Abstract
The ird5 gene was identified in a genetic screen for Drosophila immune response mutants. Mutations in ird5 prevent induction of six antibacterial peptide genes in response to infection but do not affect the induction of an antifungal peptide gene. Consistent with this finding, Escherichia coli survive 100 times better in ird5 adults than in wild-type animals. The ird5 gene encodes a Drosophila homolog of mammalian IκB kinases (IKKs). The ird5 phenotype and sequence suggest that the gene is specifically required for the activation of Relish, a Drosophila NF-κB family member.
Keywords: Innate immunity, IκB kinase, Drosophila, antimicrobial peptide, Relish, NF-κB
In both mammals and Drosophila, microbial infection activates Toll-like receptor (TLR) signaling pathways as a part of the innate host defense response (for review, see Anderson 2000). TLR-mediated signaling pathways are essential for appropriate responses to bacterial infection. In addition, mouse Tlr4 mediates septic shock associated with infection by gram-negative bacteria (Vogel 1992; Poltorak et al. 1998).
The available data indicate that different microbial cell wall components activate different Toll-like receptor signaling pathways, which regulate distinct sets of target genes. In mammals, TLR4 is the prime mediator of responses to bacterial lipopolysaccharide, while TLR2 mediates responses to bacterial peptidoglycans (Poltorak et al. 1998; Takeuchi et al. 1999; for review, see Beutler 2000). The best-studied aspect of the Drosophila innate immune response is the rapid transcriptional induction of antimicrobial peptide genes in response to infection (Hultmark 1993; Hoffmann 1995). Infection by different classes of microorganisms leads to the preferential induction of particular subsets of antimicrobial peptides (Lemaitre et al. 1997), indicating that different microbial components activate different signaling pathways.
At least two Toll-related signaling pathways are required for the activation of the Drosophila antimicrobial peptide genes. The Toll pathway itself, which was first identified because of its essential role in Drosophila embryonic patterning (Anderson et al. 1985), is essential for the induction of an antifungal peptide gene, drosomycin, although the antibacterial peptide genes are still induced in Toll pathway mutants (Lemaitre et al. 1996). Another Drosophila member of the Toll family, 18-wheeler, is required for the normal induction of attacin, an antibacterial peptide gene, but mutations in 18-wheeler do not prevent the induction of other antibacterial peptides (Williams et al. 1997). The imd gene is important for the induction of Diptericin and other antibacterial peptides (Lemaitre et al. 1995a; Corbo and Levine 1996) and, therefore, appears to be a component of a third signaling pathway activated by infection, but its biochemical function is not known.
Each of the three Drosophila signaling pathways activated by infection leads to activation of NF-κB/Rel dimers, just as the mammalian TLRs activate NF-κB. All three Drosophila Rel proteins, Dorsal, Dif, and Relish, are expressed in the fat body cells that produce the antimicrobial peptides, and all three are activated within 30 min after infection by translocation from the cytoplasm to the nuclei (Ip at al. 1993; Lemaitre et al. 1995b; Stöven et al. 2000). Adults that lack Dif fail to induce Drosomycin, an antifungal peptide, and Defensin, which is active against gram-positive bacteria, but the other antimicrobial peptide genes are induced normally (Manfruelli et al. 1999; Meng et al. 1999; Rutschmann et al. 2000). Animals that lack Dorsal show normal induction of the antimicrobial peptide genes in response to infection (Lemaitre et al. 1995b), although Dorsal may act redundantly with Dif in larvae (Manfruelli et al. 1999; Rutschmann et al. 2000). Relish is a compound protein with an N-terminal Rel domain and a C-terminal IκB-like domain, similar to mammalian p100 and p105 (Dushay et al. 1996). Relish is activated by signal-dependent proteolysis, which liberates the N-terminal Rel domain, allowing it to translocate into nuclei (Stöven et al. 2000). Adults that lack Relish completely fail to induce the antibacterial peptides Diptericin and Cecropin and show reduced induction of the other antimicrobial peptides (Hedengren et al. 1999).
The signaling pathway that activates Relish and controls induction of the antibacterial peptide genes has not been defined. We carried out a genetic screen to identify EMS-induced mutations on the Drosophila third chromosome that affect the antibacterial signaling pathway (Wu and Anderson 1998). A large number of mutants were identified and named ird (immune response deficient) mutants. This screen identified two alleles of the ird5 gene on the basis of the failure of homozygous mutant larvae to induce a diptericin-lacZ reporter gene in response to infection. Here we show that the ird5 gene is essential for antibacterial responses and encodes a Drosophila homolog of mammalian IκB kinases.
