP1) was verified by Southern blotting.
Strain MZ101 (SUB2/sub2::TRP1) was transformed with pMZ11, and four isogenic haploids, MZ102a, MZ102b, MZ102c, and MZ102d, were obtained. MZ102b (sub2::TRP1/pMZ11) was used for all the subsequent gene-shuffling experiments.
hUAP56 and yUAP/Sub2p expression strain MZ103 (sub2::TRP1, trp3::GAL–hUAP56 HIS3, pMZ11) and MZ104 (sub2::TRP1, trp3::GAL–yUAP/Sub2p HIS, pMZ11) were constructed by integration of linearized pMZ13 and pMZ14 into MZ102b at the TRP3 locus as described (Walker et al. 1997). For conditional yUAP/Sub2p expression, plasmid pMZ19 was transformed into MZ102b, and pMZ11 was shuffled out to generate MZ105 (sub2::TRP1, pMZ19). To construct strains conditionally expressing yUAP/Sub2p mutants, linearized plasmids pMZ14, pMZ15, pMZ16, pMZ17, and pMZ18 were integrated into strain MZ102b or W303a at the TRP3 locus. The resulting strains are: MZ104, MZ106 (sub2::TRP1, trp3::GAL–yUAP/Sub2p–GNT HIS3, pMZ11), MZ107 (sub2::TRP1, trp3::GAL–yUAP/Sub2p–EECD HIS3, pMZ11), MZ108 (sub2::TRP1, trp3::GAL–yUAP/Sub2p–LAT HIS3, pMZ11), MZ109 (sub2::TRP1, trp3::GAL–yUAP/Sub2p–DEAD HIS3, pMZ11), MZ117 (trp3::GAL–yUAP/Sub2p HIS3), MZ110 (trp3::GAL–yUAP/Sub2p–GNT HIS3), MZ111 (trp3::GAL- yUAP/Sub2p–EECD HIS3), MZ112 (trp3::GAL–yUAP/Sub2p–LAT HIS3), and MZ113 (trp3::GAL–yUAP/Sub2p–DEAD HIS3).
Splicing and complex assembly assay
Splicing extracts were prepared according to Liao et al. (1992). For preparation of depleted extracts, cells were harvested after shifting from galactose- to glucose-containing media for 16 h. The pre-mRNA substrate WT-Δ2 (Liao et al. 1992) was used in all experiments. Splicing and native gel analysis of splicing complex formation was carried out as described previously (Seraphin and Rosbash 1989).
Preparation of GST–yUAP/Sub2p
yUAP/Sub2p coding sequence was cloned into plasmid pPS892 (a gift from P. Silver, Harvard Medical School) in frame, and the resulting plasmid pMZ20 (GAL1–GST–yUAP/Sub2p, URA3) was transformed into strain MZ105 to generate strain MZ114 (sub2::TRP1, pMZ20). Strain MZ114 was grown in galacotose-containing media, which induces GST–yUAP/Sub2p expression, to OD600 2–3, and whole-cell extracts were prepared. GST–yUAP/Sub2p was purified on glutathione-agrose beads and dialyzed against buffer D.
Isolation of a yUAP/Sub2p temperature-sensitive mutant
Mutagenized yUAP/Sub2p fragments were created using error-prone PCR (Leung et al. 1989) and subcloned into plasmid pMZ12 at HpaI and XbaI to replace the wild-type yUAP/Sub2p fragment. All the Escherichia coli transformants (∼10,000) were collected to generate a library of yUAP/Sub2p mutants. This library was subsequently transformed into yeast strain MZ102b. To screen temperature-sensitive yUAP/Sub2p mutants, ∼4000 transformants were patched onto His− plates and then replica-plated to 5FOA media at 30°C and 37°C. Approximately 30% of the transformants did not grow on 5FOA plates, indicating lethal mutations in yUAP/Sub2p. Colonies that grew at 30°C but not at 37°C were restreaked to verify the temperature-sensitive phenotype. Plasmids were recovered from temperature-sensitive strains and retransformed into a SUB2 deletion strain to confirm that the temperature-sensitive phenotype was caused by mutated yUAP/Sub2p.
