Abstract
Hepatitis C virus (HCV) establishes persistent infections and leads to chronic liver disease. It only recently became possible to study the entire HCV life cycle due to the ability of a unique cloned patient isolate (JFH-1) to produce infectious particles in tissue culture. However, despite efficient RNA replication, yields of infectious virus particles remain modest. This presents a challenge for large-scale tissue culture efforts, such as inhibitor screening. Starting with a J6/JFH-1 chimeric virus, we used serial passaging to generate a virus with substantially enhanced infectivity and faster infection kinetics compared to the parental stock. The selected virus clone possessed seven novel amino acid mutations. We analyzed the contribution of individual mutations and identified three specific mutations, core K78E, NS2 W879R, and NS4B V1761L, which were necessary and sufficient for the adapted phenotype. These three mutations conferred a 100-fold increase in specific infectivity compared to the parental J6/JFH-1 virus, and media collected from cells infected with the adapted virus yielded infectious titers as high as 1 × 108 50% tissue culture infective doses (TCID50)/ml. Further analyses indicated that the adapted virus has longer infectious stability at 37°C than the wild type. Given that the adapted phenotype resulted from a combination of mutations in structural and nonstructural proteins, these data suggest that the improved viral titers are likely due to differences in virus particle assembly that result in significantly improved infectious particle stability. This adapted virus will facilitate further studies of the HCV life cycle, virus structure, and high-throughput drug screening.
INTRODUCTION
Hepatitis C virus (HCV) is an enveloped positive-strand RNA virus that has only recently been adapted to tissue culture (22, 41, 46). The full-length genome of isolate JFH-1 was demonstrated to be competent for viral particle production in tissue culture (22, 41, 46) by using Huh-7-derived cell lines that are permissive to HCV infection and replication (2, 20). Several of these HCV cell culture (HCVcc) systems have been described, the most robust of which are based on chimeric J6/JFH-1 viruses or tissue culture-adapted strains of JFH-1 (1, 3, 4, 17, 18, 26, 36, 47). However, the quantity of infectious virions these systems can produce is limited, presumably due to currently unidentified constraints on infectious virus particle production in tissue culture (5, 16, 26, 47). After passage of tissue culture-grown J6/JFH-1 virus in animals, the resultant viruses exhibited a higher specific infectivity (23). Similarly, passage of HCV in primary human hepatocytes yielded virus with higher specific infectivity (33). Although Huh-7 cells are currently the most efficient system for culturing of HCV, these data suggest that virus particle production is suboptimal in these cells compared to that in bona fide hepatocytes.
Serial passage of HCV in cell culture has yielded virus isolates with increased viral titers (9, 10, 32, 37, 44). Interestingly, these studies revealed complex genetic interactions between viral structural and nonstructural proteins that influence the efficiency of virus particle production. Specifically, interactions between core and NS5A proteins or between NS2, NS3, and the viral envelope glycoproteins have been demonstrated to be important for virus production (25, 31). These viral genetic studies demonstrate interplay between structural and nonstructural proteins and suggest that an appropriate set of adaptive mutations in HCV proteins might improve viral replication and/or assembly.
In this study, we generated an adapted HCV with a specific infectivity that is approximately 100-fold greater than that of wild-type HCV. This adapted phenotype is conferred by the mutations core K78E, NS2 W879R, and NS4B V1761L. Collectively, these 3 mutations increase specific infectivity by increasing the stability of infectious viral particles. The adapted viral replication, assembly, and egress were not significantly enhanced relative to those of the wild type. The generation of this adapted HCV enables the pursuit of studies that until now have been difficult to conduct. These include antibody production, cryo-electron microscopy, drug resistance selection, and high-throughput screening for novel HCV inhibitors.
MATERIALS AND METHODS
Cell culture.
Huh-Lunet and Lunet-CD81 cells were maintained in Dulbecco's modified Eagle medium (DMEM-Glutamax) supplemented with nonessential amino acids and 10% fetal bovine serum (FBS) (DMEM complete medium). Huh-Lunet cells (20) were obtained from ReBLikon GmbH (Mainz, Germany). Media and supplements were purchased from Gibco-BRL, Life Technologies Ltd. (Madison, WI). All cell lines were maintained in humidified incubators at 37°C and 5% CO2.
Generation of stable Lunet-CD81 cell line.
pOTBR-human CD81, a plasmid carrying the human CD81 gene, was obtained from ATCC (Manassas, VA). A lentivirus encoding human CD81 was generated by using the ViraPower lentivirus expression system (Invitrogen, Carlsbad, CA) per the manufacturer's instructions. Huh-Lunet cells were transduced with the human-CD81 lentivirus and subsequently selected in complete medium supplemented with blasticidin (5 μg/ml). Stably transduced cells were stained with a CD81-specific monoclonal antibody (JS-81; Becton Dickinson Biosciences, San Diego, CA) and sorted for high CD81 expression at the Stanford University fluorescence-activated cell sorter (FACS) facility (Palo Alto, CA). Subsequently, cells were transduced and sorted a second time to generate a homogenous cell line that stably expresses high levels of CD81. This cell line was designated Lunet-CD81.
Plasmid construction.