Results and Discussion
ird5 mutations block the antibacterial but not the antifungal immune response
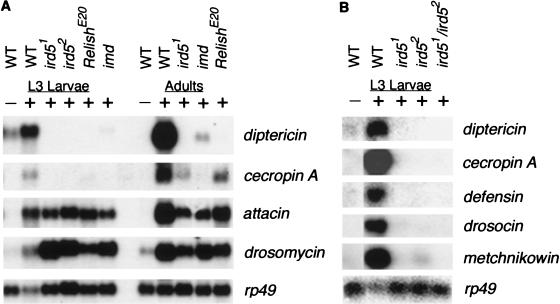
Mutations in ird5 prevent induction of a diptericin-lacZ reporter gene in response to infection and also prevent transcriptional induction of the endogenous diptericin gene (Wu and Anderson 1998). We examined E. coli-induced expression of all seven classes of antimicrobial peptide genes in ird5 mutant larvae by RNA blot hybridization, including the genes encoding antibacterial (Diptericin, Cecropin A, Defensin, Attacin, Drosocin, and Metchnikowin) and antifungal (Drosomycin) peptides (Fig. 1A,B). In wild-type larvae, all the antimicrobial genes were strongly induced after bacterial challenge. In contrast, in larvae homozygous for either ird5 allele, there was no detectable induction of the diptericin, cecropin A, defensin, drosocin, or metchnikowin genes. The attacin gene was induced in the mutants to ∼30% of normal levels, while drosomycin was induced to normal levels (Fig. 1; Table 1). The same effects on the induction of antimicrobial peptide genes were seen in ird51/Df and ird52/Df animals (data not shown), suggesting that both alleles cause a complete loss of gene function.
Figure 1.
Expression of the antimicrobial peptide genes in wild-type and mutant animals. The induction of antimicrobial peptide genes was assayed by Northern blot. (A) Induction of diptericin, cecropin, drosomycin, and attacin in wild-type, ird5, Relish, and imd mutants in response to infection at L3 larval and adult stages. (B) Induction of the battery of antimicrobial peptides in wild-type and ird5 mutants at L3 larval stage. Each lane contains 30 μg of total RNA from untreated third-instar larvae (−) or 2–3 h after injection with Escherichia coli (+). rp49 was used as loading control.
Table 1.
Induction of antimicrobial peptides in mutants
| Antimicrobial peptides
|
L3 Larvae
|
Adults
|
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Wild type
|
Wild type
|
ird51
|
ird52
|
Relish
|
imd
|
Wild type
|
Wild type
|
ird51
|
Relish
|
imd
|
|
| Uninfected
|
Infected
|
Uninfected
|
Infected
|
||||||||
| diptericin | 0 | 100 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 2 | 0 | 100 | 1 ± 1 | 0 ± 0 | 13 ± 7 |
| cecropin | 0 | 100 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 ± 0 | 0 | 100 | 14 ± 2 | 30 ± 7 | 5 ± 3 |
| attacin | 0 | 100 | 33 ± 20 | 29 ± 16 | 43 ± 28 | 20 ± 15 | 0 | 100 | 35 ± 17 | 64 ± 30 | 47 ± 26 |
| drosomycin | 0 | 100 | 125 ± 21 | 54 ± 39 | 46 ± 28 | 76 ± 30 | 0 | 100 | 40 ± 15 | 71 ± 18 | 90 ± 8 |
Induction of antimicrobial peptides in response to infection in mutant animals, expressed as the percentage of the induction seen in wild type. Northern blots (including the data in Fig. 1) were analyzed by phosphorimager and normalized relative to the rp49 loading control. The average and standard deviation in three experiments are shown.
Mutations in three other genes, imd, Relish, and Dredd, have been shown to prevent normal induction of antibacterial peptide genes in adult Drosophila (Hedengren et al. 1999; Elrod-Erickson et al. 2000; Leulier et al. 2000). We compared the pattern of antimicrobial peptide gene induction in ird5 mutants with that in imd and Relish mutants in both larvae and adults (Fig. 1A; Table 1). Mutations in all three genes had very similar effects on antimicrobial gene induction in larvae: diptericin and cecropin A were not induced; attacin induction was reduced and drosomycin induction was normal. In adult animals, the antimicrobial gene expression phenotype of ird5 and Relish mutants were very similar: diptericin induction was blocked, cecropin A and attacin induction was reduced, and drosomycin induction was normal. The antimicrobial gene expression phenotype of imd adults was slightly different, with some residual diptericin expression. Mutations in Dredd, a Drosophila caspase, prevent normal induction of diptericin and attacin and allow induction of drosomycin (Elrod-Erickson et al. 2000; Leulier et al. 2000). These comparisons suggest that ird5, Dredd, Relish, and probably imd act in the same signaling pathway to control the induction of antibacterial peptide genes in response to infection.
