Abstract
Replication-associated histone genes encode the only metazoan mRNAs that lack polyA tails, ending instead in a conserved 26-nt sequence that forms a stem–loop. Most of the regulation of mammalian histone mRNA is posttranscriptional and mediated by this unique 3′ end. Stem–loop–binding protein (SLBP) binds to the histone mRNA 3′ end and is thought to participate in all aspects of histone mRNA metabolism, including cell cycle regulation. To examine SLBP function genetically, we have cloned the gene encoding Drosophila SLBP (dSLBP) by a yeast three-hybrid method and have isolated mutations in dSLBP. dSLBP function is required both zygotically and maternally. Strong dSLBP alleles cause zygotic lethality late in development and result in production of stable histone mRNA that accumulates in nonreplicating cells. These histone mRNAs are cytoplasmic and have polyadenylated 3′ ends like other polymerase II transcripts. Hypomorphic dSLBP alleles support zygotic development but cause female sterility. Eggs from these females contain dramatically reduced levels of histone mRNA, and mutant embryos are not able to complete the syncytial embryonic cycles. This is in part because of a failure of chromosome condensation at mitosis that blocks normal anaphase. These data demonstrate that dSLBP is required in vivo for 3′ end processing of histone pre-mRNA, and that this is an essential function for development. Moreover, dSLBP-dependent processing plays an important role in coupling histone mRNA production with the cell cycle.
Keywords: Drosophila, cell cycle, SLBP, histone, replication, RNA processing
The replication-dependent histone mRNAs are the only metazoan mRNAs that do not have polyA tails. Instead, they end in a conserved 26-nt sequence that contains a 16-nt stem–loop (Marzluff 1992; Dominski and Marzluff 1999). The only processing reaction necessary for formation of mature histone mRNA is an endonucleolytic cleavage to form the 3′ end (Gick et al. 1986). This reaction is directed by two sequence elements, the stem–loop sequence (Vasserot et al. 1989; Melin et al. 1992) and a conserved region 3′ of the stem–loop that binds to the U7 snRNP (Mowry and Steitz 1987; Cotten et al. 1988; Soldati and Schumperli 1988). The protein that binds the stem–loop, SLBP, has previously been cloned from vertebrates (Wang et al. 1996b; Martin et al. 1997). SLBP is required for histone pre-mRNA processing in vitro (Dominski et al. 1999) and accompanies the histone mRNA to the cytoplasm, where it remains as part of the histone mRNP (Dominski et al. 1995; Hanson et al. 1996).
In cultured mammalian cells, accumulation of histone mRNA occurs only during S phase and requires ongoing DNA replication (Schumperli 1986; Osley 1991). Even though this much has been known for many years, it remains unclear how histone mRNA production is coupled to other regulatory events in the cell cycle. Much of the control of histone mRNA levels is mediated by the 3′ end of histone mRNA (Stauber et al. 1986; Luscher and Schumperli 1987; Stauber and Schumperli 1988; Harris et al. 1991). Both pre-mRNA processing and mRNA half-life (Pandey and Marzluff 1987) are regulated by the sequence at the 3′ end of the histone mRNA. SLBP protein also only accumulates during S phase of the mammalian cell cycle and is destroyed at entry into G2 phase, suggesting that regulation of SLBP is one of the major factors responsible for cell cycle regulation of histone mRNA in mammalian cells (Whitfield et al. 2000).
In early embryonic development of many species (e.g., frogs, sea urchins, and fruit flies), there are very high rates of DNA replication and, hence, a massive demand for histone proteins. During these cycles, histone mRNA is not cell cycle regulated and is present continuously, although these histone mRNAs end in the same stem–loop sequence as the cell cycle–regulated mRNAs expressed later in development. In frogs and Drosophila, histone mRNA is synthesized during oogenesis and deposited in large amounts in the egg (Woodland and Adamson 1977; Anderson and Lengyel 1980, 1984). Histone mRNA levels remain constant throughout the initial zygotic cycles when there is no transcription and then increase when transcription is activated at the midblastula transition.
The failure to coordinate histone mRNA accumulation with DNA synthesis during the initial rapid embryonic cell cycles is likely related to the differences in cell cycle regulation between early and late development. In many animals, the very first cell cycles lack gap phases and consist simply of a series of alternating S and M phases. The key regulators of the cell cycle, the cyclin-cdk complexes, are also differentially regulated between early and late development. For instance, in Drosophila, cyclin E/cdk2 is present constitutively during the first 16 embryonic cell cycles, and periodic activation of cyclin E/cdk2 does not occur until interphase 17, when G1/S control is introduced into the cell cycle (Richardson et al. 1993; Duronio and O'Farrell 1995; Sauer et al. 1995). At this stage, there is robust cell cycle–regulated histone expression, with histone mRNA accumulating only in replicating cells (Hassan and Vaessin 1997; R.J. Duronio, unpubl.).
We have initiated genetic analyses in Drosophila to examine the role played by SLBP in histone mRNA regulation during development. Here we report the isolation and characterization of mutations in the single Drosophila dSLBP gene. Our results demonstrate that production of the normal histone mRNA 3′ end in vivo is essential for development and requires dSLBP function. Moreover, dSLBP plays a major role in restricting histone mRNA accumulation to S phase.
Results
Isolation of Drosophila SLBP cDNA
We cloned the Drosophila stem–loop–binding protein (dSLBP) using the same three-hybrid strategy that was successful for cloning the vertebrate SLBPs (Wang et al. 1996a; Martin et al. 1997). Five positive clones were obtained from screening a Drosophila 0–4-h embryonic cDNA library (Dahanukar et al. 1999). Each of these clones encoded a fusion protein containing a region that was 75% identical to the 72–amino acid RNA binding domain (RBD) of the vertebrate proteins (Fig. 1B). Inspection of the Drosophila cDNA database resulted in the identification of ESTs that overlapped with the fusion protein clones, although the available sequences did not include the RBD. These cDNAs were obtained and sequenced, allowing identification of the predicted translation product (Fig. 1A). dSLBP is a 276–amino acid protein, slightly larger than the vertebrate SLBPs. The end of the dSLBP RBD is located 17 amino acids from the COOH terminus. Hydrophobic and basic residues in the RBD of vertebrate SLBP that are required for binding the stem–loop (Dominski et al. 2001) are conserved in dSLBP (Fig. 1B). There are few obvious similarities between dSLBP and the vertebrate or worm SLBPs other than the RBD.
Figure 1.
Cloning of dSLBP. (A) Predicted translation product of Drosophila SLBP cDNA. The 71–amino acid RNA binding domain is shown in bold. (B) Alignment of human, Xenopus, Drosophila, and Caenorhabditis elegans SLBP RNA-binding domains. Note that two SLBP proteins have been identified in Xenopus. The asterisks indicated the conserved hydrophobic and basic residues in vertebrate SLBP known to be required for RNA binding (Dominski et al. 2001).
dSLBP binds the histone mRNA 3′UTR stem–loop
Gel mobility shift assays were performed to test whether dSLBP is the major protein in Drosophila extracts that is capable of binding the histone RNA stem loop. When extract prepared from Drosophila ovaries (not shown), cultured SL2 cells (Fig. 2A, lanes 3,4), or 0–20-h Drosophila embryos (Fig. 2A, lane 5) was incubated with a radiolabeled stem–loop RNA probe, a single gel-shifted complex was detected. This complex specifically bound the stem–loop, as unlabeled wild-type stem–loop RNA could effectively compete for binding (Fig. 2A, lane 6) but a reversed stem–loop RNA sequence could not (Fig. 2A, lane 7). The complex comigrates with that formed between in vitro translated dSLBP and the stem–loop RNA (Fig. 2A, lane 1), suggesting that the complex detected in extracts contains dSLBP. To test this further, we performed supershift experiments. Polyclonal antibodies raised against recombinant dSLBP supershift the complex formed in Drosophila embryo extracts, whereas nonspecific antibodies have no effect (Fig 2B, cf. lanes 3,4). Addition of purified dSLBP protein to the reaction effectively competes with antibody binding and blocks formation of the supershifted complex (Fig. 2B, lane 5). These results indicate that virtually all of the gel-shift activity in the embryo extracts is caused by the dSLBP we have cloned.
Figure 2.
dSLBP binds the histone mRNA stem loop. (A) Detection of dSLBP by gel-shift assay. Incubation of a radiolabeled histone stem–loop probe (SL Probe) with in vitro–translated dSLBP (lane 1) retards the mobility of the free probe (lane 2). An identical gel shift is observed with SL2 cell cytoplasmic extract (lane 3), SL2 cell nuclear extract (lane 4), or 0–20-h embryo extract (EE; lane 5). Addition of unlabeled wild-type stem–loop RNA (WT) to the reaction effectively competes gel shift activity (lane 6), whereas addition of a reversed stem–loop sequence (RS) does not (lane 7). (B) Anti-dSLBP antibodies supershift the dSLBP/stem loop complex. Addition of a nonspecific antibody (αH) to the binding reaction (lane 3) has no effect on the mobility of the embryonic dSLBP/stem–loop complex (lane 2). Addition of affinity purified anti-dSLBP antibody (αD) reduces the mobility of the dSLBP/stem–loop complex (lane 4, asterisk). Formation of the supershifted complex is prevented by inclusion of excess purified dSLBP protein in the reaction (lane 5). (C) Expression of dSLBP in Xenopus oocytes. Endogenous Xenopus SLBP can be detected by incubation of the stem loop probe (lane 1) with extract from uninjected oocytes (lane 2, asterisk). Whole-cell extracts made from oocytes (Oo-W) injected with a synthetic dSLBP mRNA result in the appearance of a new complex (lane 3, arrow) that has the same mobility as the complex formed with a Drosophila embryo extract (lane 6). Nuclear (lane 4) and cytoplasmic (lane 5) fractions of dSLBP-injected oocytes each contain gel-shift activity, indicating that dSLBP can enter the nucleus.