All constructs were verified by DNA sequencing (Tacgen, Richmond, CA). Restriction enzymes were purchased from New England BioLabs (Ipswich, MA). A chimeric HCV genome consisting of the J6 sequence (genotype 2a) from core through NS2 (genotype 2a) followed by the JFH-1 sequence (genotype 2a) of the 5′ untranscribed region (UTR) and then from NS3 through the 3′ UTR with a FLAG tag inserted in the NS5A coding sequence was called J6/JFH-1 Rluc and was previously described (30). To create a nonreporter version of this genome, the Rluc gene was removed by using the flanking MluI restriction sites. The religated product did not contain the Rluc gene, and the resulting plasmid is referred to as pJ6/JFH-1. The individual mutations K78E (core), S401N (E2), W879R (NS2), Y904H (NS2), V1761L (NS4B), D2173G (NS5A), and V2417A (NS5A) were introduced into the pJ6/JFH-1 plasmid by using the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA.). Mutated regions were then exchanged into wild-type pUC19-J6/JFH-1 plasmid by using the restriction enzyme sets EcoRI/NsiI (for the region containing K78E), NsiI/MluI (for the region containing S401), MluI/SpeI (for the region containing W879R and Y904H), SpeI/BamHI (for the region containing V1761L), BamHI/RsrII (for the region containing D2173G), or RsrII/BsrGI (for the region containing V2417A). Plasmids containing a combination of mutations were created by using a QuikChange Multi kit (Stratagene) and up to three mutagenesis primers at once. A region containing both NS2 mutations (W879R and Y904H) was synthesized (GeneOracle, Mountain View, CA) with incorporated 5′ MluI and 3′ SpeI restriction sites and then subcloned into the virus cDNA by using these enzymes. The pJ6/JFH-All 7 construct containing all 7 mutations was synthesized through a combination of ligation and mutagenesis techniques. Single reversion mutants were created by reverting individual mutations in pJ6/JFH-All 7 back to the wild type using the primer sets described.
RNA transcription and electroporation.
In vitro transcripts were generated as previously described (30). Briefly, plasmids were linearized with XbaI and purified by using a MinElute column (Qiagen, Valencia, CA). RNA was transcribed from 1 to 5 μg of purified template by using a T7 Megascript kit (Ambion, La Jolla, CA). Reaction mixtures were incubated at 37°C for 2 h and then digested with 3 U of DNase I (Ambion) for 15 min. RNA was purified by using an RNeasy kit (Qiagen) and quantified by absorbance at 260 nm. RNA integrity was determined by agarose gel electrophoresis and SYBR staining (Invitrogen). Each transcribed RNA (5 μg) was mixed with a suspension of trypsinized Huh-Lunet cells (7.5 × 106 cells/ml in a volume of 400 μl). Cells were then electroporated at 260 V and 950 μF in a Gene Pulser II apparatus (Bio-Rad, Hercules, CA). Electroporated cells were allowed to recover for 10 min at room temperature prior to addition of cell culture medium and plating in T75 flasks or 12-well plates.
Adaptation of wild-type J6/JFH-1 to Lunet-CD81 cells.
Five 96-well plates of Lunet-CD81 cells were infected by limiting dilution (one to four dilutions, from a multiplicity of infection [MOI] of 0.8 to 0.0008) of wild-type-J6/JFH-1-containing supernatant. Following the initial infection, supernatants containing virus were passed onto naive Lunet-CD81 cells. Supernatant passages were performed by collecting supernatant 72 h postinfection and transferring 100 μl virus-containing supernatant onto naive Lunet-CD81 cells, seeded at 3,500 cells per well in 100 μl of complete DMEM medium, so that cells in each well were maintained in a total volume of 200 μl. Overall, eight supernatant passages were performed during the initial adaptation. During the adaptation process, the percentage of cells infected at each passage was quantified by indirect immunofluorescence (see below). Following the 96-well supernatant passages, supernatants from wells consistently exhibiting >50% infected cells were passed into 24-well plates for two passages and then 6-well plates for four passages, T25 and T75 flasks for two passages, and finally a T225 flask. During the process, the percentage of cells infected was continuously monitored by indirect immunofluorescence, and virus supernatants producing greater than 80% infected cells were passed into larger vessels. Cell culture supernatants were assessed for infectious virus via indirect NS5A immunofluorescence or TCID50 assay, and cells were used for preparation of total RNA (see below).
Virus collection.
Virus-containing supernatants were collected and filtered through 0.22-μm filters (Corning, Union City, CA). Following filtration, HEPES buffer, pH 7.5 (Gibco), was added to a final concentration of 10 mM. Viral supernatants were aliquoted and stored immediately at −80°C. Large stocks of virus were prepared by seeding 20 T225 flasks of Lunet-CD81 cells at 2.5 × 106 cells per flask. After cell attachment (>6 h postseeding), medium was removed and 5 to 7 ml of virus-containing supernatants were added at an MOI of >0.3. The next morning, 45 ml of complete DMEM was added to each flask. Three days later, supernatants were collected from subconfluent cells and fresh medium was added. Thirty hours later, supernatants were collected again. For high-titer virus stock collection, virus infection was the same as above. However, virus-containing supernatant was collected at 4-h intervals three times per day 2 to 5 days postinfection.
Preparation of total RNA, amplification of viral RNA by reverse transcription-PCR (RT-PCR), and cloning of amplified DNA fragments.
Total RNA was isolated from T75 flasks of confluent Lunet-CD81 cells infected with the adapted virus by using the QiaShredder (Qiagen) and RNeasy (Qiagen) kits or from infected cell supernatants by using the virus RNA QIAamp kit (Qiagen) as recommended by the manufacturer. One microgram total RNA and 50 pmol primer were used for cDNA synthesis by using the SuperScript III RT-PCR system (Invitrogen) as recommended by the manufacturer. Sequence analysis was performed with a set of primers covering the complete HCV genome (Tacgen).
NS3-4A protease assay.
The presence of viral NS3-4A protease was indirectly measured by using a europium-labeled NS3-4A protease substrate as described previously with slight modifications (43). Briefly, virus-infected cells were grown in 96-well plates and lysed in 1× lysis buffer (Promega) containing 150 mM NaCl and 150 nM europium-labeled NS3 protease substrate (AnaSpec, Fremont, CA). Time-resolved fluorescence was measured by using a BioTek Synergy 2 instrument (Winooski, VT).
Indirect immunofluorescence.