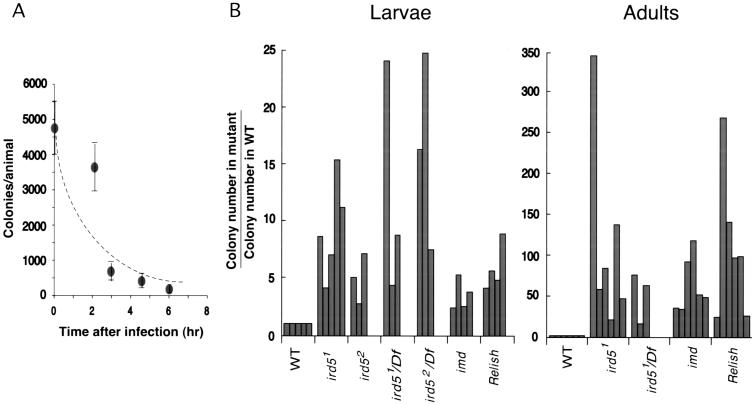
To assess the importance of the ird5 gene in controlling the growth of invading bacteria, we compared bacterial survival and growth in wild-type, ird5, imd, and Relish animals (see Materials and Methods). In wild-type larvae, most of the E. coli injected into the animal were killed by 6 h after infection (Fig. 2A). At this same time point, there were four to 15 times as many E. coli in ird5 mutant larvae as in wild-type animals (Fig. 2B). The effects of the ird5 mutations were more striking in experiments with adults: at 24 h after infection, there were 20–350 times as many bacteria per animal in ird5 mutants compared to wild type. The bacterial growth phenotype of ird5 mutants was similar to that seen in Relish mutant larvae and adults and somewhat stronger than that of imd mutants (Fig. 2B). This is consistent with the stronger effects of ird5 and Relish on the antimicrobial peptide genes: Mutations in either ird5 or Relish prevent normal induction of diptericin, cecropin, drosocin, attacin, and metchnikowin (Fig. 1; Hedengren et al. 1999), while induction of metchnikowin is induced in imd mutants (Levashina et al. 1998).
Figure 2.
Analysis of bacterial growth after Escherichia coli infection. (A) Wild-type larvae kill E. coli within hours after infection. The number of ampicillin-resistant E. coli present per animal at time points after injection is shown. The data shown are the average of two experiments, with three to five animals per time point in each experiment. (B) Bacterial growth in wild-type, ird5, Relish, and imd mutant larvae and adults after infection, indicated as the ratio of the number of colony-forming units (cfu) per animal in the mutant relative to the number of cfu in the wild-type control in the same experiment. Each bar represents an independent experiment. For larvae, the number of bacteria per animal was assayed at 6 h after infection to ensure that the animals had not begun to pupate. For adults, the number of bacteria per animal was assayed at 24 h after infection. Df: Df(3R)sbd45.
ird5 encodes a Drosophila IκB kinase
We mapped the mutations responsible for the failure to induce the diptericin-lacZ reporter gene for both ird5 alleles between the visible markers cu (86D1-4) and sr (90D2-F7) on the right arm of the third chromosome. The deficiency Df(3R)sbd45(89B4-10) failed to complement the immune response defect of either ird5 allele. Further deficiency-complementation tests and male recombination mapping narrowed the ird5 interval to 89B4-9, between pannier and Stubble (data not shown). Two molecular-defined genes in this interval were considered as candidates responsible for the ird5 phenotype, Akt and a gene defined by an EST that was related to mammalian IκB kinases (IKKs; Fig. 3A). Mutant alleles of Akt cause recessive lethality (Staveley et al. 1998), and ird5/Akt(l(3)89Bq) heterozygous animals were viable and showed normal induction of the diptericin-lacZ reporter gene (data not shown), indicating that the ird5 phenotypes were not caused by mutations in Akt.
Figure 3.
The wild-type DmIkkβ gene and the ird5 mutations. (A) The DmIkkβ genomic region in the polytene map. Based on deficiency mapping, ird5 lies in 84B4-9, proximal to Sb; male recombination mapping placed ird5 distal to pannier (pnr; data not shown). In genomic DNA, DmIkkβ lies between pnr and mini spindles (msps; Cullen et al. 1999); Akt is 50 kb distal to DmIkkβ. Comparison of the genomic and cDNA sequences indicated that DmIkkβ has five exons that produce a 2.7-kb transcript, shown relative to the restriction map of genomic DNA. (B) Comparison of domains of ird5/DmIkkβ and human IKKβ indicating the position of the ird51 stop codon. The homology between the Drosophila and human genes is greatest in the kinase domain (34% identity to human IKKβ); there is weak homology to the leucine zipper (LZ) and helix–loop–helix (HLH) motifs in the Drosophila gene (dotted boxes). (C) DmIkkβ expression in wild-type and ird5 mutants, as assayed by Northern blot. Two transcript sizes, 2.7 and 4.2 kb, are present in wild-type animals; the smaller transcript corresponds to the cDNAs we have isolated. Based on phosphorimager analysis, both DmIkkβ transcripts are induced 1.5-fold at 2 h after infection compared with uninfected animals. rp49 was the loading control. (−), uninfected; (+), 2 h after Escherichia coli infection; L3, third-instar larvae; F, adult female; Df, Df(3R)sbd45.