Injection of Xenopus oocytes with synthetic mRNA encoding SLBP results in the synthesis of active SLBP that can be detected by gel-shift analyses using extracts prepared from the injected oocytes (Wang et al. 1999). When dSLBP is expressed in frog oocytes, there is accumulation of a large amount of a new complex that has the same mobility as the complex detected in Drosophila embryo extract (Fig. 2C, cf. lanes 3 and 6). There is an equal amount of the dSLBP-RNA complex in nuclear (Fig. 2C, lane 4) and cytoplasmic (Fig. 2C, lane 5) fractions, indicating that dSLBP is imported into the nucleus. dSLBP is also detected in extract from both cytoplasmic and nuclear fractions of SL2 cells (Fig. 2A, lanes 3,4). These data are consistent with dSLBP being involved in histone pre-mRNA processing in the nucleus.
dSLBP mRNA is expressed in all stages of the Drosophila life cycle
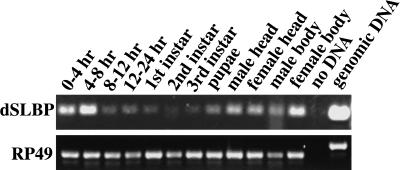
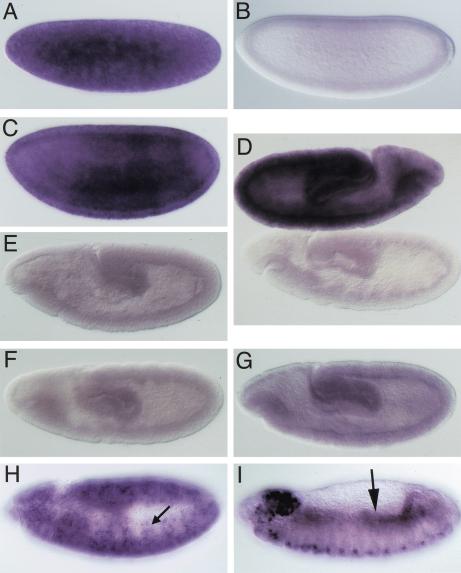
We assayed a panel of first-strand cDNAs isolated from a variety of Drosophila tissues and developmental stages (Rapid Scan, ORIGEN) by PCR to get an estimate of the timing of expression of dSLBP mRNA (Fig. 3). The primers used amplify a 181-nt fragment from dSLBP mRNA. As a control, the RP49 ribosomal protein mRNA was assayed. The primers used for the RP49 mRNA generate a different size fragment with Drosophila genomic DNA because of the presence of a small intron. This confirms that the Rapid Scan panels are free of pre-mRNA or DNA contamination. dSLBP mRNA is expressed in all stages of the Drosophila life cycle (Fig. 3), suggesting that it may function in all cycling cells. The mRNA is most abundant in early (0–8-h) embryos and adult females. This is consistent with storage of maternal dSLBP mRNA in the developing oocyte to provide the dSLBP protein necessary for the accumulation of large amounts of histone mRNA required during the rapid cycles of early embryogenesis. Whole-mount in situ hybridization was used to examine embryonic dSLBP expression in more detail (Fig. 4). dSLBP RNA is indeed supplied maternally (Fig. 4A), but this RNA is destroyed by the end of the syncytial cycles (Fig. 4B) and replaced by ubiquitous, zygotically expressed dSLBP RNA at the cellular blastoderm stage (Fig. 4C). During early germ-band extended stages, when rapid cell division occurs throughout the embryo, ubiquitous dSLBP expression is maintained (Fig. 4D, top). At later stages, dSLBP expression is down-regulated in G1 arrested cells (e.g., the epidermis) and restricted to proliferating (e.g., CNS and PNS) and endoreduplicating (e.g., midgut) cell populations (Fig. 4H,I). Taken together with the gel-shift analyses and the fact that there is not another SLBP-like sequence in the Drosophila genome, these expression data suggest that we have cloned the only histone mRNA stem–loop binding protein in Drosophila.
Figure 3.
dSLBP is expressed throughout Drosophila development. The accumulation of dSLBP transcripts was monitored by PCR using Origen's Rapid-Scan Gene Expression Panels, which contain first-strand cDNAs derived from different tissues and developmental stages. The top panel shows the 181-nt dSLBP specific amplicon, and the bottom panel shows the 433-nt RP49-specific amplicon. Note that the RP49 primers span a small intron, which generates a larger amplicon from genomic DNA than from cDNA.
Figure 4.
dSLBP mRNA expression in wild-type and dSLBP mutant embryos. RNA was detected by in situ hybridization of whole embryos using a dSLBP probe. (A–C,D [top], H,I) yw67 wild-type embryos; (D [bottom], E–G) dSLBP mutant embryos. All embryos are oriented with anterior to the left and dorsal to the top, except the wild-type embryo in panel D, which has anterior to the right. dSLBP RNA is supplied maternally to the embryo (A). By the cellular blastoderm stage, maternal dSLBP RNA is destroyed (B) and subsequently replaced by ubiquitous zygotic dSLBP expression (C), which is maintained during germ band extension (D, top). During stage 11 (H), dSLBP RNA becomes restricted to cells that are replicating, including those of the central and peripheral nervous system (arrow in H). Replication-associated dSLBP expression continues in dividing cells of the CNS (ventral part of the embryo in I) and in endoreduplicating midgut cells (arrow in I) of stage 13, germ-band-retracted embryos. G1-arrested epidermal cells do not express dSLBP. This program of gene expression is very similar to other genes encoding factors required for S phase (Duronio and O'Farrell 1994). (D–G) Little to no dSLBP RNA accumulates in embryos that are genotypically dSLBP12/dSLBP12 (E), dSLBP15/dSLBP15 (D, bottom), or dSLBP19/dSLBP19 (F). In contrast, dSLBP RNA is detected in dSLBP10/dSLBP10 embryos (G) but at a level much reduced relative to their phenotypically wild-type siblings. Note that phenotypically wild-type and mutant sibling embryos of identical stage are shown within the same photomicrograph in panel D.
Generation of null and hypomorphic dSLBP mutations
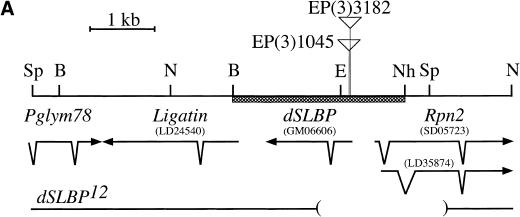
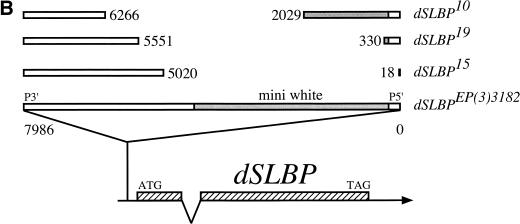
The Berkeley Drosophila Genome Project (BDGP) maintains a collection of P-element insertion lines from which flanking chromosomal DNA has been isolated and sequenced to determine the precise location of the insertion. BLAST searching of this database revealed two independent P-element insertions into dSLBP. EP(3)1045 and EP(3)3182 are inserted in the 5′ UTR 54 and 68 nt upstream of the dSLBP initiator methionine, respectively (Fig. 5A,B). Both P-element lines are viable when homozygous or in trans to a large deficiency (Df(3R)3450) that deletes the entire dSLBP region. Although viable, these flies are female sterile (but male fertile). Because of the critical role SLBP plays in histone mRNA accumulation in vertebrates, we were somewhat surprised to find that both P-element insertions in the 5′ UTR of dSLBP were homozygous viable. A possible explanation for this result is that SLBP function is not essential in Drosophila. Alternatively, these alleles may be hypomorphic and not completely devoid of dSLBP function, which is a common occurrence for P-element insertions into a noncoding region. We therefore mobilized the EP(3)3182 and EP(3)1045 P-element lines to isolate additional alleles of dSLBP via imprecise excision (see Materials and Methods for details). Of the new dSLBP excision alleles that we recovered, four (dSLBP10, dSLBP12, dSLBP15, dSLBP19) were subjected to detailed analysis.
Figure 5.
The dSLBP locus located at 98F/99A on the right arm of chromosome 3. (A) Restriction map of a 6.5-kb fragment containing dSLBP. The direction and intron/exon structure of dSLBP and surrounding transcripts are indicated below the restriction map. Within this 6.5 kb of sequence, three genes in addition to dSLBP were identified: RPN2, encoding the 110-kD proteasome subunit (Adams et al. 2000); Ligatin, encoding a neuronal phosphoglycoprotein receptor (Jakoi et al. 1989); and Pglym78, encoding the glycolytic enzyme phosphoglycerate mutase (Currie and Sullivan 1994). cDNA clones LD24540 (Ligatin), GM06606 (dSLBP), and LD35874 (Rpn2) were sequenced in their entirety. The indicated splice variants of Rpn2 were identified by comparing the sequence of clone LD35874 to the 5′ EST sequence from clone SD05723 obtained by the BDGP. These transcripts utilize the same AUG codon but generate proteins that differ in primary sequence near the amino terminus. The location of the EP(3)3182 and EP(3)1045 P elements is indicated by triangles. The BamHI-NheI restriction fragment used in the transgene that complements dSLBP mutant alleles is indicated by a hatched box. The bottom line indicates the location of the deletion breakpoints in the dSLBP12 excision allele. B = BamHI, E = EcoRI, Sp = SphI, N = NcoI, Nh = NheI. (B) Diagram of dSLBP excision alleles. The dSLBP transcript and coding region (hatched box) is shown at the bottom. The 7986-bp EP(3)3182 P element is located in the 5′ UTR 68 bp upstream of the AUG. The breakpoints of each excision allele are indicated relative to the full-length EP P element. Note that the miniwhite gene (shaded) has been disrupted in each w− excision.