Infected cells were grown in 96-well plates and fixed with 50 μl/well glacial methanol-acetone (1:1) at room temperature for 20 min. Cells were then washed three times with phosphate-buffered saline (PBS). Immunostaining of NS5A was performed by using a mouse monoclonal antibody (9E10; Apath, Brooklyn, NY) at a dilution of 1:10,000 in PBS with 3% bovine serum albumin (BSA) for 1 h at room temperature. After two washes with PBS, bound primary antibodies were detected by using a mouse antibody conjugated to Alexa-Fluor 555 at a dilution of 1:3,000 in PBS containing 3% BSA for 20 min in the dark at 4°C. DNA was stained with 4′,6′-diamidino-2-phenylindole dihydrochloride (Molecular Probes, Madison, WI) for 10 min at 4°C in the dark. Finally, cells were washed three times with PBS and imaged by using a Zeiss microscope with fluorescence capabilities (Thornwood, NY). The percentage of infected cells was quantified by visual inspection and counting. To further quantify the percentage of cells infected, images were instead acquired using the ImageXpress Micro system (Molecular Devices, Sunnyvale, CA). Following immunofluorescence with anti-NS5A antibody, Hoechst-stained cells were imaged in the Cy3 and 4′,6-diamidino-2-phenylindole (DAPI) channels, respectively, at magnification ×4 with a minimum of 4 images per well. Infected cells were distinguished from noninfected cells using the Cell Scoring module in the MetaXpress 2.0 software program (Molecular Devices).
Determination of virus titers in cell culture supernatants.
Virus titers were determined as described elsewhere, with slight modifications (21). Lunet-CD81 target cells were seeded at a concentration of 5,000 cells per well in 96-well plates in a total volume of 100 μl complete DMEM/well. The next day, one to four serial dilutions of virus-containing supernatant were prepared and 100 μl of each dilution was added, with four wells per dilution. Three days later, cells were fixed according to the indirect immunofluorescence protocol described above. Virus titers (TCID50/ml) were calculated based on the method of Reed and Muench (35).
Determining specific infectivity.
Specific infectivity was calculated as extracellular TCID50/ml at 72 h posttransfection/packaged extracellular RNA copies at 72 h posttransfection. Packaged extracellular RNA levels were measured by collecting 1 ml of extracellular medium from transfected Huh7-Lunet cell cultures growing on a 12-well plate 72 h posttransfection. In this cell line, viral spread is limited (20). Aliquots of extracellular medium were separated by using equilibrium density gradient ultracentrifugation according to the method of Lindenbach et al. (22). Briefly, 1 ml of extracellular medium was loaded at the top of 10 ml 10 to 40% iodixanol gradients. Iodixanol gradients were prepared by using 10% and 40% iodixanol solutions that each contained 10 mM HEPES, pH 7.5, and 0.02% BSA. In addition, the 10% iodixanol solution contained 125 mM NaCl and the 40% iodixanol solution contained 50 mM NaCl to maintain iso-osmolarity. Samples were centrifuged through the gradients at 40,000 rpm for 16 h at 4°C in an SW41 Ti swinging bucket rotor (Beckman, Brea, CA). Subsequently, each gradient was divided into 26 0.4-ml fractions, and each fraction was assayed for relative infectivity by using the NS3-4A protease assay described above. The packaged extracellular RNA was isolated from the peak infectious fraction (fraction 15) (data not shown) in each case using a QIAamp viral RNA isolation kit (Qiagen). Viral RNA was quantified by using quantitative RT-PCR (qRT-PCR).
Determining infectious HCV stability.
Infectious HCV stability was determined by incubating 1-ml aliquots of extracellular HCV in cell culture medium at 37°C and freezing 100-μl aliquots at −80°C at various times. After collecting samples for all of the time points, naive cells were infected with each sample for 72 h and the relative levels of infectivity were measured using the NS3-4A protease assay described above. The level of infectivity was normalized to 100% at time zero for each viral strain tested. The data were plotted as percent infectivity versus time (hours) and fit to a first-order decay equation by using the SigmaPlot software program (Systat, Chicago, IL) to determine the slope (−k). The half-lives for each infectious HCV strain (t1/2) were calculated in hours by using ln 2/k (7).
Statistical analysis.
Statistical analysis was performed using the Prism 4.0 software program (GraphPad, La Jolla, CA). All comparisons were made with two-tailed t tests. A P value of <0.05 is reported as significant.
RESULTS
Adapted J6/JFH-1 virus infects a greater percentage of cells in culture and produces significantly higher extracellular titers than wild-type J6/JFH-1 virus.
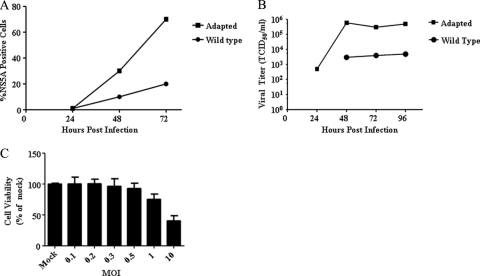
We passaged wild-type J6/JFH-1 virus several times (see Materials and Methods) and characterized the resulting virus population to determine if it was adapted for more efficient growth and infection in cell culture. We compared the percentages of cells that the wild-type and adapted viruses are able to infect over time by infecting naive Lunet-CD81 cells with adapted virus or wild-type virus at an MOI of 0.5. Infection and virus spread were followed by NS5A immunofluorescence and titration of infectious virus from cell culture supernatants. Three days after infection, the adapted virus infected ∼80% of the Lunet-CD81 cells, while the wild-type virus infected only ∼20% of the cells (Fig. 1A). The difference was confirmed by the ImageExpress Micro system (see Materials and Methods) (data not shown). Correspondingly, cells infected at an MOI of 0.5 with the adapted virus released over 1 × 105 TCID50/ml at 3 and 4 days postinfection, whereas wild-type titers were 30 to 100 times lower (Fig. 1B). In a separate experiment in which cells were infected at an MOI of 1, the adapted virus reached 1 × 108 TCID50/ml after 5 days, while the wild-type virus titer remained at ∼1 × 104 TCID50/ml (data not shown).
Fig. 1.
The adapted virus infects a greater percentage of cells and reaches higher viral titers than the wild-type virus. (A) Naive Lunet-CD81 cells were infected at an MOI of 0.5, and the infections were monitored over time using NS5A staining. Data are representative curves. (B) Naive Lunet-CD81 cells were infected at an MOI of 0.5, and viral titers were followed over time. A representative time course is shown. Data are representative curves. (C) Naive Lunet-CD81 cells were infected at various multiplicities of infection, and the percentage of viable cells was estimated 3 days postinfection. Error bars indicate the standard deviations of data from three assays.