Mammalian IKKβ is required for activation of NF-κB in response to inflammatory signals such as TNF-α and IL-1 (Q. Li et al. 1999; Z. Li et al. 1999); the IKK homolog was therefore considered as a candidate gene for ird5. We cloned a full-length cDNA for the IKK homolog, which we call DmIkkβ (Fig. 3B). The same gene was also identified molecularly as encoding a kinase activated by LPS in a Drosophila cell line (Kim et al. 2000; Medzhitov and Janeway 2000; Silverman et al. 2000). Based on genomic DNA sequence, DmIkkβ is located between pannier and mini-spindles (Fig.3A; Experimental Procedures). We detected two size classes of transcripts, 2.7 and 4.2 kb, from the DmIkkβ gene (Fig. 3C); the cDNA corresponded to the 2.7-kb transcript. Both transcripts were expressed at higher levels after infection (Fig. 3C). Similar induction of other genes that encode components of the immune response machinery has been observed previously (Dushay et al. 1996; Lemaitre et al. 1996). We sequenced the complete open reading frame of DmIkkβ from the ird51 and ird52 chromosomes. We identified a single C-to-T nucleotide substitution in ird51 that would change a glutamine codon (CAA) at amino acid 266 of the open reading frame to a stop codon (TAA) within the conserved kinase domain (Fig. 3). No sequence changes were identified in the open reading frame in ird52; however, neither DmIkkβ transcript was detectable in ird52 homozygotes (Fig. 3). This analysis indicates that both ird5 alleles are associated with mutations that should abolish DmIkkβ activity.
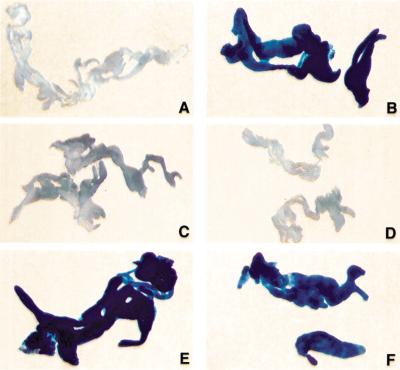
To confirm that ird5 is the same as the DmIkkβ gene, we tested the ability of a DmIkkβ transgene to rescue the immune response defect of ird5 flies. Neither a UAS-DmIkkβ cDNA transgene nor a fat-body GAL4 driver alone rescued the immunity phenotype of ird51 (Fig. 4). However, in ird51 diptericin-lacZ/Df(3R)sbd45 larvae carrying both a UAS-DmIkkβ cDNA transgene and a transgene that expressed GAL4 in the fat body, the diptericin-lacZ reporter was activated after E. coli infection (Fig. 4). These results demonstrate that DmIkkβ is the gene responsible for the ird5 mutant phenotype.
Figure 4.
Expression of the DmIkkβ cDNA rescues the ird5 phenotype. Each panel shows histochemical staining of fat body β-galactosidase activity in third-instar larvae carrying the diptericin-lacZ reporter gene. Wild-type uninfected (A) and infected (B) larvae, showing the normal induction of the reporter after infection. (C) Fat body from infected w; Gal-c564/+; ird51, diptericin-lacZ, e/Df(3R)sbd45, e larvae. These ird5 animals carrying the GAL4 driver failed to induce the reporter gene, just as do ird5 animals. (D) Fat body from infected w; UAS-DmIkkβ-cDNA (6–4)/+; ird51 diptericin-lacZ, e/ ird51 diptericin-lacZ, e larvae. These ird5 animals carrying the UAS-cDNA failed to induce the reporter gene, like ird5 mutants. (E) and (F) ird5 homozygous larvae carrying both the GAL4 driver and the UAS-DmIkkβ transgene expressed the reporter gene after infection. (E) Fat body from infected w; UAS-DmIkkβ-cDNA (6–4)/Gal-c564 ; ird51 diptericin-lacZ, e/Df(3R)sbd45, e larvae. (F) Infected larvae carrying a different transgene insertion site also express the reporter: fat body from infected w; UAS-DmIkkβ-cDNA (9–9)/Gal-c564; ird51 diptericin-lacZ, e/Df(3R)sbd45 animals.
Ird5/DmIkkβ is not a component of the Toll/cactus pathway
The ird5 immune response phenotype showed striking specificity: All of the antibacterial peptide genes were strongly affected by the ird5 mutations, but the antifungal peptide gene drosomycin was induced normally in ird5 mutants. The specific immune response phenotype of ird5/DmIkkβ in vivo contrasts with the global effects on antimicrobial peptide genes seen in cell lines when a dominant negative form of the same gene was expressed in cultured cells (Kim et al. 2000). The ird5/DmIkkβ mutant phenotype implies that, in vivo, ird5 is not an essential component of the Toll pathway, which is required for the induction of drosomycin. The ird5/DmIkkβ gene is therefore a component of an independent signaling pathway, which could be activated by another member of the Drosophila Toll-like receptor family (Tauszig et al. 2000).