Acknowledgments
We thank Michael Rosbash for providing yeast strains and Wu-Cheng Shen for comments on the manuscript. We are especially grateful to Amy Kistler, Christine Guthrie, and Domenico Libri for communicating results before publication, providing reagents, and insightful discussions. M.R.G. is an investigator of the Howard Hughes Medical Institute. This work was supported by a grant from NIH to M.R.G.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL michael.green@umassmed.edu; FAX (508) 856-5473.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.851701.
References
- Burge CB, Tuschl TH, Sharp PA. Splicing of precursors to mRNAs by the spliceosome. In: Gesteland RF, et al., editors. The RNA world. 2nd ed. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 1999. pp. 525–560. [Google Scholar]
- Dalbadie-McFarland G, Abelson J. PRP5: A helicase-like protein required for mRNA splicing in yeast. Proc Natl Acad Sci. 1990;87:4236–4240. doi: 10.1073/pnas.87.11.4236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes & Dev. 1997;11:1864–1872. doi: 10.1101/gad.11.14.1864. [DOI] [PubMed] [Google Scholar]
- Guthrie C, Fink GR. Guide to yeast genetics and molecular biology. New York: Academic Press; 1991. [Google Scholar]
- Higuchi R, Krummel B, Saiki RK. A general method of in vitro preparation and specific mutagenesis of DNA fragments: Study of protein and DNA interactions. Nucleic Acids Res. 1988;16:7351–7367. doi: 10.1093/nar/16.15.7351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jamison SF, Crow A, Garcia-Blanco MA. The spliceosome assembly pathway in mammalian extracts. Mol Cell Biol. 1992;12:4279–4287. doi: 10.1128/mcb.12.10.4279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for Sub2, an essential spliceosomal ATPase. Genes & Dev. 2001;15:42–49. doi: 10.1101/gad.851301. (This issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kramer A. The structure and function of proteins involved in mammalian pre-mRNA splicing. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- Laggerbauer B, Achsel T, Luhrmann R. The human U5–200kD DEXH-box protein unwinds U4/U6 RNA duplices in vitro. Proc Natl Acad Sci. 1998;95:4188–4192. doi: 10.1073/pnas.95.8.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung DW, Chen E, Goeddel DV. A method for random mutagenesis of a defined DNA segment using a modified polymerase chain reaction. Technique. 1989;1:11–15. [Google Scholar]
- Liao XC, Colot HV, Wang Y, Rosbash M. Requirements for U2 snRNP addition to yeast pre-mRNA. Nucleic Acids Res. 1992;20:4237–4245. doi: 10.1093/nar/20.16.4237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Libri D, Graziani N, Saguez C, Boulay J. Multiple roles for the yeast SUB2/yUAP56 gene in splicing. Genes & Dev. 2001;15:36–41. doi: 10.1101/gad.852101. (This issue). [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michaud S, Reed R. An ATP-independent complex commits pre-mRNA to the mammalian spliceosome assembly pathway. Genes & Dev. 1991;5:2534–2546. doi: 10.1101/gad.5.12b.2534. [DOI] [PubMed] [Google Scholar]
- Noble SM, Guthrie C. Transcriptional pulse-chase analysis reveals a role for a novel snRNP-associated protein in the manufacture of spliceosomal snRNPs. EMBO J. 1996;15:4368–4379. [PMC free article] [PubMed] [Google Scholar]
- O'Day CL, Dalbadie-McFarland G, Abelson J. The Saccharomyces cerevisiae Prp5 protein has RNA-dependent ATPase activity with specificity for U2 small nuclear RNA. J Biol Chem. 1996;271:33261–33267. doi: 10.1074/jbc.271.52.33261. [DOI] [PubMed] [Google Scholar]