Because the adapted virus produced high titers of extracellular virus, we wondered if it had altered cytopathic effects on target cells. We observed that cells infected with the adapted virus at an MOI of 10 showed a significant decrease in viability relative to a mock infection. Specifically, less than 50% of target Lunet-CD81 cells were viable after 3 days of infection at a MOI of 10 compared to results for mock-infected cells (Fig. 1C). However, greater than 80% of the target cells were viable 3 days after infection with adapted virus at an MOI of 1 or less (Fig. 1C). Thus, the adapted virus showed cytopathic effects at high multiplicities of infection. This is consistent with prior work showing that HCV induces cytopathic effects in long-term cultures (38, 47).
Mutations in core, NS2, and NS4B collectively reproduce the adapted viral phenotype.
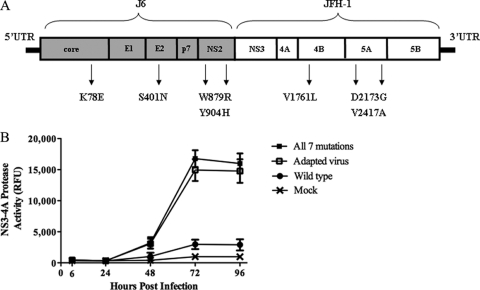
To identify the mutations responsible for the enhanced virus production, the entire HCV genome isolated from supernatants of infected cells was reverse transcribed, PCR amplified, and sequenced. We determined that the adapted virus contained seven mutations relative to the wild-type virus (Fig. 2A). The mutations were located in the structural (core [K78E] and E2 [S401N]) and nonstructural (NS2 [W879R, Y904H], NS4B [V1761L], and NS5A [D2173G, V2417A]) genes. Five silent mutations were also identified (data not shown).
Fig. 2.
The adapted virus contains 7 mutations relative to the wild type. (A) A cartoon map showing the J6/JFH-1 construct, where J6 is core-NS2 and JFH-1 is NS3-NS5B. Indicated are the locations of the 7 mutations found in the adapted virus. (B) The All-7 virus replicates in cells similarly to the adapted virus. NS3-4A protease activity levels for wild-type virus, adapted virus, All-7 virus, and a mock infection control are indicated. Error bars indicate standard deviations of data from 3 assays.
To determine if the seven mutations identified from genotyping were sufficient to confer the adaptive phenotype, we engineered them into the wild-type viral construct by using site-directed mutagenesis, transcribing viral RNAs, and transfecting them into Lunet-CD81 cells to generate viral stocks. We performed time course experiments to compare the kinetics of infectious virus production after infecting Lunet-CD81 cells at an MOI of 0.5 with wild-type virus, the adapted virus population, or J6/JFH-1 engineered to contain all seven mutations (All-7 virus). At each time point, virus replication was monitored by measuring the relative level of NS3-4A serine protease activity expressed in target cells. The quantitative protease expression assay is an alternative to monitoring intracellular viral replication via NS5A immunostaining (see Materials and Methods). The All-7 virus had infection kinetics identical to that of the adapted virus (Fig. 2B), indicating that the 7 mutations were sufficient to confer cell culture adaptation. This phenotype included a greater than 100-fold increase in virus production (TCID50/ml) relative to that of wild-type virus at 72 h after infection (Table 1) and a greater than 6-fold increase in NS3-4A protease accumulation in cells (Fig. 2B and Table 1) compared to that of the wild-type virus.
Table 1.
Effects of reversion of single mutations on infectivity posttransfection and postinfection
| Virus descriptiona | Protein | Result at 72 h postinfection (MOI = 0.5) |
|
|---|---|---|---|
| NS3 accumulation (% of All-7)b | Virus titer (TCID50/ml)c | ||
| Wt | 15.3 ± 10.6** | 1.30 × 102 | |
| All-7 | 100.0 ± 11.6 | 1.40 × 104 | |
| Rev K78E | Core | 30.8 ± 9.1** | 2.00 × 103 |
| Rev S401N | E2 | 104.6 ± 13.5 | 2.50 × 104 |
| Rev W879R | NS2 | 46.2 ± 8.9** | 2.00 × 103 |
| Rev Y904H | NS2 | 110.4 ± 22.2 | 2.50 × 104 |
| Rev V1761L | NS4B | 67.5 ± 18.6* | 4.60 × 103 |
| Rev D2173G | NS5A | 132.4 ± 17.9 | 1.40 × 104 |
| Rev V2417A | NS5A | 119.3 ± 19.9 | 1.40 × 104 |
Wt, wild type. “Rev” indicates reversion of the mutation to the wild-type residue.
Standard deviations of data from three independent experiments are given (two-tailed t test relative to data for All-7 clone (*, P < 0.05; **, P < 0.005).
Virus titers calculated from one of three representative experiments.
To identify which mutations were necessary for the adapted phenotype, we reverted each of the mutations individually back to its wild-type residue in the All-7 construct. These constructs were then transfected into Huh7-Lunet cells, viral supernatants were collected, and titers were determined at 72 h after transfection. We then infected Lunet-CD81 cells with each of the reversion mutants as well as the wild-type and All-7 viruses at an MOI of 0.5. To assess viral replication and spread, we measured NS3-4A protease activity in infected cell lysates 72 h after infection (Table 1). These assays indicated that reversion of the mutation core K78E, NS2 W879R, or NS4B V1761L reduced the replication and spread of the virus compared to those of All-7. However, reversion of S401N (E2), Y904H (NS2), D2173G (NS5A), or V2417A (NS5A) did not have significant effects on viral replication and spread. We also determined the titers of virus produced by the revertants at 72 h after infection (Table 1). In agreement with the NS3-4A protease activity assay, reversion of K78E (core), W879R (NS2), and V1761L (NS4B) reduced the viral titer to levels below that of the All-7 virus while the remaining four reversions did not. Importantly, no single reversion mutation reduced viral replication, spread, or production to wild-type viral levels, indicating that a combination of mutations is required for full adaptation. Collectively, these results indicate that the mutations core K78E, NS2 W879R, and NS4B V1761L are primarily responsible for the adapted viral phenotype.