The precise breakpoints of these four excision alleles were determined by PCR analysis and direct sequencing of PCR products. The structure of each mutant is diagrammed in Figure 5. Three of the mutants (dSLBP10, dSLBP19, and dSLBP15) have internal deletions within the P element, and the fourth (dSLBP12) contains a small genomic deletion. The dSLBP12 deletion removes the entire P element, the promoter of dSLBP and the adjacent Rpn2 gene, and a portion of the dSLBP and Rpn2 coding region (Fig. 5A). Consequently, dSLBP12 is a molecular null for both genes. As three of the four excision alleles do not delete any dSLBP coding sequence, we performed in situ hybridization on embryos collected from heterozygous parents to determine if the mutations reduce expression of dSLBP. Little if any dSLBP RNA was detected in homozygous dSLBP15, dSLBP12, and dSLBP19 germ-band extended embryos (Fig. 4D bottom, 4E, and 4F, respectively). This is consistent with the observation that maternal dSLBP RNA is quantitatively destroyed before the start of zygotic gene expression (Fig. 4B). Moreover, dSLBP15 and dSLBP19 homozygous embryos are indistinguishable from dSLBP12 embryos, which contain a molecular null for dSLBP. In contrast, a low but detectable level of dSLBP RNA was observed in homozygous dSLBP10 mutant embryos (Fig. 4G), indicating that this allele is hypomorphic.
These observations were confirmed by immunoblot analysis of homozygous mutant embryos. In SL2 nuclear and cytoplasmic extract, a protein that comigrates with in vitro–translated dSLBP is detected with anti-dSLBP antibodies (Fig. 6A). This protein can be specifically precipitated by a biotinylated stem–loop RNA bound to streptavidin agarose but not by control reverse stem–loop RNA (Fig. 6B). Thus, we can measure dSLBP abundance by Western blotting. In addition, we can obtain pure collections of homozygous mutant embryos by hand sorting using GFP expressing balancer chromosomes (see Materials and Methods). dSLBP is readily detected in late stage (i.e., 13–16-h AED) wild-type embryos (Fig. 6C, lane 1). In contrast, equivalently staged hemizygous dSLBP15/Df(3R)3450 mutant embryos contain very little if any detectable dSLBP (Fig. 6C, lane 2). Similar to the in situ hybridization results, a low amount of dSLBP is observed in dSLBP10/dSLBP10 and dSLBP10/Df(3R)3450 mutant embryos (Fig. 6C, lane 3). These molecular data strongly indicate that the P-element excision mutations are simple loss of function alleles of dSLBP.
Figure 6.
dSLBP protein expression is reduced in dSLBP mutant embryos. Each panel shows a Western blot using affinity purified dSLBP antibodies. (A) The antibody recognizes a protein in SL2 cell nuclear (lane 2) and cytoplasmic (lane 3) extracts that comigrates with the protein translated in vitro from dSLBP cDNA (lane 1). Note that the antibody cross reacts with several unknown polypeptides, including one at 53 kD that appears in both embryo and SL2 cell extracts and one at 50 kD (asterisk in C) that appears only in embryo extracts. One cross-reacting protein is found in nuclear but not cytoplasmic SL2 cell extract (A), indicating successful fractionation. (B) RNA/protein complexes were precipitated from SL2 nuclear extract incubated with wild-type (WT) biotinylated stem–loop RNA or biotinylated RNA containing a reverse stem–loop sequence (RS). The protein in the extract that comigrates with in vitro translated dSLBP (lane 1) but not the cross-reacting proteins, is specifically precipitated by the wild-type (WT) stem–loop RNA (lane 3) and not by the control RS RNA (lane 2). (C) Extracts were made from hand-selected 13–16-h yw67 wild-type embryos (lane 1) or dSLBP homozygous mutant embryos (dSLBP15/Df(3R)3450, lane 2; dSLBP10/Df(3R)3450, lane 4). Mutant embryos were distinguished from their phenotypically wild-type siblings using balancer chromosomes that express GFP. Little if any dSLBP is produced by the dSLBP15 allele, whereas drastically reduced but detectable levels of dSLBP are produced by the hypomorphic dSLBP10 allele. Anti-armadillo antibodies were used as a loading control (Arm).
Genetic analysis of the four excision alleles corroborates the molecular studies. We performed reciprocal crosses among each of the four excision alleles and the Df(3R)3450 deficiency (Table 1). dSLBP10 behaves much like the EP(3)3182 chromosome from which it was derived: dSLBP10/Df flies are viable but female sterile and male fertile (Table 1). Unlike dSLBP10, dSLBP19and dSLBP15 have a dramatic effect on viability. Adult dSLBP15/Df flies are very rarely recovered (1/2621; Table 1), while 7%–18% of the expected number of dSLBP19/Df flies are recovered. For both alleles, the lethal phase is very late in development, with many flies dying at eclosion. The few surviving dSLBP19/Df individuals have no overt morphological defects but are female sterile and male fertile. The dSLBP12 allele is fully lethal in trans to Df(3R)3450 and behaved like the null Df(3R)3450 deletion in each cross, as expected (Table 1). Together, the molecular and genetic data suggest that the four excision mutations represent an allelic series for dSLBP function, with dSLBP10 > dSLBP19 > dSLBP15 ≥ dSLBP12 = Df(3R)3450 = null. The precise reason why internal deletion of the EP P element affects dSLBP mRNA accumulation is not known. One possibility is that these alleles remove cryptic splice and/or transcription start sites from within the P element, thereby reducing production of dSLBP mRNA.
Table 1.
Complementation testing of dSLBP excision alleles
| Female1
|
Male1
|
Number of heterozygous flies
|
Number of homozygous mutant flies2
|
Percentage of expected if fully viable3
|
|---|---|---|---|---|
| Df(3R)3450 | dSLBP10 | 462 | 115 | 60 |
| dSLBP10 | Df(3R)3450 | 709 | 282 | 86 |
| Df(3R)3450 | dSLBP12 | 279 | 0 | 0 |
| dSLBP12 | Df(3R)3450 | 1715 | 0 | 0 |
| Df(3R)3450 | dSLBP15 | 455 | 0 | 0 |
| dSLBP15 | Df(3R)3450 | 596 | 1 | 0.5 |
| Df(3R)3450 | dSLBP19 | 404 | 10 | 7 |
| dSLBP19 | Df(3R)3450 | 675 | 42 | 18 |
| dSLBP10 | dSLBP12 | 853 | 145 | 44 |
| dSLBP12 | dSLBP10 | 653 | 157 | 58 |
| dSLBP10 | dSLBP15 | 870 | 216 | 60 |
| dSLBP15 | dSLBP10 | 640 | 128 | 50 |
| dSLBP10 | dSLBP19 | 714 | 104 | 38 |
| dSLBP19 | dSLBP10 | 777 | 94 | 32 |
| dSLBP12 | dSLBP15 | 819 | 0 | 0 |
| dSLBP15 | dSLBP12 | 751 | 0 | 0 |
| dSLBP12 | dSLBP19 | 679 | 28 | 12 |
| dSLBP19 | dSLBP12 | 847 | 28 | 10 |
| dSLBP15 | dSLP19 | 802 | 1 | 0.3 |
| dSLBP19 | dSLBP15 | 1009 | 30 | 9 |
| Rescue crosses: | ||||
| Df(3R)3450 | P{w+, dSLBP}/+; | 358 | 1264 | 78 |
| dSLBP15/TM3 Sb | ||||
| Df(3R)3450 | P{w+, dSLBP}/+; | 156 | 0 | 0 |
| dSLBP12/TM3 Sb | ||||
| dSLBP12/TM3 Sb | P{w+, dSLBP}/+; | 278 | 0 | 0 |
| Df(3R)3450/TM3 Sb |
Balancer chromosome is TM3 Ser P[w+; act-GFP] for all flies, unless otherwise indicated.
All viable flies are female sterile.
Expected is 33% of the total eclosed flies, since balancer homozygotes die.
Female fertile.
We tested whether a P-element transgene containing only the full-length dSLBP transcription unit would rescue the mutant phenotypes. A BamHI–NheI genomic restriction fragment containing dSLBP (Fig. 5A) complemented the lethality of dSLBP15/Df(3R)3450 flies (Table 1) and restored fertility to dSLBP10/Df(3R)3450 adult females. Therefore, these alleles do not disrupt the function of the neighboring genes but only mutate dSLBP. In contrast, the same transgene could not rescue the lethality of dSLBP12/Df(3R)3450 flies, which lack both dSLBP and Rpn2 (Table 1). This demonstrates that Rpn2 is also an essential gene.