- Pause A, Sonenberg N. Mutational analysis of a DEAD box RNA helicase: The mammalian translation initiation factor eIF-4A. EMBO J. 1992;11:2643–2654. doi: 10.1002/j.1460-2075.1992.tb05330.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pause A, Methot N, Sonenberg N. The HRIGRXXR region of the DEAD box RNA helicase eukaryotic translation initiation factor 4A is required for RNA binding and ATP hydrolysis. Mol Cell Biol. 1993;13:6789–6798. doi: 10.1128/mcb.13.11.6789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Plumpton M, McGarvey M, Beggs JD. A dominant negative mutation in the conserved RNA helicase motif ‘SAT’ causes splicing factor PRP2 to stall in spliceosomes. EMBO J. 1994;13:879–887. doi: 10.1002/j.1460-2075.1994.tb06331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raghunathan PL, Guthrie C. RNA unwinding in U4/U6 snRNPs requires ATP hydrolysis and the DEIH-box splicing factor Brr2. Curr Biol. 1998;8:847–855. doi: 10.1016/s0960-9822(07)00345-4. [DOI] [PubMed] [Google Scholar]
- Ruby SW, Chang TH, Abelson J. Four yeast spliceosomal proteins (PRP5, PRP9, PRP11, and PRP21) interact to promote U2 snRNP binding to pre-mRNA. Genes & Dev. 1993;7:1909–1925. doi: 10.1101/gad.7.10.1909. [DOI] [PubMed] [Google Scholar]
- Schmid SR, Linder P. Translation initiation factor 4A from Saccharomyces cerevisiae: Analysis of residues conserved in the D-E-A-D family of RNA helicases. Mol Cell Biol. 1991;11:3463–3471. doi: 10.1128/mcb.11.7.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Seraphin B, Rosbash M. Identification of functional U1 snRNA–pre-mRNA complexes committed to spliceosome assembly and splicing. Cell. 1989;59:349–358. doi: 10.1016/0092-8674(89)90296-1. [DOI] [PubMed] [Google Scholar]
- ————— The yeast branchpoint sequence is not required for the formation of a stable U1 snRNA–pre-mRNA complex and is recognized in the absence of U2 snRNA. EMBO J. 1991;10:1209–1216. doi: 10.1002/j.1460-2075.1991.tb08062.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staley JP, Guthrie C. Mechanical devices of the spliceosome: Motors, clocks, springs, and things. Cell. 1998;92:315–326. doi: 10.1016/s0092-8674(00)80925-3. [DOI] [PubMed] [Google Scholar]
- Vidal M, Brachmann RK, Fattaey A, Harlow E, Boeke JD. Reverse two-hybrid and one-hybrid systems to detect dissociation of protein–protein and DNA–protein interactions. Proc Natl Acad Sci. 1996;93:10315–10320. doi: 10.1073/pnas.93.19.10315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner JD, Jankowsky E, Company M, Pyle AM, Abelson JN. The DEAH-box protein PRP22 is an ATPase that mediates ATP-dependent mRNA release from the spliceosome and unwinds RNA duplexes. EMBO J. 1998;17:2926–2937. doi: 10.1093/emboj/17.10.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker SS, Shen WC, Reese JC, Apone LM, Green MR. Yeast TAF(II)145 required for transcription of G1/S cyclin genes and regulated by the cellular growth state. Cell. 1997;90:607–614. doi: 10.1016/s0092-8674(00)80522-x. [DOI] [PubMed] [Google Scholar]
- Wang HT, Frackman S, Kowalisyn J, Esposito RE, Elder R. Developmental regulation of SPO13, a gene required for separation of homologous chromosomes at meiosis I. Mol Cell Biol. 1987;7:1425–1435. doi: 10.1128/mcb.7.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y, Wagner JD, Guthrie C. The DEAH-box splicing factor Prp16 unwinds RNA duplexes in vitro. Curr Biol. 1998;8:441–451. doi: 10.1016/s0960-9822(98)70178-2. [DOI] [PubMed] [Google Scholar]
- Wells SE, Ares M. Interaction between highly conserved U2 small nuclear RNA structures and Prp5p, Prp9p, Prp11p, and Prp21p proteins are required to ensure integrity of the U2 small nuclear ribonucleoprotein in Saccharomyces cerevisiae. Mol Cell Biol. 1994;14:6337–6349. doi: 10.1128/mcb.14.9.6337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wells SE, Neville M, Haynes M, Wang J, Igel H, Ares M. CUS1, a supressor of cold-sensitive U2 snRNA mutations, is a novel yeast splicing factor homologous to human SAP 145. Genes & Dev. 1996;10:220–232. doi: 10.1101/gad.10.2.220. [DOI] [PubMed] [Google Scholar]
- Wiest DK, O'Day CL, Abelson J. In vitro studies of the Prp9.Prp11.Prp21 complex indicate a pathway for U2 small nuclear ribonucleoprotein activation. J Biol Chem. 1996;271:33268–33276. doi: 10.1074/jbc.271.52.33268. [DOI] [PubMed] [Google Scholar]
- Will CL, Luhrmann R. Protein functions in pre-mRNA splicing. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]