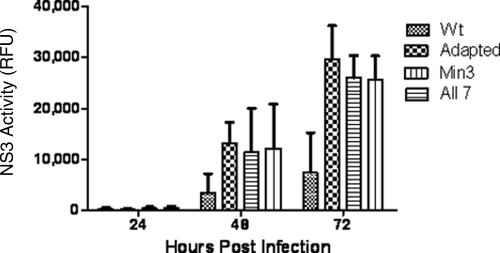
To further support the conclusion that three mutations are responsible for the adapted viral phenotype, we engineered these mutations, alone or in various combinations, into the wild-type virus. Huh7-Lunet cells were transfected with in vitro-transcribed RNA for each clone, and virus-containing media were collected 3 and 4 days posttransfection. Supernatants were then used to infect Lunet-CD81 cells at an MOI of 0.5. The single point mutation core K78E, NS2 W879R, or NS4B V1761L was not sufficient to reproduce the adapted phenotype as determined by monitoring the expression of NS3-4A protease activity over time (data not shown). However, the Min3 virus containing all three mutations reproduced the adapted phenotype (Fig. 3).
Fig. 3.
The Min3 virus replicates in cells at levels similar to those of the adapted and All-7 viruses. NS3-4A protease activity levels for wild-type virus(Wt), adapted virus, All-7 virus, and Min3 virus are indicated. Error bars indicate standard deviations of data from 3 assays.
Adaptive mutations increase specific infectivity of virus particles.
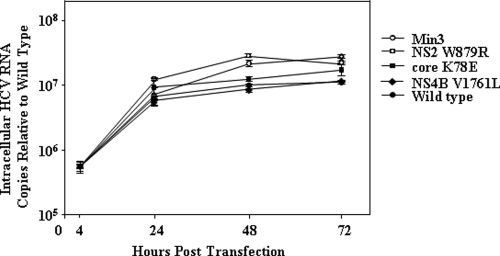
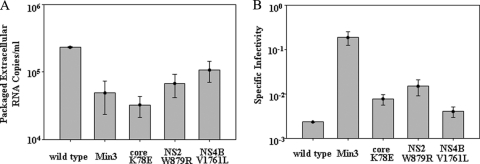
We sought to determine why the Min3 virus infected more cells and produced higher titers than wild-type virus. We therefore asked whether the core, NS2, or NS4B mutations, alone or in combination, enhanced the levels of intracellular RNA accumulation or the specific infectivity of virus particles. To do this, we transfected Huh7-Lunet cells with RNA transcripts encoding the Min3, core K78E, NS2 W879R, or NS4B V1761L viral genome. The Huh7-Lunet cell line supports efficient HCV RNA replication but due to its low expression of CD81 limits viral spread (20). This characteristic allows us to directly compare intracellular viral RNA production with the levels of packaged extracellular RNA and infectious viral particles without the bias of viral spread. The lack of significant viral spread in this cell type was confirmed by monitoring NS5A-positive cells over 4 days post-transfection with all viruses (data not shown). We then compared intracellular levels of HCV RNA at different time points following transfection. The intracellular levels of HCV RNA were similar for the wild type and core K78E and NS4B V1761L mutants and only slightly higher for the Min3 and NS2 W879R mutants (Fig. 4). When we measured both intracellular and extracellular viral titers, we observed that the wild-type, core K78E, NS2 W879R, and NS4B V1761L titers were similar over a 3-day time course (Fig. 5A and B). However, the Min3 intracellular and extracellular viral titers were dramatically higher than those of the wild type or each single mutant (Fig. 5A and B). Finally, when we compared the levels of packaged extracellular viral RNA by using density gradient equilibrium ultracentrifugation (see Materials and Methods), we observed that the wild type, Min3, and the single mutants each released roughly similar amounts of viral particles (Fig. 6A). When we calculated the specific infectivity for the wild type, Min3, and each single-mutant virus (extracellular viral titer/packaged extracellular RNA copies) (see Materials and Methods), we noted that the Min3 specific infectivity was approximately 100-fold greater than that of the wild-type virus (Fig. 6B). In addition, each single-mutant virus had a relatively low specific infectivity in comparison to that of Min3 (Fig. 6B), suggesting that none of these single mutations significantly increased the specific infectivity of virus particles on their own.
Fig. 4.
Intracellular RNA replication for wild-type HCV, Min3, and the HCV single mutants is similar following RNA transfection under conditions of limited viral spread. Intracellular HCV RNA levels for the wild type, Min3, core K78E, NS2 W879R, and NS4B V1761L are indicated. The intracellular RNA levels were normalized relative to the wild-type intracellular RNA levels at 4 h following RNA transfection. Error bars indicate standard deviations of data from 2 assays.
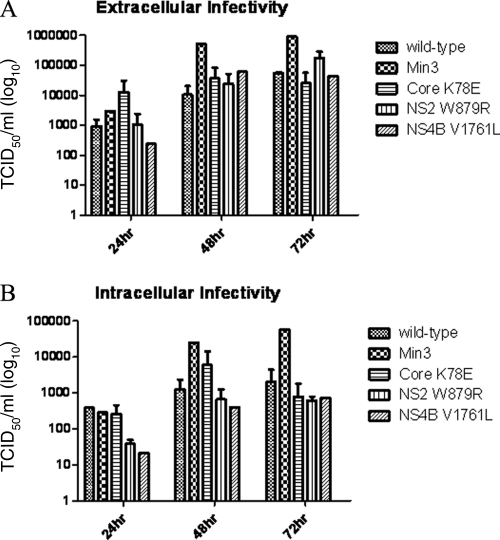
Fig. 5.
Min3 extracellular and intracellular infectious titers reach significantly higher levels than wild-type HCV or HCV single-mutant infectious titers over time. (A) Infectious titers from the extracellular milieu were measured following RNA transfection under conditions of limited viral spread. (B) The infectious titer from each intracellular fraction was determined in the same experiment shown in panel A. HCV infectious titers for the wild type, Min3, core K78E, NS2 W879R, and NS4B V1761L are indicated. Error bars indicate standard deviations of data from 2 assays.