Analysis of histone mRNA expression in dSLBP mutant embryos
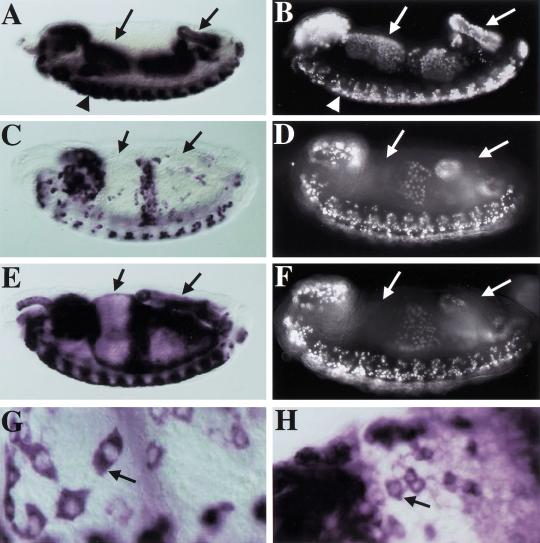
The homozygous lethality caused by the dSLBP19 and dSLBP15 mutations suggested that histone mRNA accumulation may be disrupted in the mutants. We chose to initially examine histone mRNA at embryonic stages when replication-associated histone expression is known to occur (i.e., post-germ-band retraction, cycle 17 and later). In situ hybridization of wild-type embryos with a histone H4 antisense RNA probe results in a stereotypic but dynamic pattern of expression that is essentially identical to that obtained by pulse labeling with BrdU (Fig. 7A–D). This includes dividing cells of the CNS (Fig. 7A,B, arrowheads) and endoreduplicating cells of the alimentary canal (e.g., midgut and hindgut; Fig. 7A–D, left and right arrows, respectively). We then examined H4 expression in mutant embryos. dSLBP19, dSLBP15, and dSLBP12 mutations each caused a similar, recessive phenotype, whether as homozygotes or in trans to Df(3R)3450. Contrary to our expectations for a gene product suspected of being required for production of a mature mRNA, histone H4 RNA still accumulates to high levels in cells of dSLBP mutant embryos (Fig. 7E). In addition, the pattern of staining in the mutant embryos differed from wild-type embryos at the same stage. In particular, histone transcripts were present in cells that were not replicating, as judged by pulse labeling with BrdU (Fig. 7E,F, arrows). As in wild type, the histone mRNAs in mutant embryos were present in the cytoplasm (Fig. 7G,H, arrow), consistent with their being translated at this stage. A similar aberrant accumulation of RNA was observed using a probe derived from histone H3 (not shown). The altered pattern of histone RNA accumulation suggests that endoreduplicating cells that normally down-regulate histone expression after each endocycle S phase no longer do so in the mutant (e.g., the anterior midgut and hindgut; cf. arrows in Fig. 7C,E, and 7D,F). Therefore, mutation of dSLBP uncoupled histone RNA accumulation from ongoing replication in cycling cells.
Figure 7.
Altered histone RNA expression in dSLBP mutant embryos. (A,B) Stage-13 wild-type embryos stained for histone H4 expression by in situ hybridization (A) or DNA synthesis by pulse labeling for 15 min with BrdU (B). Note that the histone expression and replication patterns are identical, indicating that histone mRNA accumulates only in cells that are synthesizing DNA. This includes proliferating cells in the CNS (arrowhead) and endoreduplicating cells in the anterior midgut (left arrow) and the hindgut (right arrow). (C) H4 expression in a stage 14 dSLBP19/+ embryo. (D) BrdU-labeled stage 14 dSLBP15/+ embryo. These heterozygous embryos are phenotypically wild type. Again, histone expression and replication are coincident, but the pattern has changed from stage 13. Anterior midgut and hindgut cells have exited the endocycle S phase and do not express histone H4 mRNA. (E) H4 expression in a dSLBP19/dSLBP19 mutant sibling of the embryo in C. (F) DNA replication in a dSLBP15/dSLBP15 mutant sibling of the embryo in D. In the mutant embryos, histone H4 mRNA persists (arrows in E) in cells that have already finished replicating (arrows in F). The histone H4 expression pattern is identical in dSLBP19 and dSLBP15 homozygous embryos (not shown). Histone H4 mRNA accumulates in the cytoplasm of cells in both dSLBP19/+ wild-type (arrow in G) and dSLBP19/dSLBP19 (arrow in H) mutant embryos.
To determine whether the accumulated histone mRNAs present are processed normally, we prepared total RNA from embryos collected from heterozygous parents and analyzed them by Northern blotting. Wild-type embryos contained only the 500-nt H3 transcript that has been described previously (Fig. 8A, lane 6). In embryos derived from stocks of mutants dSLBP10, dSLBP12, dSLBP15, dSLBP19, and dSLBP47, we detected larger histone H3 transcripts in addition to the wild-type mRNA (asterisks, Fig. 8A, lanes 1–5, respectively; see Materials and Methods for a description of dSLBP47). The largest of these was ∼1 kb longer than the wild-type transcript. A similar result was obtained using RNA extracted from dSLBP15embryos and a histone H4 probe, although the mutant transcript detected was not as long (Fig. 8C, lane 3, asterisk). To further characterize these transcripts, RNA extracted from embryo collections of dSLBP15 heterozygous parents were fractionated on oligo(dT) magnetic beads, and the polyA+ RNA was analyzed by Northern blotting. Whereas very little wild-type H3 RNA was found in the polyA+ fraction (Fig. 8B, lanes 1,2), the longer, aberrant H3 RNA species were readily detected in the polyA+ fraction (Fig. 8B, lane 1, asterisks). Similar results were obtained using H4 probes (not shown). The binding of these transcripts to oligo dT suggests that they are polyadenylated. Therefore, mutation of dSLBP resulted in the synthesis of abnormal histone mRNA, probably because of a failure to properly generate the usual 3′ end. As polyadenylated histone mRNAs are stable at the end of S phase or when DNA replication is inhibited (Levine et al. 1987; Harris et al. 1991), it is likely that the continued expression of histone mRNA in cells that have completed S phase is because of a failure to degrade much of the histone mRNA expressed in the mutant embryos. Consistent with this hypothesis, we have shown previously that accumulation of mammalian histone mRNAs containing the stem–loop internally in the 3′ untranslated region, rather than at the 3′ end, is not coupled to DNA replication (Levine et al. 1987; Pandey and Marzluff 1987).
Figure 8.
Altered RNA processing in dSLBP mutants. (A) Total RNA extracted from 7–22-h embryos collected from dSLBP10 (lane 1), dSLBP12 (lane 2), dSLBP15 (lane 3), dSLBP19 (lane 4), dSLBP47 (lane 5) heterozygous mutant stocks, or from yw67 wild-type flies (lane 6) and was blotted and probed with radiolabeled histone H3 coding region. Longer H3 transcripts are detected only in the mutant samples (asterisks). Note that three-quarters of the embryos in each preparation are phenotypically wild type, accounting for the RNA of wild-type length in the mutant lanes. (B) dSLBP15 mutant (lane 1) and yw67 wild-type (lane 2) embryonic RNA samples prepared as in (A) were fractionated with oligo dT magnetic beads, and the polyA+ RNA was subjected to Northern analysis with the H3 coding region probe, or an RP49 probe as loading control. The arrow indicates migration of the wild-type H3 mRNA. The aberrant, long H3 transcripts (asterisks) bind to oligo dT, whereas wild-type H3 mRNA does not. (C) The maternal effect lethal allele of dSLBP causes reduced deposition of maternal histone mRNA. Total RNA extracted from 0–2-h embryos collected from dSLBP10/Df(3R)3450 (lane 1) or yw67 wild-type (lane 2) females was blotted and probed with a histone H4 or H3 coding region probe or RP49 probe as loading control. Lane 3 contains total RNA prepared from 7–22-h embryos collected from dSLBP15/+ flies as a comparison. At these later embryonic stages, there is less total histone RNA expressed per embryo when compared to 0–2- h embryos, accounting for the reduced intensity of the wild-type band. Note that the longer histone transcripts (asterisks) are not nearly as abundant in the mutant maternal RNA samples (lane 1).
dSLBP is required very early in embryonic development
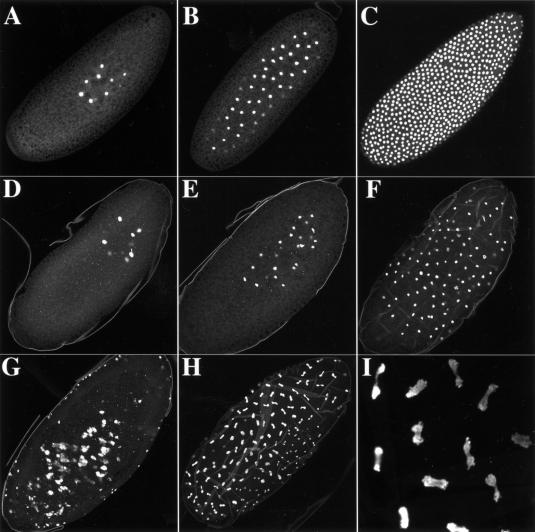
The first ∼2.5 h of Drosophila embryogenesis consist of 13 rapid, nuclear division cycles that occur in a syncytial cytoplasm (Fig. 9A–C), lack G1 and G2 phases and are driven by large stores of maternally provided cell cycle components, including histone RNA and protein. Rather than being synthesized only during S phase, maternally provided histone mRNA and protein are present continuously during these cycles (Anderson and Lengyel 1984). dSLBP mRNA is also present at this stage (Fig. 4A), suggesting that dSLBP may function during the cleavage divisions. The dSLBP10 allele is viable in trans to a deficiency but causes female sterility when crossed with wild-type males (Table 1). These females lay a normal amount of eggs that are wild type in appearance. Essentially, none of these eggs complete embryogenesis (99% failed to hatch; n = 777), suggesting that the defect occurs during the earliest syncytial cycles before the activation of the paternal genome, which would provide wild-type dSLBP. To test this, embryos collected 0–4 h AED from dSLBP10/Df(3R)3450 mothers were pulse labeled with BrdU to monitor DNA synthesis and visualize syncytial stage nuclei. The mutants had obvious disruptions of the nuclear cleavage cycles and accumulated nuclei that had aberrant morphologies (Fig. 9D–I). The earliest cycles appeared somewhat normal (Fig. 9D,E), but we could never identify a blastoderm stage embryo with the typical uniform arrangement of dividing nuclei (cf. Fig. 9C with 9F–H).
Figure 9.
The dSLBP10 maternal effect lethal allele causes disruption of the early nuclear cleavage cycles. Confocal images of syncytial stage embryos collected from yw67 wild-type (A–C) and dSLBP10/Df(3R)3450 mutant (D–I) females. These embryos were pulse labeled for 10 min with BrdU to detect replicating nuclei. A, B, and C show wild-type embryos of increasing age, with a nuclear cycle 13 syncytial blastoderm shown in panel C. Panels D–I show representative mutant embryos. Panel I is a high-magnification view of panel H. Most embryos (e.g., G) were impossible to stage for a particular nuclear cycle because of the accumulation of defects with increasing nuclear density. The vitelline membrane was inefficiently removed from the mutant embryos by our standard procedure and is, therefore, visible in these images.