Mammalian IKKα and IKKβ phosphorylate serine residues in the N-terminal domain of IκB that target IκB for degradation, thereby allowing the nuclear localization and activation of NF-κB (Chen et al. 1996; DiDonato et al. 1997; Regnier et al. 1997). The ird5/DmIkkβ sequence suggests that the protein encoded by this gene phosphorylates an IκB-like protein. There are two known Drosophila IκB-like proteins that could act as inhibitor proteins in the immune response, Cactus, and the C-terminal ankyrin repeat domain of Relish. In cactus mutants, drosomycin is expressed constitutively, but the antibacterial peptide genes are not (Lemaitre et al. 1996), which indicates that Cactus is not involved in the pathways that regulate the antibacterial peptide genes. Furthermore, ird5/DmIkkβ homozygous mutant females are fertile, demonstrating that this gene is not required for degradation of Cactus during dorsal-ventral patterning in the embryo.
Ird5/DmIkkβ and Relish act in a common pathway
The ird5/DmIkkβ phenotype is similar to the phenotype of Relish mutants (Hedengren et al. 1999; Figs. 1,2). For both genes, homozygous mutant flies are viable and fertile, indicating that the two genes are not essential for development. Mutations in either Relish or ird5/DmIkkβ completely prevent induction of diptericin and cecropin but allow some induction of attacin and drosomycin (Fig. 1). Mutations in either gene produce comparable effects on bacterial growth (Fig. 2). These results argue that ird5/DmIkkβ and Relish act in the same signaling pathway and suggest that Ird5/DmIkkβ activates Relish-containing dimers. Relish activation requires proteolytic cleavage of Relish protein into an N-terminal Rel domain that translocates to the nucleus and a C-terminal ankyrin repeat domain that remains in the cytoplasm (Stöven et al. 2000). Recent biochemical experiments have shown that DmIkkβ can phosphorylate Relish protein (Silverman et al. 2000), which is consistent with the model that phosphorylation of Relish by DmIkkβ leads to targeted proteolysis and activation of Relish.
Drosophila IκB kinases in development and immunity
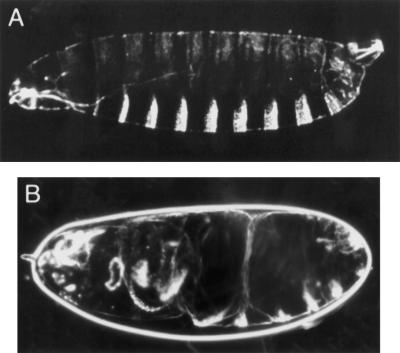
Although ird5/DmIkkβ is expressed maternally (data not shown), ird5 mutant females are fertile, demonstrating that the gene is not required for embryonic dorsal-ventral patterning. However, a small fraction of embryos (∼0.5%) produced by homozygous ird51 or ird51/ird52 females show a weakly dorsalized phenotype (Fig. 5), suggesting that ird5/DmIkkβ does have a minor role in the maternal pathway that activates Dorsal. We suggest that there is another kinase in the early embryo that is primarily responsible for phosphorylation and degradation of Cactus. The normal induction of drosomycin in ird5/DmIkkβ mutants suggests that there will also be another kinase activated by the Toll pathway in the immune response—perhaps the same kinase that acts downstream of Toll to activate Dorsal in the embryo. The genome sequence indicates that there is one additional IκB kinase gene in Drosophila. Future experiments will test whether this gene plays a role in embryonic patterning and the antifungal immune response.
Figure 5.
Weak maternal effect of ird5. (A) Dark field view of the cuticle of a wild-type larva just after hatching. Most of the embryos produced by ird5 homozygous females show this wild-type cuticle pattern. Dorsal is up. (B) Cuticle of a weakly dorsalized embryo produced by an ird51/ird51 female, showing the twisted body characteristic of embryos that have deficiencies in mesoderm, the most ventral cell type (Anderson et al. 1985).
Our data suggest that different Drosophila Rel dimers are activated by homologous but distinct signaling pathways. Given the similarities of innate immune response pathways in Drosophila and mammals, it is likely that similar pathway-specific signaling components will mediate the activities of the members of the mammalian Rel proteins.
Materials and methods
Bacterial infection and bacterial survival in infected animals
In bacterial growth experiments, E. coli were introduced into animals by injection, as described previously (Wu and Anderson 1998). For larvae, E. coli containing an Ampr plasmid were diluted 10-fold from an OD600 0.5 culture in PBS and mixed with 1:200-fold diluted India ink. Wandering third instar larvae of the appropriate genotype were injected, with the ink as indicator of the volume injected, transferred to apple juice agar plates, and incubated at 25°C for 6–7 h. Because many of the injected larvae pupated shortly thereafter, longer time points were not assayed. For adults, the E. coli culture was diluted 100-fold from an OD600 0.5 culture in PBS; a lower concentration of E. coli was used in the adult experiments because adults do not control bacterial growth as efficiently as larvae. Adult flies (1–2 d old) of the appropriate genotype were injected and incubated at 29°C for 24 h. For both larvae and adults, three to five animals at the appropriate time after E. coli injections were homogenized in LB media and spread on LB plates containing Ampicillin (50μg/mL).