Fig. 6.
Min3 specific infectivity is significantly greater than that of wild-type HCV. (A) The relative levels of packaged extracellular HCV RNA are shown for wild-type HCV, Min3, and the HCV single mutants. Packaged extracellular RNA levels were determined as described in Materials and Methods. (B) The specific infectivity values (extracellular TCID50/ml/packaged extracellular RNA levels) for wild-type HCV, Min3, and the HCV single mutants are compared. Error bars indicate standard deviations of data from 2 assays.
Adaptive mutations increase the stability of infectious virus particles.
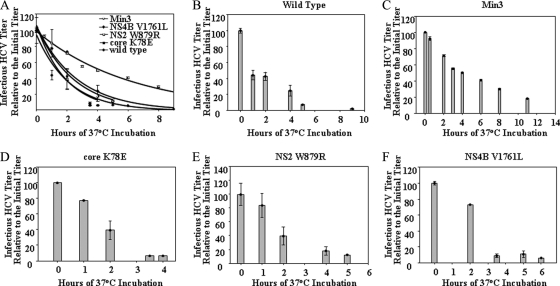
The Min3 virus exhibited significantly higher extracellular titers of infectious virus but released a relatively similar number of RNA-containing viral particles (Fig. 5A and 6A). Thus, we sought to determine if Min3 viral infectivity is longer lived than that of the wild-type or single mutant viruses. We compared the stability of infectious wild-type, Min3, and single-mutant viruses by incubating samples of each virus at 37°C and freezing aliquots at various times over the course of several hours. Subsequently, we compared the infectivity in each sample by infecting naive cells and measuring the NS3-4A protease activity levels in each case (see Materials and Methods). The infectivity of the wild-type and single-mutant viruses decayed with a half-life of approximately 1.5 h (Fig. 7). This infectious stability is similar to what others have previously observed with HCV JFH-1 (8, 29, 39). However, Min3 infectivity decayed with a half-life of approximately 4 h under the same conditions (Fig. 7). Thus, the adaptive phenotype strongly correlated with the increased infectious stability of virus particles.
Fig. 7.
Infectious stability of Min3 is significantly greater than that of wild-type HCV or the HCV single mutants. Infectivities at various times for the wild type, Min3, core K78E, NS2 W879R, and NS4B V1761L are indicated. Infectious stability (t1/2) was calculated as described in Materials and Methods. For wild-type HCV, t1/2 = 1.5 ± 0.3 h; for Min3, t1/2 = 4.0 ± 0.5 h; for core K78E, t1/2 = 1.3 ± 0.2 h; for NS2 W879R, t1/2 = 1.7 ± 0.3 h; and for NS4B V1761L, t1/2 = 1.9 ± 0.5 h. (B to F) Infectious stability data are shown individually for the wild type, Min3, core K78E, NS2 W879R, and NS4B V1761L virus as bar graphs. Error bars indicate standard deviations of data from 2 assays.
DISCUSSION
The adaptive mutations in core, NS2, and NS4B may alter infectious particle assembly.
In this work we observed that single mutations in core, NS2, and NS4B collectively increase HCV specific infectivity and stability of infectious virus particles. These results reveal a unique mechanism of cell culture adaptation and highlight the interplay between viral structural and nonstructural proteins during virus particle assembly.
We believe that the increase in specific infectivity (almost 100-fold) and higher viral titers observed with the adapted virus relative to those with the wild type are dependent to a great extent on the observed ∼3-fold difference in infectious stability. The increase in specific infectivity for the adapted virus was due primarily to an increase in the extracellular infectious titer relative to that of the wild-type (Fig. 5A) and not to a significant increase in the number of excreted virions (Fig. 6A). An ∼3-fold difference in infectious stability can mean the difference, for example, between an initial virus population having 16 h to infect cells in a culture versus 48 h (i.e., assuming 2,500 infectious particles are used to infect 5,000 cells [MOI = 0.5] as in Fig. 1A) before all virions lose infectivity. In addition, this increase in infectious stability means that infected cells will already be adding newly synthesized infectious virions to the extracellular population before much of the earlier-synthesized extracellular virions have lost infectivity. In this manner, it is possible that higher viral titers may be achieved.
HCV core is thought to encapsidate the viral RNA genome within virus particles (27). The lysine residue at position 78 in core is conserved among 58% (33/57) of the genotype 2 sequences listed in the Los Alamos HCV sequence database, with other major variations being glutamine (Q) or arginine (R). K78 has been shown to be important for core oligomerization in the early process of virus assembly (27). Deletion or alanine replacement of a region of core including residue 78 abolishes infectious virus production (27, 28). Importantly, residue 78 is located within the RNA binding region and near the region implicated in homotypic core-core interactions (19). Thus, we speculate that the K78E substitution in core may improve viral RNA packaging.
NS2 has been implicated in virus particle assembly, although its precise role in this process is not understood (6, 9, 12, 13, 32, 37, 42, 45). The W879 residue in NS2 is conserved in 100% of the genotype 2 and genotype 3 NS2 sequences listed in the Los Alamos HCV database. This residue is methionine (M) in genotype 5 sequences and either isoleucine (I) or leucine (L) in all other genotypes. Residue 879 is predicted to be in the third transmembrane domain of NS2 (31). Therefore, this residue could interact with core or another viral or host protein involved in assembly. Such an interaction between core and NS5A for production of infectious particles has been reported previously (25). Recently, Stapleford and Lindenbach showed that NS2 coordinates virus particle assembly through interactions with the viral E1-E2 glycoprotein and NS3-4A enzyme complexes and that this region of NS2 helps mediate interactions between NS2 and the glycoprotein complex that are essential for virus particle assembly (40). We speculate that the NS2 W879R mutation alters NS2's function as a scaffold for virus particle assembly and may help to promote the production of infectious particles with increased stability. It was notable that the NS2 adaptive mutation gave rise to a small but reproducible increase in intracellular viral RNA levels (Fig. 4). Given that NS2 is not necessary for viral genome replication (24), these results could reveal a previously unknown role for NS2 in this process. Alternatively, this moderate increase in intracellular HCV RNA levels could have been due to an increase in nascent virus particles, although Fig. 5B implies that these particles were not yet infectious. Further experiments will be needed to discern between these possibilities.