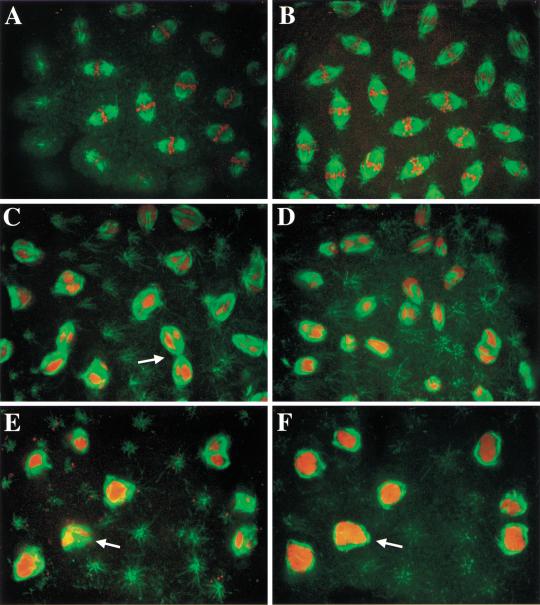
These nuclear cycle defects appear to result, at least in part, from improper chromosome condensation during mitosis. This was revealed by simultaneous detection of microtubules with anti β-tubulin antibodies and DNA with propidium iodide (Fig. 10). While mitotic nuclei in wild-type embryos contain fully condensed chromosomes that align at the metaphase plate (Fig. 10A,B), embryos with a similar nuclear density collected from dSLBP10/Df(3R)3450 mutant females never formed normal metaphase spindles (Fig. 10C–F). Microtubules assemble around improperly condensed DNA (Fig. 10E,F), causing the formation of spindles with dramatically altered morphologies (Fig. 10C,D). Spindle-pole fusion is also frequently observed (Fig. 10C, arrow). Improper chromosome condensation prevents anaphase, and core histones are needed to recruit condensation factors (Saka et al. 1994; Strunnikov et al. 1995; Sutani et al. 1999; Wei et al. 1999; de la Barre et al. 2000; Freeman et al. 2000). A failure of anaphase could contribute to the large chromosome masses sometimes observed (Fig. 9G and 10 E,F). Indeed, dramatic anaphase bridges were detected in some embryos (Fig. 9H,I). As such nuclei were labeled with BrdU, the condensation failure did not block DNA synthesis.
Figure 10.
Aberrant spindle morphology caused by dSLBP10 maternal effect allele. Syncytial stage embryos were collected from yw67 wild-type (A,B) or dSLBP10/Df(3R)3450 mutant females (C–F), fixed, and stained with anti-β-tubulin antibodies to detect mitotic spindles (green) and propidium iodide to detect chromosomes (red). Each panel shows a merged confocal image. Note the regular array of spindles with properly condensed chromosomes that have congressed to the metaphase plate in wild type (A,B). The normal regular array of mitotic spindles is lost in mutant embryos (C–F). Nuclear DNA fails to condense into discrete chromosomes (best seen in F), but microtubules nevertheless form around the improperly condensed DNA generating misshapen spindles. These spindles sometimes appear fused to one another (e.g., arrow in C). Panels E and F show different focal planes of the same visual field to illustrate how the spindle microtubules encapsulate the DNA (arrow).
One possible explanation for the failure of proper chromosome condensation is insufficient histone biosynthesis. We therefore examined histone H3 and H4 RNA levels in the maternally mutant embryos by Northern blot analysis. Mutant embryos contained about 10-fold less histone H3 and H4 RNA than wild-type embryos (Fig. 8C, cf. lanes 1 and 2). Unlike embryos mutant for zygotic dSLBP function (Fig. 8C, lane 3), very little of the longer histone mRNAs were detected in eggs laid by dSLBP10/Df(3R)3450 females. This suggests that the mechanism for forming these longer transcripts was not as efficient during oogenesis and that any unprocessed histone mRNA is degraded during oogenesis. Perhaps in the embryo all the controls of histone mRNA production are not yet in place, allowing polyadenylation of the unprocessed histone pre-mRNA. Production of lower amounts of histone mRNA suggests that insufficient synthesis of histone protein prevents proper chromatin structure, leading to the observed defects in chromosome condensation.
Discussion
The replication-associated histone mRNAs are unique in that they are the only metazoan mRNAs lacking polyadenylated tails. Their mature 3′ end contains a sequence that forms a stem–loop to which SLBP binds. Here we have shown that in Drosophila SLBP is required for proper cell cycle regulation of histone mRNA in vivo. Mutation of dSLBP causes the synthesis of histone mRNAs with aberrant 3′ ends that accumulate in nonreplicating cells. This is not well tolerated, as both maternal and zygotic dSLBP function is essential for development.
The role of dSLBP in histone mRNA production
Before cell cycle 16, histone H4 expression in dSLBP mutant embryos appears like wild type (not shown), suggesting that it is directed by maternal supplies of dSLBP protein. We first observed a defect in histone mRNA expression during cell cycle 17 of dSLBP mutant embryos, which display an altered pattern of histone mRNA accumulation that is most consistent with expression of a stabilized histone message: The mRNA accumulates in cells that are replicating during an endocycle S phase but fails to disappear when the cells exit S phase. At the end of S phase, histone mRNA is normally destabilized, and this requires the 3′ end of the mature message (Harris et al. 1991), which is thought to provide a substrate for exonucleases (Ross et al. 1986, 1987). Consistent with this model, the 3′ end of histone mRNA is altered in the dSLBP mutant. The transcripts are longer than normal and bind to oligo(dT), suggesting that they are polyadenylated. In the absence of sufficient dSLBP, it is likely that transcription continues past the normal 3′ end of the histone mRNA, resulting in subsequent processing at cryptic polyA sites (Akhmanova et al. 1997). In mammalian cells, artificially polyadenylated histone mRNA has an increased half-life and is not subject to normal posttranscriptional regulation (Pandey and Marzluff 1987). We therefore conclude that dSLBP is required for efficient processing of the histone pre-mRNA in vivo. This is consistent with the known role of mammalian SLBP in processing histone pre-mRNA substrates in a reconstituted in vitro processing assay (Dominski et al. 1999) and following injection of histone genes into frog oocytes (Wang et al. 1999).
We do not know the efficiency of forming the polyadenylated 3′ ends in the dSLBP mutants. In mammalian cells, histone mRNAs are transcribed in G1 phase, but the pre-mRNAs are not processed and are rapidly degraded (DeLisle et al. 1983; Stauber and Schumperli 1988). A similar situation is seen when one mutates the 3′ end of histone mRNA; the genes are still transcribed, but no histone mRNA accumulates (Pandey et al. 1994). This suggests that mammalian cells have an efficient mechanism for destroying unprocessed histone pre-mRNA. Perhaps Drosophila embryos do not have such a mechanism.
The role of SLBP in coupling histone mRNA production with the cell cycle
Changes in the rate of pre-mRNA processing play a major role in coupling the synthesis of mature histone mRNA with the cell cycle. There is evidence that histone pre-mRNA processing is activated at the G1–S transition in mammalian cells (Luscher and Schumperli 1987; Stauber and Schumperli 1988; Harris et al. 1991). If one introduces a gene that contains both a histone 3′ end followed by a strong polyadenylation site, then polyadenylated histone mRNA accumulates in G1 cells, while mRNAs ending in the stem–loop end accumulate in S-phase cells (Harris et al. 1991). Recent data suggest that this increase in processing is probably a result of increased synthesis of SLBP at the G1/S transition. SLBP is cell cycle regulated in mammalian cells, accumulating just before the accumulation of histone mRNA and disappearing at the end of S phase (Whitfield et al. 2000). Thus, SLBP could be the limiting component for histone pre-mRNA processing in vivo both in G1 and G2, with production of SLBP at the G1/S transition stimulating processing and destruction of SLBP at the end of S phase inactivating processing (Whitfield et al. 2000). We postulate that SLBP provides a direct link between the cell cycle machinery and histone mRNA biosynthesis. When cells are blocked at the G1/S transition by DNA synthesis inhibitors, there are normal S-phase levels of SLBP, but very little histone mRNA accumulates (Whitfield et al. 2000). Therefore, while the levels of mammalian histone mRNA are coupled to the rate of DNA replication and not only to the phase of the cell cycle, SLBP levels are coupled to the phase of the cell cycle. We are currently investigating whether dSLBP protein is cell cycle regulated in Drosophila.
Cell cycle regulation of histone mRNA accumulation also is a result of an increased rate of histone gene transcription as cells enter S phase (DeLisle et al. 1983; Heintz et al. 1983). A number of elements in histone gene promoters have been implicated in this transcriptional increase (LaBella et al. 1988; Heintz 1991), and a sequence shared among all the histone-coding regions has also been implicated in histone gene transcription (Hurt et al. 1991; Bowman and Hurt 1995) through the transcription factor YY1 both in mammals (Eliassen et al. 1998) and in Drosophila (K. Eliassen, M. Hurt, and D. Garza, pers. comm.). Moreover, recent evidence suggests that the mammalian cyclin E/cdk2 substrate NPAT directly couples the G1–S cell cycle machinery with histone gene transcriptional activation (Ma et al. 2000; Zhao et al. 2000). Transcription of Drosophila histone genes appears to be unaffected by mutation of dSLBP. As in wild type, dSLBP mutant embryos express histone mRNA only in tissues that contain cycling cells (either dividing, like the CNS, or endoreduplicating, like the midgut) and do not express histone mRNA in G1-arrested cells (e.g., the epidermis). Thus, transcriptional regulation alone is insufficient to provide the tight coupling of histone mRNA accumulation with S phase.