Molecular characterization of DmIkkβ
A full-length cDNA corresponding to the 2.7-kb RNA was isolated from an embryonic cDNA library, using the ESTs GM10440 and LD09214 (Genome Systems) as probes. According to the NetStart 1.0 program, the open reading frame is likely to be translated starting at a methionine 20 amino acids upstream of that reported previously (Kim et al. 2000; Medzhitov and Janeway 2000); the position of the stop codon in ird51 at position 266 is numbered using this upstream methionine.
We did not isolate cDNAs corresponding to the larger (4.2-kb) RNA, but RT–PCR analysis of RNA from infected larvae revealed that transcripts exist with additional 5′ and 3′ sequences. One transcript has an alternative 3′ coding exon of five amino acids and a different 3′ untranslated region than present in the cDNA.
For sequencing, genomic DNA from wild-type and homozygous ird5 mutant flies was amplified by PCR. Two independent clones of each PCR product were subcloned into pGEM-T vector (Promega) and were sequenced by the Cornell sequencing facility. For Northern analysis of the transcript, RNA was prepared using the RNA-STAT60 (Tel-Test) reagent. Poly(A)+ RNA was purified using the Oligotex (QIAGEN) kit. Four micrograms of poly(A)+ RNA was electrophoresed and transferred to Hybond N+ nylon filters.
Northern analysis
To assay antimicrobial peptide gene induction, E. coli were injected into wandering third-instar larvae and adult flies, as described previously (Wu and Anderson 1998). DNA from the complete DmIkkβ cDNA and antimicrobial peptide cDNAs were labeled by random priming (Boehringer) and used as probes.
Embryonic phenotypes
Cuticle preparations were made as described previously (Wieschaus and Nüsslein-Volhard 1986).
Transgene production and rescue of the ird5 phenotype
The 2.7-kb DmIkkβ cDNA was cloned into the pUAST (w+) transformation vector (Brand and Perrimon 1993). The construct was introduced into y w flies by P element–mediated transformation (Spradling 1986). w+ adults were used to establish several independent lines with insertions located on X, second, and third chromosomes. Several lines with insertions on the second chromosome were crossed with flies carrying ird51 mutant allele to obtain flies with the following genotype: w; UAS-DmIkkβ (cDNA); ird51 diptericin-lacZ, e/T(2;3) CyO; TM6B Tb (strain A). The second-chromosome GAL4 line Gal-c564, which is expressed in the fat body (Harrison et al. 1995), was used to drive expression of the UAS construct. We confirmed that Gal-c564 is expressed in the fat body by crossing to a UAS-GFP transgenic line. Flies carrying Gal-c564 were crossed to flies carrying a deficiency uncovering ird5 to obtain flies with the following genotype: w; Gal-c564; Df(3R)sbd45, e/ T(2;3) CyO; TM6B Tb (strain B). Strains A and B flies were then crossed and the progeny Tb+ larvae were selected and tested for the ability to induce the diptericin-lacZ reporter gene in response to infection. Escherichia coli infection and β-galactosidase activity analysis were performed as described previously (Wu and Anderson 1998), but the staining reaction was carried out for 30–60 min at 37°C.
Acknowledgments
We thank Joe Delaney and David Schneider for help with the design of the bacterial growth experiments. We thank Tim Bestor and Katie Brennan for helpful comments on the manuscript. We thank Pascal Heitzler for deficiency stocks, Armen Manoukian for the Akt allele, Norbert Perrimon for the GAL4 lines (which were generated by K. Kaiser), the Drosophila Stock Centers in Bloomington and Umeå for stocks, and the Berkeley Drosophila Genome Project for EST sequences and cDNA library. This work was supported by NIH AI45149 and the Lita Annenberg Hazen Foundation. L.P.W. is a Leukemia Society of America Special Fellow.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL k-anderson@ski.mskcc.org; FAX (212) 717-3623.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.856901.