NS4B is a transmembrane protein that is thought to have roles in both viral replication and assembly (11, 14). NS4B residue V1761 is 100% conserved in genotype 2a, 6a, and 7 sequences. This residue is leucine (L) in genotype 1, 2b-2i, 3, and 4 and alanine (A) in genotype 5. This residue lies in the second putative amphipathic helix of NS4B, a region that has been implicated in RNA replication (11). However, the NS4B V1761L mutation had no effect on RNA replication of the JFH-1 subgenomic replicon (data not shown). These data agree with the RNA measurements we performed with viral constructs transfected into Huh-Lunet cells, which also indicated that transfection efficiency and RNA replication levels were similar between wild-type and adapted viruses (Fig. 4). Interestingly, a mutation similar to V1761L was identified in a noncytopathic strain of bovine viral diarrhea virus (BVDV), suggesting that the NS4B mutation could help maintain a noncytopathic phenotype necessary for long-term HCVcc passaging (34). Additionally, given the predicted role of NS4B in viral assembly (14), we speculate that NS4B V1761L mutation functions in combination with the core K78E and NS2 W879R mutations to promote the assembly of virus particles.
Adapted virus as a tool for future research.
This adapted virus should prove to be a useful tool for future research. Previously reported adaptive mutations did not overcome a limit of ≈1 × 106 TCID50/ml (9, 10, 15, 18, 37, 47). By using our adapted virus from this study and collecting virus multiple times each day, we routinely obtained virus stocks with titers between 5 × 107 and 1 × 108 TCID50/ml. These titers were 50- to 100-fold higher than those reported in the literature for other cell culture-adapted HCVcc isolates (10, 32). Generating virus titers in tissue culture that are comparable to those generated in vivo is a significant step forward for HCV studies. We believe that this will enable new and previously unobtainable studies of HCV, including cryo-electron microscopy and the development of high-quality assays for antiviral screening.
ACKNOWLEDGMENTS
We thank Brett Lindenbach, Jane McKeating, Thomas Pietschmann, and Charles Rice for important scientific discussions and Serena Constantino, Amy Herstek, and Brett Lindenbach for critical reading of the manuscript.
Footnotes
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Bartenschlager R. 2006. Hepatitis C virus molecular clones: from cDNA to infectious virus particles in cell culture. Curr. Opin. Microbiol. 9:416–422 [DOI] [PubMed] [Google Scholar]
- 2. Bartenschlager R., Pietschmann T. 2005. Efficient hepatitis C virus cell culture system: what a difference the host cell makes. Proc. Natl. Acad. Sci. U. S. A. 102:9739–9740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bungyoku Y., et al. 2009. Efficient production of infectious hepatitis C virus with adaptive mutations in cultured hepatoma cells. J. Gen. Virol. 90:1681–1691 [DOI] [PubMed] [Google Scholar]
- 4. Delgrange D., et al. 2007. Robust production of infectious viral particles in Huh-7 cells by introducing mutations in hepatitis C virus structural proteins. J. Gen. Virol. 88:2495–2503 [DOI] [PubMed] [Google Scholar]
- 5. Deng L., et al. 2008. Hepatitis C virus infection induces apoptosis through a Bax-triggered, mitochondrion-mediated, caspase 3-dependent pathway. J. Virol. 82:10375–10385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dentzer T. G., Lorenz I. C., Evans M. J., Rice C. M. 2009. Determinants of the hepatitis C virus nonstructural protein 2 protease domain required for production of infectious virus. J. Virol. 83:12702–12713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Eisenberg D. C., Crothers D. M. 1979. Physical chemistry with applications to life sciences. Addison Wesley Longman, Reading, MA [Google Scholar]
- 8. Farquhar M. J., et al. 2008. Protein kinase A-dependent step(s) in hepatitis C virus entry and infectivity. J. Virol. 82:8797–8811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gottwein J. M., et al. 2007. Robust hepatitis C genotype 3a cell culture releasing adapted intergenotypic 3a/2a (S52/JFH1) viruses. Gastroenterology 133:1614–1626 [DOI] [PubMed] [Google Scholar]
- 10. Gottwein J. M., et al. 2009. Development and characterization of hepatitis C virus genotype 1-7 cell culture systems: role of CD81 and scavenger receptor class B type I and effect of antiviral drugs. Hepatology 49:364–377 [DOI] [PubMed] [Google Scholar]
- 11. Gouttenoire J., Penin F., Moradpour D. 2010. Hepatitis C virus nonstructural protein 4B: a journey into unexplored territory. Rev. Med. Virol. 20:117–129 [DOI] [PubMed] [Google Scholar]
- 12. Jirasko V., et al. 2008. Structural and functional characterization of nonstructural protein 2 for its role in hepatitis C virus assembly. J. Biol. Chem. 283:28546–28562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Jones C. T., Murray C. L., Eastman D. K., Tassello J., Rice C. M. 2007. Hepatitis C virus p7 and NS2 proteins are essential for production of infectious virus. J. Virol. 81:8374–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jones D. M., Patel A. H., Targett-Adams P., McLauchlan J. 2009. The hepatitis C virus NS4B protein can trans-complement viral RNA replication and modulates production of infectious virus. J. Virol. 83:2163–2177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Kato T., et al. 2008. Hepatitis C virus JFH-1 strain infection in chimpanzees is associated with low pathogenicity and emergence of an adaptive mutation. Hepatology 48:732–740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Kato T., et al. 2007. Production of infectious hepatitis C virus of various genotypes in cell cultures. J. Virol. 81:4405–4411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kaul A., Woerz I., Meuleman P., Leroux-Roels G., Bartenschlager R. 2007. Cell culture adaptation of hepatitis C virus and in vivo viability of an adapted variant. J. Virol. 81:13168–13179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Kaul A., Worz I., Bartenschlager R. 2009. Adaptation of the hepatitis C virus to cell culture. Methods Mol. Biol. 510:361–372 [DOI] [PubMed] [Google Scholar]