The role of SLBP in early embryonic development
Early embryonic development is one exception to the tight coupling between histone mRNA levels and S phase. Histone mRNA persists at a constant level during the initial, rapid S–M cycles of Drosophila embryos (Anderson and Lengyel 1980, 1984). Our results indicate that dSLBP function is required maternally for these cycles. Eggs laid by females hemizygous for a viable, hypomorphic allele (dSLBP10) display defects in the syncytial nuclear division cycles. Rather than the rapid exponential increase in the number of nuclei characteristic of normal development, there is production of a reduced number of nuclei that are abnormal in arrangement and morphology. The amount of wild-type histone H4 and H3 mRNA is drastically reduced in these embryos. Consequently, while replication still occurs, the DNA probably cannot be properly packaged into normal chromatin because of limited histone biosynthesis. This likely causes the observed failure of chromosome condensation during mitosis, leading to impaired nuclear division. Thus, the demand for dSLBP function appears to be highest in the earliest stages of embryogenesis. However, we have not yet formally determined whether this represents a requirement for dSLBP during oogenesis (e.g., for production of mature histone mRNA) and/or a requirement during the cleavage cycles themselves (e.g., for translation of the maternally provided histone mRNA). Perhaps the ∼100 copies of the tandemly repeated Drosophila histone genes are only needed to provide the stored histone mRNA in the oocyte and for production of histones in the early embryo. In subsequent developmental stages there will be a lower demand for histone mRNA as the cell cycles are slower (and S phase is longer), and the reduced level of dSLBP produced in the hypomorphic mutant is sufficient to support development.
What is the purpose of restricting histone synthesis to S phase?
Replication-associated histone gene expression is evolutionarily conserved, occurring from yeast to metazoans (although in yeast the regulation is primarily at the transcriptional level; Osley 1991). This conservation of regulation presumably has important functional significance. The tight regulation may have evolved to ensure production of histones when they are needed, to achieve the proper stoichiometry of each histone protein, and to prevent production of histones when they are not needed. Proper regulation is likely to be important for normal nucleosome formation, and hence, proper chromosome function. Consequently, the overaccumulation of individual histone proteins, or even all histones, when cells are not synthesizing DNA may be deleterious. Thus, the lethality of dSLBP mutants is likely caused by the misexpression of replication-associated histone genes, although we cannot rule out that dSLBP functions in the regulation of other mRNAs, the absence of which could cause the deleterious effects.
Misexpression of histone genes could disrupt development in many ways. For instance, in Saccharomyces cerevisiae, the overexpression of H2A/H2B or H3/H4 histone dimer pairs causes an increased frequency of chromosome loss (Meeks-Wagner and Hartwell 1986), and changes in the dose of histone H2B alter transcription of certain genes (Clark-Adams et al. 1988). Chromosome segregation during cell division may also be affected. There are several variant histone proteins that are expressed at low levels throughout the cell cycle (Wells and Kedes 1985; Fretzin et al. 1991; van Daal and Elgin 1992). These variants substitute for the core histones to form specialized nucleosomes at particular loci that perform specific functions. In dSLBP mutant cells, the overexpression or mistimed expression of replication-associated histones during the cell cycle may affect the formation of these specialized nucleosomes. For instance, the Drosophila H3-like protein Cid accumulates in nucleosomes only at centromeric DNA, presumably as a critical component of centromere identity and function (Henikoff et al. 2000). Perhaps continuous overexpression of H3 during the cell cycle in dSLBP mutants competes with Cid for centromere binding, thereby disrupting centromere function and chromosome transmission during cell division. Whatever the basis is for the lethality, dSLBP mutants survive until very late in development, long after the first evidence of misexpressed histone mRNA. Therefore, any deleterious affects from histone misexpression are not acute but, rather, may eventually be harmful by causing the accumulation of defects throughout development. Our current efforts are focused on addressing many of these issues.
Materials and methods
Yeast three-hybrid screen
Yeast strains expressing either wild-type or control bait that does not bind SLBP have been described previously (Wang et al. 1996b). Thirty-seven His+ positives that also activated the lacZ reporter were identified from ∼3 × 106 colonies obtained from a 0–4-h embryonic cDNA library (gift of A. Dahanukar and R. Wharton, Duke University). Only five of these required the hybrid RNA expressing plasmid for growth in His− medium. Plasmid DNA was isolated from each of these colonies, and all five contained clones with dSLBP sequence. One corresponding cDNA, GM06606, was sequenced in its entirety (GenBank AF258617).
Protein expression and antibody production
The coding region of dSLBP was engineered by PCR to contain 5′ NcoI and 3′ XbaI sites and expressed in Sf9 insect cells using the Bac-TO-Bac baculovirus expression system (GIBCO BRL). His-tagged full-length dSLBP was purified by affinity chromatography on Ni-NTA agarose (QIAGEN) and used as an antigen to generate polyclonal antibodies in rabbits (Pocono Rabbit Farms). Anti-dSLBP antibody was affinity purified using nitrocellulose-mobilized full-length dSLBP protein as described (Robinson et al. 1988).
Gel shift assays
Mobility shift assays were carried out as previously described (Dominski et al. 1999). dSLBP was synthesized in vitro with a TnT kit (Promega) using cDNA subcloned into the pSP64T vector. Embryo extract (gift of K. Williamson and L. Searles, University of North Carolina) for gel-shift experiments was made as previously described (Rio 1988).
Preparation of Drosophila cell extract and RNA-pull down of dSLBP
SL2 cells were grown in suspension at room temperature using Shield's and Sung M3 medium (Sigma) supplemented with 12.5% heat-inactivated fetal bovine serum and streptomycin/penicillin mix. Nuclear and cytoplasmic extracts from 5 L of culture (1 × 106 cells/mL) were prepared as previously described (Dominski et al. 1995, 1999). dSLBP was precipitated from the SL2 nuclear extract by binding to synthetic histone stem–loop RNA biotinylated at the 5′ end (5′BiWT RNA; 5′ Biotin-18s-18s-CCAAAAAGGGUCUUUUCAGAGCCACCCA 3′, where 18s refers to an 18-atom spacer) as described in Dominski et al. (1995). Protein bound to 5′BiWT RNA was recovered from streptavidin agarose beads with SDS–PAGE loading buffer and analyzed by immunoblotting with anti-dSLBP antibody. A non-SLBP-binding biotinylated RNA (5′BiRS; 5′-Biotin-18s-18s-CCAAAAACCGAGAUUUCUCUCGGACCCA-3′) was used to monitor nonspecific binding to the agarose beads. Both 5′BiWT and 5′BiRS RNAs were synthesized by Dharmacon Research.
Expression of dSLBP in Xenopus oocytes
Healthy stage V–VI oocytes were injected with 45 nL of a solution containing 0.5 μg/μL of synthetic dSLBP mRNA transcribed from a modified version of pSP64T (Promega) as previously described (Wang et al. 1999). Whole-cell, nuclear, and cytoplasmic extracts were prepared from injected ooctyes that had been incubated for 48 h at 18°C. dSLBP RNA-binding activity in the extracts was detected by gel mobility-shift assays as described above.
Cosmid analysis
Cosmid clones 144B6, 36E5, and 1H12 kindly provided by EDGP were confirmed to contain dSLBP by Southern blotting. A 4.5-kb NcoI fragment and a 5.3-kb SphI fragment from cosmid 144B6 that hybridized to the probe were isolated in pGEM5Zf (Promega). DNA sequence analysis indicated that a 6465-nt region containing the entire dSLBP gene is contained in these overlapping subclones (shown in Fig. 5A). Database searches using this sequence identified ESTs corresponding to the Ligatin and Rpn2 transcription units on either side of dSLBP. The insert of a single cDNA clone representing each of these genes was sequenced and the data deposited in GenBank: AF260235 for Ligatin and AF312231 for Rpn2. To generate the dSLBP-complementing transgene, a 2.3-kb NheI–BamHI genomic dSLBP fragment was cloned into SpeI/BamHI cut pCaSpeR4 transformation vector.
P element excision mutagenesis
Male flies of the genotype w1118/Y; EP(3)1045 or EP(3)3182/TM6B were crossed to Sb, P[ry+, Δ 2, 3]/TM6, P[ry+, Δ 2, 3] females, and the resulting EP(3)/TM6, P[ry+, Δ 2, 3] male progeny were subsequently crossed to yw67/yw67; D/TM3, Sb females. w− revertant male flies of genotype Δ EP(3)/TM3, Sb were identified and backcrossed to yw67/yw67; D/TM3, Sb to create a balanced stock of each P-element excision chromosome having the genotype yw67/yw67; Δ EP(3)/TM3, Sb. Five homozygous lethal chromosomes (four from EP(3)3182 and one from EP(3)1045) that affect dSLBP were identified from among 64 w− lines by PCR, Southern blot, and inter se complementation analysis. Two probes were used for Southern analysis of genomic DNA prepared from flies heterozygous for the excision chromosome: the EcoRI–BamHI fragment containing only dSLBP and the SphI–BamHI fragment containing dSLBP and the 5′ part of Rpn2 (see Fig. 5A). The dSLBP19, dSLBP15, and dSLBP12 mutations were identified this way. Two lethal lines (dSLBP35 and dSLBP47) within the dSLBP complementation group showed no changes by Southern analysis. These lines may have suffered a deletion that extends beyond the region we probed into the Rpn2 gene (i.e., past the SphI site). dSLBP10 was identified because it is female sterile over Df(3R)3450 and each of the other members of the dSLBP complementation group. A series of primers within the P element and in the adjacent genes were used to identify the breakpoints of excision alleles. Because these were large PCR products (2.5–10 kb), we used Boehringer Mannheim's Expand Long Template PCR System and high–molecular weight genomic DNA. Once fragments were successfully amplified, they were sequenced directly. Detailed PCR conditions for all experiments are available on request.