References
- Anderson KV. Toll signaling pathways in the innate immune response. Curr Opin Immunol. 2000;12:13–19. doi: 10.1016/s0952-7915(99)00045-x. [DOI] [PubMed] [Google Scholar]
- Anderson KV, Jürgens G, Nüsslein-Volhard C. Establishment of dorsal-ventral polarity in the Drosophila embryo: Genetic studies on the role of the Toll gene product. Cell. 1985;42:779–789. doi: 10.1016/0092-8674(85)90274-0. [DOI] [PubMed] [Google Scholar]
- Beutler B. Tlr4: Central component of the sole mammalian LPS sensor. Curr Opin Immunol. 2000;12:20–26. doi: 10.1016/s0952-7915(99)00046-1. [DOI] [PubMed] [Google Scholar]
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. doi: 10.1242/dev.118.2.401. [DOI] [PubMed] [Google Scholar]
- Chen ZJ, Parent L, Maniatis T. Site-specific phosphorylation of IκBα by a novel ubiquitination-dependent protein kinase activity. Cell. 1996;84:853–862. doi: 10.1016/s0092-8674(00)81064-8. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M. Characterization of an immunodeficiency mutant in Drosophila. Mech Dev. 1996;55:211–220. doi: 10.1016/0925-4773(96)00506-0. [DOI] [PubMed] [Google Scholar]
- Cullen CF, Deak P, Glover DM, Ohkura H. mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiDonato JA, Hayakawa M, Rothwarf DM, Zandi E, Karin M. A cytokine-responsive IκB kinase that activates the transcription factor NF-κB. Nature. 1997;388:548–554. doi: 10.1038/41493. [DOI] [PubMed] [Google Scholar]
- Dushay MS, Asling B, Hultmark D. Origins of immunity: Relish, a compound Rel-like gene in the antibacterial defense of Drosophila. Proc Natl Acad Sci. 1996;93:10343–10347. doi: 10.1073/pnas.93.19.10343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elrod-Erickson M, Mishra S, Schneider D. Interactions between the cellular and humoral immune responses in Drosophila. Curr Biol. 2000;10:781–784. doi: 10.1016/s0960-9822(00)00569-8. [DOI] [PubMed] [Google Scholar]
- Harrison DA, Binari R, Nahreini TS, Gilman M, Perrimon N. Activation of a Drosophila Janus kinase (JAK) causes hematopoietic neoplasia and developmental defects. EMBO J. 1995;14:2857–2865. doi: 10.1002/j.1460-2075.1995.tb07285.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hedengren M, Asling B, Dushay MS, Ando I, Ekengren S, Wihlborg M, Hultmark D. Relish, a central factor in the control of humoral but not cellular immunity in Drosophila. Mol Cell. 1999;4:827–837. doi: 10.1016/s1097-2765(00)80392-5. [DOI] [PubMed] [Google Scholar]
- Hoffmann JA. Innate immunity of insects. Curr Opin Immunol. 1995;7:4–10. doi: 10.1016/0952-7915(95)80022-0. [DOI] [PubMed] [Google Scholar]
- Hultmark D. Immune reactions in Drosophila and other insects: A model for innate immunity. Trends Genet. 1993;9:178–183. doi: 10.1016/0168-9525(93)90165-e. [DOI] [PubMed] [Google Scholar]
- Ip YT, Reach M, Engström Y, Kadalayil L, Cai H, Gonzalez-Crespo S, Tatei K, Levine M. Dif, a dorsal-related gene that mediates an immune response in Drosophila. Cell. 1993;75:753–763. doi: 10.1016/0092-8674(93)90495-c. [DOI] [PubMed] [Google Scholar]
- Kim YS, Han SJ, Ryu JH, Choi KH, Hong YS, Chung YH, Perrot S, Raibaud A, Brey PT, Lee WJ. Lipopolysaccharide-activated kinase, an essential component for the induction of the antimicrobial peptide genes in Drosophila melanogaster cells. J Biol Chem. 2000;275:2071–2079. doi: 10.1074/jbc.275.3.2071. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Kromer-Metzger E, Michaut L, Nicolas E, Meister M, Georgel P, Reichhart JM, Hoffmann JA. A recessive mutation, immune deficiency (imd), defines two distinct control pathways in the Drosophila host defense. Proc Natl Acad Sci. 1995a;92:9465–9469. doi: 10.1073/pnas.92.21.9465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Meister M, Govind S, Georgel P, Steward R, Reichhart JM, Hoffmann JA. Functional analysis and regulation of nuclear import of Dorsal during the immune response in Drosophila. EMBO J. 1995b;14:536–545. doi: 10.1002/j.1460-2075.1995.tb07029.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86:973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- Lemaitre B, Reichhart JM, Hoffmann JA. Drosophila host defense: Differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc Natl Acad Sci. 1997;94:14614–14619. doi: 10.1073/pnas.94.26.14614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leulier F, Rodriguez A, Khush RS, Abrams JM, Lemaitre B. The Drosophila caspase Dredd is required to resist Gram-negative bacterial infection. EMBO Rep. 2000;1:353–358. doi: 10.1093/embo-reports/kvd073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levashina EA, Ohresser S, Lemaitre B, Imler JL. Two distinct pathways can control expression of the gene encoding the Drosophila antimicrobial peptide metchnikowin. J Mol Biol. 1998;278:515–527. doi: 10.1006/jmbi.1998.1705. [DOI] [PubMed] [Google Scholar]