- 19. Klein K. C., Dellos S. R., Lingappa J. R. 2005. Identification of residues in the hepatitis C virus core protein that are critical for capsid assembly in a cell-free system. J. Virol. 79:6814–6826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Koutsoudakis G., Herrmann E., Kallis S., Bartenschlager R., Pietschmann T. 2007. The level of CD81 cell surface expression is a key determinant for productive entry of hepatitis C virus into host cells. J. Virol. 81:588–598 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lindenbach B. D. 2009. Measuring HCV infectivity produced in cell culture and in vivo. Methods Mol. Biol. 510:329–336 [DOI] [PubMed] [Google Scholar]
- 22. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 23. Lindenbach B. D., et al. 2006. Cell culture-grown hepatitis C virus is infectious in vivo and can be recultured in vitro. Proc. Natl. Acad. Sci. U. S. A. 103:3805–3809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Lohmann V., et al. 1999. Replication of subgenomic hepatitis C virus RNAs in a hepatoma cell line. Science 285:110–113 [DOI] [PubMed] [Google Scholar]
- 25. Masaki T., et al. 2008. Interaction of hepatitis C virus nonstructural protein 5A with core protein is critical for the production of infectious virus particles. J. Virol. 82:7964–7976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Mateu G., Donis R. O., Wakita T., Bukh J., Grakoui A. 2008. Intragenotypic JFH1 based recombinant hepatitis C virus produces high levels of infectious particles but causes increased cell death. Virology 376:397–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Murray C. L., Jones C. T., Tassello J., Rice C. M. 2007. Alanine scanning of the hepatitis C virus core protein reveals numerous residues essential for production of infectious virus. J. Virol. 81:10220–10231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nakai K., et al. 2006. Oligomerization of hepatitis C virus core protein is crucial for interaction with the cytoplasmic domain of E1 envelope protein. J. Virol. 80:11265–11273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Paintsil E., He H., Peters C., Lindenbach B. D., Heimer R. 2010. Survival of hepatitis C virus in syringes: implication for transmission among injection drug users. J. Infect. Dis. 202:984–990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Paulson M. S., et al. 2009. Comparison of HCV NS3 protease and NS5B polymerase inhibitor activity in 1a, 1b and 2a replicons and 2a infectious virus. Antiviral Res. 83:135–142 [DOI] [PubMed] [Google Scholar]
- 31. Phan T., Beran R. K., Peters C., Lorenz I. C., Lindenbach B. D. 2009. Hepatitis C virus NS2 protein contributes to virus particle assembly via opposing epistatic interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 83:8379–8395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pietschmann T., et al. 2006. Construction and characterization of infectious intragenotypic and intergenotypic hepatitis C virus chimeras. Proc. Natl. Acad. Sci. U. S. A. 103:7408–7413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Podevin P., Carpentier A., Pene V., Aoudjehane L., Carriere M., Zaidi S., Hernandez C., Calle V., Meritet J. F., Scatton O., Dreux M., Cosset F. L., Wakita T., Bartenschlager R., Demignot S., Conti F., Rosenberg A. R., Calmus Y. 2010. Production of infectious hepatitis C virus in primary cultures of human adult hepatocytes. Gastroenterology 139:1355–1364 [DOI] [PubMed] [Google Scholar]
- 34. Qu L., McMullan L. K., Rice C. M. 2001. Isolation and characterization of noncytopathic pestivirus mutants reveals a role for nonstructural protein NS4B in viral cytopathogenicity. J. Virol. 75:10651–10662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Reed L. J., Muench H. 1938. A simple method of estimating 50 percent end-points. Am. J. Hyg. 27:493–497 [Google Scholar]
- 36. Russell R. S., et al. 2008. Advantages of a single-cycle production assay to study cell culture-adaptive mutations of hepatitis C virus. Proc. Natl. Acad. Sci. U. S. A. 105:4370–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Scheel T. K., et al. 2008. Development of JFH1-based cell culture systems for hepatitis C virus genotype 4a and evidence for cross-genotype neutralization. Proc. Natl. Acad. Sci. U. S. A. 105:997–1002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sekine-Osajima Y., et al. 2008. Development of plaque assays for hepatitis C virus-JFH1 strain and isolation of mutants with enhanced cytopathogenicity and replication capacity. Virology 371:71–85 [DOI] [PubMed] [Google Scholar]
- 39. Song H., et al. 2010. Thermal stability and inactivation of hepatitis C virus grown in cell culture. Virol. J. 7:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stapleford K. A., Lindenbach B. 2011. Hepatitis C virus NS2 coordinates virus particle assembly through physical interactions with the E1-E2 glycoprotein and NS3-NS4A enzyme complexes. J. Virol. 85:1706–1717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Welbourn S., et al. 2009. Investigation of a role for lysine residues in non-structural proteins 2 and 2/3 of the hepatitis C virus for their degradation and virus assembly. J. Gen. Virol. 90:1071–1080 [DOI] [PubMed] [Google Scholar]
- 43. Yang H., Delaney IV W. E. 2006. A novel fluorescence-based protease assay using the endogenous NS3/4A protease activity present in the total cell lysates of HCV replicon cells. J. Clin. Virol. 36:S109 [Google Scholar]
- 44. Yi M., Ma Y., Yates J., Lemon S. M. 2007. Compensatory mutations in E1, p7, NS2, and NS3 enhance yields of cell culture-infectious intergenotypic chimeric hepatitis C virus. J. Virol. 81:629–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Yi M., Ma Y., Yates J., Lemon S. M. 2009. Trans-complementation of an NS2 defect in a late step in hepatitis C virus (HCV) particle assembly and maturation. PLoS Pathog. 5:e1000403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhong J., et al. 2005. Robust hepatitis C virus infection in vitro. Proc. Natl. Acad. Sci. U. S. A. 102:9294–9299 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhong J., et al. 2006. Persistent hepatitis C virus infection in vitro: coevolution of virus and host. J. Virol. 80:11082–11093 [DOI] [PMC free article] [PubMed] [Google Scholar]