RNA analysis
Total RNA was prepared from 0–2-h- or 7–22-h-old fly embryos collected at 25°C using the TRIzol Reagent (GIBCO BRL) protocol. The only modification was the need to homogenize the flies in a frosted glass homogenizer (10 strokes). The RNA was fractionated into polyA+ and polyA− fractions using Promega's PolyATtract mRNA isolation system. An expression profile for dSLBP mRNA was obtained by the PCR-based Rapid-Scan System (Origene Technologies). Hybridization probes were generated from a clone (gift of T. Shermoen, University of California, San Francisco) containing the coding region of the H3 and H4 genes from the histone gene cluster (GenBank X14215).
Embryo analysis
In situ hybridization was done as previously described (Duronio and O'Farrell 1994) using probes containing the coding region of H3, H4, and dSLBP. Embryos were collected at room temperature and aged at 18°C or 25°C (for histone experiments) or room temperature (for dSLBP experiments) until the desired stage. The dSLBP mutant histone expression phenotype was the same at 18° and 25°C. BrdU pulse labeling was performed exactly as described (Duronio and O'Farrell 1994). Mutant germ-band-extended embryos were distinguished from wild-type siblings with a balancer chromosome expressing β-galactosidase (TM3 P[Ubx-lacZ]). Microtubules were detected in embryos fixed for 5 min in 37% formaldehyde using anti-β-tubulin mouse monoclonal (Amersham) and goat antimouse Oregon Green (Molecular Probes) antibodies. Live mutant embryos were sorted from their wild-type siblings using stocks containing a TM3 Ser P[act-GFP] balancer chromosome. At room temperature, 1–2-h egg collections were aged for 13–14 h, dechorionated, and then hand selected by visualization using a Zeiss stereomicroscope equipped with epifluorescence. Hemizygous embryos (Fig. 6, lanes 2,3) were obtained using Df(3R)3450/TM3 P[act-GFP] as the mothers. Batches of hand-sorted embryos were frozen at −80°C and then combined and solublized in SDS–PAGE gel loading buffer (at a ratio of 1 μL/embryo) by boiling for 5 min. For Westerns, 20 embryo equivalents were loaded per lane.
Acknowledgments
We thank Lillie Searles, Kelly Williamson, Steve Crews, Tony Shermoen, and Robin Wharton for reagents; the EDGP for cosmid clones; the Bloomington Drosophila Stock Center and BDGP for fly strains; Prem Fort for help with embryo sorting; Dan Garza, and Myra Hurt for communicating unpublished results; and Steve Crews and Mark Peifer for comments on the manuscript. This work was supported by NIH grants GM58921 to W.F.M and GM57859 to R.J.D., an NSF Career Award to R.J.D., and the Cancer Research Fund of the Damon Runyon-Walter Winchell Foundation Award, DRS-10, to R.J.D. T.C.I. was supported by a postdoctoral fellowship from the Mellon Foundation. Z.D. was supported by a grant from the North Carolina Biotechnology Center.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL duronio@med.unc.edu; FAX (919) 962-8472.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.862801.
References
- Adams MD, Celniker SE, Holt RA, Evans CA, Gocayne JD, Amanatides PG, Scherer SE, Li PW, Hoskins RA, Galle RF. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. doi: 10.1126/science.287.5461.2185. [DOI] [PubMed] [Google Scholar]
- Akhmanova A, Miedema K, Kremer H, Hennig W. Two types of polyadenated mRNAs are synthesized from Drosophila replication-dependent histone genes. Eur J Biochem. 1997;244:294–300. doi: 10.1111/j.1432-1033.1997.00294.x. [DOI] [PubMed] [Google Scholar]
- Anderson KV, Lengyel JA. Changing rates of histone mRNA synthesis and turnover in Drosophila embryos. Cell. 1980;21:717–727. doi: 10.1016/0092-8674(80)90435-3. [DOI] [PubMed] [Google Scholar]
- ————— . Histone gene expression in Drosophila development: Multiple levels of gene regulation. In: Stein G, et al., editors. Histone genes: Structure, organization and regulation. New York: Wiley; 1984. pp. 135–162. [Google Scholar]
- Bowman TL, Hurt MM. The coding sequences of mouse H2A and H3 histone genes contains a conserved seven nucleotide element that interacts with nuclear factors and is necessary for normal expression. Nucleic Acid Res. 1995;23:3083–3092. doi: 10.1093/nar/23.16.3083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark-Adams CD, Norris D, Osley MA, Fassler JS, Winston F. Changes in histone gene dosage alter transcription in yeast. Genes & Dev. 1988;2:150–159. doi: 10.1101/gad.2.2.150. [DOI] [PubMed] [Google Scholar]
- Cotten M, Gick O, Vasserot A, Schaffner G, Birnstiel ML. Specific contacts between mammalian U7 snRNA and histone precursor RNA are indispensable for the in vitro 3′ RNA processing reaction. EMBO J. 1988;7:801–808. doi: 10.1002/j.1460-2075.1988.tb02878.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Currie PD, Sullivan DT. Structure, expression and duplication of genes which encode phosphoglyceromutase of Drosophila melanogaster. Genetics. 1994;138:352–363. [PMC free article] [PubMed] [Google Scholar]
- Dahanukar A, Walker JA, Wharton RP. Smaug, a novel RNA-binding protein that operates a translational switch in Drosophila. Mol Cell. 1999;4:209–218. doi: 10.1016/s1097-2765(00)80368-8. [DOI] [PubMed] [Google Scholar]
- de la Barre AE, Gerson V, Gout S, Creaven M, Allis CD, Dimitrov S. Core histone N-termini play an essential role in mitotic chromosome condensation. EMBO J. 2000;19:379–391. doi: 10.1093/emboj/19.3.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeLisle AJ, Graves RA, Marzluff WF, Johnson LF. Regulation of histone mRNA production and stability in serum-stimulated mouse 3T6 fibroblasts. Mol Cell Biol. 1983;3:1920–1929. doi: 10.1128/mcb.3.11.1920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Marzluff WF. Formation of the 3′ end of histone mRNA. Gene. 1999;239:1–14. doi: 10.1016/s0378-1119(99)00367-4. [DOI] [PubMed] [Google Scholar]
- Dominski Z, Sumerel J, Hanson RJ, Marzluff WF. The polyribosomal protein bound to the 3′ end of histone mRNA can function in histone pre-mRNA processing. Rna. 1995;1:915–923. [PMC free article] [PubMed] [Google Scholar]
- Dominski Z, Zheng LX, Sanchez R, Marzluff WF. Stem–loop binding protein facilitates 3′-end formation by stabilizing U7 snRNP binding to histone pre-mRNA. Mol Cell Biol. 1999;19:3561–3570. doi: 10.1128/mcb.19.5.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dominski, Z., Erkmann, J.A., Greenland, J.A., and Marzluff, W.F. 2001. Mutations in the RNA binding domain of SLBP define separable requirements for RNA binding and for histone pre-mRNA processing. Mol. Cell. Biol. (In press). [DOI] [PMC free article] [PubMed]
- Duronio RJ, O'Farrell PH. Developmental control of a G1-S transcriptional program in Drosophila. Development. 1994;120:1503–1515. doi: 10.1242/dev.120.6.1503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Developmental control of the G1 to S transition in Drosophila: Cyclin E is a limiting downstream target of E2F. Genes & Dev. 1995;9:1456–1468. doi: 10.1101/gad.9.12.1456. [DOI] [PubMed] [Google Scholar]
- Eliassen KA, Baldwin A, Sikorski EM, Hurt MM. Role for a YY1-binding element in replication-dependent mouse histone gene expression. Mol Cell Biol. 1998;18:7106–7118. doi: 10.1128/mcb.18.12.7106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman L, Aragon-Alcaide L, Strunnikov A. The condensin complex governs chromosome condensation and mitotic transmission of rDNA. J Cell Biol. 2000;149:811–824. doi: 10.1083/jcb.149.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fretzin S, Allan BD, van Daal A, Elgin SC. A Drosophila melanogaster H3.3 cDNA encodes a histone variant identical with the vertebrate H3.3. Gene. 1991;107:341–342. doi: 10.1016/0378-1119(91)90337-b. [DOI] [PubMed] [Google Scholar]
- Gick O, Kramer A, Keller W, Birnstiel ML. Generation of histone mRNA 3′ ends by endonucleolytic cleavage of the pre-mRNA in a snRNP-dependent in vitro reaction. EMBO J. 1986;5:1319–1326. doi: 10.1002/j.1460-2075.1986.tb04362.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson RJ, Sun J, Willis DG, Marzluff WF. Efficient extraction and partial purification of the polyribosome-associated stem–loop binding protein bound to the 3′ end of histone mRNA. Biochemistry. 1996;35:2146–2156. doi: 10.1021/bi9521856. [DOI] [PubMed] [Google Scholar]
- Harris ME, Bohni R, Schneiderman MH, Ramamurthy L, Schumperli D, Marzluff WF. Regulation of histone mRNA in the unperturbed cell cycle: Evidence suggesting control at two posttranscriptional steps. Mol Cell Biol. 1991;11:2416–2424. doi: 10.1128/mcb.11.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hassan B, Vaessin H. Daughterless is required for the expression of cell cycle genes in peripheral nervous system precursors of Drosophila embryos. Dev Genet. 1997;21:117–122. doi: 10.1002/(SICI)1520-6408(1997)21:2<117::AID-DVG1>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- Heintz N. The regulation of histone gene expression during the cell cycle. Biochim Biophys Acta. 1991;1088:327–339. doi: 10.1016/0167-4781(91)90122-3. [DOI] [PubMed] [Google Scholar]