- Li Q, Van Antwerp D, Mercurio F, Lee KF, Verma IM. Severe liver degeneration in mice lacking the IκB kinase 2 gene. Science. 1999;284:321–325. doi: 10.1126/science.284.5412.321. [DOI] [PubMed] [Google Scholar]
- Li ZW, Chu W, Hu Y, Delhase M, Deerinck T, Ellisman M, Johnson R, Karin M. The IKKβ subunit of IκB kinase (IKK) is essential for nuclear factor κB activation and prevention of apoptosis. J Exp Med. 1999;189:1839–1845. doi: 10.1084/jem.189.11.1839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfruelli P, Reichhart JM, Steward R, Hoffmann JA, Lemaitre B. A mosaic analysis in Drosophila fat body cells of the control of antimicrobial peptide genes by the Rel proteins Dorsal and DIF. EMBO J. 1999;18:3380–3391. doi: 10.1093/emboj/18.12.3380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Janeway C. Innate immune recognition: Mechanisms and pathways. Immun Rev. 2000;173:89–97. doi: 10.1034/j.1600-065x.2000.917309.x. [DOI] [PubMed] [Google Scholar]
- Meng X, Khanuja BS, Ip YT. Toll receptor-mediated Drosophila immune response requires Dif, an NF-κB factor. Genes & Dev. 1999;13:792–797. doi: 10.1101/gad.13.7.792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poltorak A, He X, Smirnova I, Liu MY, Huffel CV, Du X, Birdwell D, Alejos E, Silva MM, Galanos C, et al. Defective LPS signaling in C3H/HeJ and C57BL/10ScCr mice: Mutations in Tlr4 gene. Science. 1998;282:2085–2088. doi: 10.1126/science.282.5396.2085. [DOI] [PubMed] [Google Scholar]
- Regnier CH, Song HY, Gao X, Goeddel DV, Cao Z, Rothe M. Identification and characterization of an IκB kinase. Cell. 1997;90:373–383. doi: 10.1016/s0092-8674(00)80344-x. [DOI] [PubMed] [Google Scholar]
- Rutschmann S, Jung AC, Hetru C, Reichhart J-M, Hoffmann JA, Ferrandon D. The Rel protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity. 2000;12:569–580. doi: 10.1016/s1074-7613(00)80208-3. [DOI] [PubMed] [Google Scholar]
- Silverman N, Zhou R, Stöven S, Pandey N, Hultmark D, Maniatis T. A Drosophila IκB kinase complex required for Relish cleavage and antibacterial immunity. Genes & Dev. 2000;14:2461–2471. doi: 10.1101/gad.817800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spradling AC. P element-mediated transformation. In: Roberts DM, editor. Drosophila: A practical approach. Oxford: IRL; 1986. pp. 175–197. [Google Scholar]
- Staveley BE, Ruel L, Jin J, Stambolic V, Mastronardi FG, Heitzler P, Woodgett JR, Manoukian AS. Genetic analysis of protein kinase B (AKT) in Drosophila. Curr Biol. 1998;8:599–602. doi: 10.1016/s0960-9822(98)70231-3. [DOI] [PubMed] [Google Scholar]
- Stöven S, Ando I, Kadalayil L, Engström Y, Hultmark D. Activation of the Drosophila NF-κB factor Relish by rapid endoproteolytic cleavage. EMBO Rep. 2000;1:347–352. doi: 10.1093/embo-reports/kvd072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeuchi O, Hoshino K, Kawai T, Sanjo H, Takada H, Ogawa T, Takeda K, Akira S. Differential roles of TLR2 and TLR4 in recognition of gram-negative and gram-positive bacterial cell wall components. Immunity. 1999;11:443–451. doi: 10.1016/s1074-7613(00)80119-3. [DOI] [PubMed] [Google Scholar]
- Tauszig S, Jouanguy E, Hoffmann JA, Imler JL. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc Natl Acad Sci. 2000;97:10520–10525. doi: 10.1073/pnas.180130797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel SN. The Lps gene. Insights into the genetic and molecular basis of LPS responsiveness and macrophage differentiation. In: Beutler B, editor. Tumor necrosis factors: The molecules and their emerging role in medicine. New York: Raven; 1992. pp. 485–513. [Google Scholar]
- Wieschaus E, Nüsslein-Volhard C. Looking at embryos. In: Roberts DM, editor. Drosophila: A practical approach. Oxford: IRL; 1986. pp. 199–227. [Google Scholar]
- Williams MJ, Rodriguez A, Kimbrell DA, Eldon ED. The 18-wheeler mutation reveals complex antibacterial gene regulation in Drosophila host defense. EMBO J. 1997;16:6120–6130. doi: 10.1093/emboj/16.20.6120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu LP, Anderson KV. Regulated nuclear import of Rel proteins in the Drosophila immune response. Nature. 1998;392:93–97. doi: 10.1038/32195. [DOI] [PubMed] [Google Scholar]