- Heintz N, Sive HL, Roeder RG. Regulation of human histone gene expression: Kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983;3:539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, van Steensel B. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt MM, Bowman TL, Marzluff WF. A common transcriptional activator is located in the coding region of two replication-dependent mouse histone genes. Mol Cell Biol. 1991;11:2929–2936. doi: 10.1128/mcb.11.6.2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakoi ER, Brown AL, Ho YS, Snyderman R. Molecular cloning of the cDNA for ligatin. J Cell Sci. 1989;93:227–232. doi: 10.1242/jcs.93.2.227. [DOI] [PubMed] [Google Scholar]
- LaBella F, Sive HL, Roeder RG, Heintz N. Cell-cycle regulation of a human histone H2b gene is mediated by the H2b subtype-specific consensus element. Genes & Dev. 1988;2:32–39. doi: 10.1101/gad.2.1.32. [DOI] [PubMed] [Google Scholar]
- Levine BJ, Chodchoy N, Marzluff WF, Skoultchi AI. Coupling of replication type histone mRNA levels to DNA synthesis requires the stem–loop sequence at the 3′ end of the mRNA. Proc Natl Acad Sci. 1987;84:6189–6193. doi: 10.1073/pnas.84.17.6189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luscher B, Schumperli D. RNA 3′ processing regulates histone mRNA levels in a mammalian cell cycle mutant: A processing factor becomes limiting in G1-arrested cells. EMBO J. 1987;6:1721–1726. doi: 10.1002/j.1460-2075.1987.tb02423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma T, Van Tine BA, Wei Y, Garrett MD, Nelson D, Adams PD, Wang J, Qin J, Chow LT, Harper JW. Cell cycle–regulated phosphorylation of p220(NPAT) by cyclin E/Cdk2 in cajal bodies promotes histone gene transcription. Genes & Dev. 2000;14:2298–2313. doi: 10.1101/gad.829500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Schaller A, Eglite S, Schumperli D, Muller B. The gene for histone RNA hairpin binding protein is located on human chromosome 4 and encodes a novel type of RNA binding protein. EMBO J. 1997;16:769–778. doi: 10.1093/emboj/16.4.769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marzluff WF. Histone 3′ ends: Essential and regulatory functions. Gene Expr. 1992;2:93–97. [PMC free article] [PubMed] [Google Scholar]
- Meeks-Wagner D, Hartwell LH. Normal stoichiometry of histone dimer sets is necessary for high fidelity of mitotic chromosome transmission. Cell. 1986;44:43–52. doi: 10.1016/0092-8674(86)90483-6. [DOI] [PubMed] [Google Scholar]
- Melin L, Soldati D, Mital R, Streit A, Schumperli D. Biochemical demonstration of complex formation of histone pre-mRNA with U7 small nuclear ribonucleoprotein and hairpin binding factors. EMBO J. 1992;11:691–697. doi: 10.1002/j.1460-2075.1992.tb05101.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mowry KL, Steitz JA. Identification of the human U7 snRNP as one of several factors involved in the 3′ end maturation of histone premessenger RNA's. Science. 1987;238:1682–1687. doi: 10.1126/science.2825355. [DOI] [PubMed] [Google Scholar]
- Osley MA. The regulation of histone synthesis in the cell cycle. Annu Rev Biochem. 1991;60:827–861. doi: 10.1146/annurev.bi.60.070191.004143. [DOI] [PubMed] [Google Scholar]
- Pandey NB, Marzluff WF. The stem–loop structure at the 3′ end of histone mRNA is necessary and sufficient for regulation of histone mRNA stability. Mol Cell Biol. 1987;7:4557–4559. doi: 10.1128/mcb.7.12.4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey NB, Williams AS, Sun JH, Brown VD, Bond U, Marzluff WF. Point mutations in the stem–loop at the 3′ end of mouse histone mRNA reduce expression by reducing the efficiency of 3′ end formation. Mol Cell Biol. 1994;14:1709–1720. doi: 10.1128/mcb.14.3.1709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson HE, O'Keefe LV, Reed SI, Saint R. A Drosophila G1-specific cyclin E homolog exhibits different modes of expression during embryogenesis. Development. 1993;119:673–690. doi: 10.1242/dev.119.3.673. [DOI] [PubMed] [Google Scholar]
- Rio DC. Accurate and efficient pre-mRNA splicing in Drosophila cell-free extracts. Proc Natl Acad Sci. 1988;85:2904–2908. doi: 10.1073/pnas.85.9.2904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson PA, Anderton BH, Loviny TL. Nitrocellulose-bound antigen repeatedly used for the affinity purification of specific polyclonal antibodies for screening DNA expression libraries. J Immunol Methods. 1988;108:115–122. doi: 10.1016/0022-1759(88)90409-7. [DOI] [PubMed] [Google Scholar]
- Ross J, Peltz SW, Kobs G, Brewer G. Histone mRNA degradation in vivo: The first detectable step occurs at or near the 3' terminus. Mol Cell Biol. 1986;6:4362–4371. doi: 10.1128/mcb.6.12.4362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J, Kobs G, Brewer G, Peltz SW. Properties of the exonuclease activity that degrades H4 histone mRNA. J Biol Chem. 1987;262:9374–9381. [PubMed] [Google Scholar]
- Saka Y, Sutani T, Yamashita Y, Saitoh S, Takeuchi M, Nakaseko Y, Yanagida M. Fission yeast cut3 and cut14, members of a ubiquitous protein family, are required for chromosome condensation and segregation in mitosis. EMBO J. 1994;13:4938–4952. doi: 10.1002/j.1460-2075.1994.tb06821.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauer K, Knoblich JA, Richardson H, Lehner CF. Distinct modes of cyclin E/cdc2c kinase regulation and S-phase control in mitotic and endoreduplication cycles of Drosophila embryogenesis. Genes & Dev. 1995;9:1327–1339. doi: 10.1101/gad.9.11.1327. [DOI] [PubMed] [Google Scholar]
- Schumperli D. Cell-cycle regulation of histone gene expression. Cell. 1986;45:471–472. doi: 10.1016/0092-8674(86)90277-1. [DOI] [PubMed] [Google Scholar]
- Soldati D, Schumperli D. Structural and functional characterization of mouse U7 small nuclear RNA active in 3′ processing of histone pre-mRNA. Mol Cell Biol. 1988;8:1518–1524. doi: 10.1128/mcb.8.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C, Schumperli D. 3′ processing of pre-mRNA plays a major role in proliferation-dependent regulation of histone gene expression. Nucleic Acids Res. 1988;16:9399–9414. doi: 10.1093/nar/16.20.9399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber C, Luscher B, Eckner R, Lotscher E, Schumperli D. A signal regulating mouse histone H4 mRNA levels in a mammalian cell cycle mutant and sequences controlling RNA 3′ processing are both contained within the same 80-bp fragment. EMBO J. 1986;5:3297–3303. doi: 10.1002/j.1460-2075.1986.tb04643.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strunnikov AV, Hogan E, Koshland D. SMC2, a Saccharomyces cerevisiae gene essential for chromosome segregation and condensation, defines a subgroup within the SMC family. Genes & Dev. 1995;9:587–599. doi: 10.1101/gad.9.5.587. [DOI] [PubMed] [Google Scholar]
- Sutani T, Yuasa T, Tomonaga T, Dohmae N, Takio K, Yanagida M. Fission yeast condensin complex: Essential roles of non-SMC subunits for condensation and Cdc2 phosphorylation of Cut3/SMC4. Genes & Dev. 1999;13:2271–2283. doi: 10.1101/gad.13.17.2271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Daal A, Elgin SC. A histone variant, H2AvD, is essential in Drosophila melanogaster. Mol Biol Cell. 1992;3:593–602. doi: 10.1091/mbc.3.6.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasserot AP, Schaufele FJ, Birnstiel ML. Conserved terminal hairpin sequences of histone mRNA precursors are not involved in duplex formation with the U7 RNA but act as a target site for a distinct processing factor. Proc Natl Acad Sci. 1989;86:4345–4349. doi: 10.1073/pnas.86.12.4345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ZF, Krasikov T, Frey MR, Wang J, Matera AG, Marzluff WF. Characterization of the mouse histone gene cluster on chromosome 13: 45 histone genes in three patches spread over 1 Mb. Genome Res. 1996a;6:688–701. doi: 10.1101/gr.6.8.688. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Whitfield ML, Ingledue TC, III, Dominski Z, Marzluff WF. The protein that binds the 3′ end of histone mRNA: A novel RNA-binding protein required for histone pre-mRNA processing. Genes & Dev. 1996b;10:3028–3040. doi: 10.1101/gad.10.23.3028. [DOI] [PubMed] [Google Scholar]
- Wang ZF, Ingledue TC, Dominski Z, Sanchez R, Marzluff WF. Two Xenopus proteins that bind the 3′ end of histone mRNA: Implications for translational control of histone synthesis during oogenesis. Mol Cell Biol. 1999;19:835–845. doi: 10.1128/mcb.19.1.835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei Y, Yu L, Bowen J, Gorovsky MA, Allis CD. Phosphorylation of histone H3 is required for proper chromosome condensation and segregation. Cell. 1999;97:99–109. doi: 10.1016/s0092-8674(00)80718-7. [DOI] [PubMed] [Google Scholar]
- Wells D, Kedes L. Structure of a human histone cDNA: Evidence that basally expressed histone genes have intervening sequences and encode polyadenylylated mRNAs. Proc Natl Acad Sci. 1985;82:2834–2838. doi: 10.1073/pnas.82.9.2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitfield ML, Zheng LX, Baldwin A, Ohta T, Hurt MM, Marzluff WF. Stem–loop binding protein, the protein that binds the 3′ end of histone mRNA, is cell cycle regulated by both translational and posttranslational mechanisms. Mol Cell Biol. 2000;20:4188–4198. doi: 10.1128/mcb.20.12.4188-4198.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woodland HR, Adamson ED. The synthesis and storage of histones during the oogenesis of Xenopus laevis. Dev Biol. 1977;57:118–135. doi: 10.1016/0012-1606(77)90359-1. [DOI] [PubMed] [Google Scholar]
- Zhao J, Kennedy BK, Lawrence BD, Barbie DA, Matera AG, Fletcher JA, Harlow E. NPAT links cyclin E-cdk2 to the regulation of replication-dependent histone gene transcription. Genes & Dev. 2000;14:2283–2297. [PMC free article] [PubMed] [Google Scholar]