Abstract
In mouse embryonic fibroblasts (MEFs), the bovine rotavirus (UK strain) but not the simian rhesus rotavirus (RRV) robustly triggers beta interferon (IFN-β) secretion, resulting in an IFN-dependent restriction of replication. We now find that both rotavirus strains trigger antiviral transcriptional responses early during infection and that both transcriptional responses and IFN-β secretion are completely abrogated in MAVS/IPS-1−/− MEFs. Replication of UK virus could be rescued in MAVS/IPS-1−/− MEFs, and synthesis of viral RNA significantly increased early during virus infection. UK virus induced IFN-β secretion and transcription of IFN-stimulated genes (ISGs) in both RIG-I−/− and MDA-5−/− MEFs, and neither receptor was essential by itself for the antiviral response to UK rotavirus. However, when receptors RIG-I and MDA-5 were depleted using RNA interference, we found that both contribute to the magnitude of the IFN response. IRF3 was found to be essential for MAVS/IPS-1-directed ISG transcription and IFN-β secretion during rotavirus infection. Interestingly, absence of the double-stranded RNA-dependent protein kinase PKR led to a profound defect in the capacity of host cells to secrete IFN-β in response to virus. Both PKR and IRF3 restricted the early replication of UK as indicated by significant increases in viral RNA in fibroblasts lacking either gene. Despite the loss in IFN-β secretion in PKR−/− MEFs, we did not observe decreased IRF3- or NF-κB-dependent early ISG transcription in these cells. Levels of transcripts encoding IFN-α4, IFN-α5, and IFN-β were high in infected PKR−/− MEFs, indicating that during rotavirus infection, PKR functions at a stage between IFN gene transcription and subsequent IFN-β secretion. These findings reveal that activation of the antiviral response by rotavirus is dependent on MAVS/IPS-1 and IRF3 and involves both RIG-I and MDA-5 and that IFN-β secretion during rotavirus infection is regulated by PKR.
INTRODUCTION
Rotaviruses are etiological agents of severe dehydrating diarrhea in infants and young children and cause nearly 600,000 deaths globally every year (34). Rotaviruses are nonenveloped icosahedral members of the family Reoviridae and contain 11 segments of genomic double-stranded RNA (dsRNA) within a triple-layered particle. Several rotavirus vaccines have been developed to reduce rotavirus-associated mortalities (26); some are based on a modified Jennerian principle of rotavirus attenuation in the heterologous host. This phenomenon is termed host range restriction and is manifested by poor replication of rotaviruses in heterologous hosts compared to that in the homologous host (1). For example, bovine, lapine, and simian rotaviruses are all restricted for replication in humans. We are interested in understanding the mechanisms underlying host range restriction of rotaviruses in order to improve our basic knowledge of viral pathogenesis and because of the importance of these mechanisms in rational attenuation of vaccine candidates.
In primary mouse embryonic fibroblasts (MEFs), several heterologous (nonmurine) rotavirus strains, including the bovine UK strain, are replication restricted by the host type I interferon (IFN) response (17, 53). In contrast, replication of homologous murine EW virus and the heterologous simian rhesus rotavirus (RRV) strain is insensitive to the presence of the interferon system. In a mouse model of rotavirus infection, RRV replication in gallbladder epithelia correlates with its ability to suppress the IFN response (N. Feng et al., submitted for publication). The rotavirus gene segment encoding the nonstructural protein 1 (NSP1) is an important determinant of host restriction of viral replication both in vitro (17, 53) and in vivo (9). Specifically, we along with others have reported a role for NSP1 in regulating the ability of rotavirus to efficiently cause diarrhea in mice (9), spread from animal to animal (9), replicate in vivo and in vitro (4, 15–17), and antagonize IFN (4, 5, 17, 24, 25, 53). Thus, the host innate immune response is an important determinant of rotavirus host range restriction and depends on both the virus strain and host cell or tissue type.
Mammalian innate immunity to viral infection is critically dependent on a successful type I IFN response (49). The early IFN response involves initial recognition of virus within infected cells by activation of host pattern recognition receptors (PRRs) by pathogen-associated molecular patterns (PAMPs) such as viral RNA (60). The two major types of host PRRs that recognize viral RNA in the cytoplasm of nonplasmacytoid dendritic cells (non-pDCs) are the cytosolic retinoic acid-inducible gene-like receptors (RLRs) and membrane-bound Toll-like receptors (TLRs). Studies using knockout (KO) mice indicate that RLRs, but not the TLR system, are required for the antiviral IFN response in most cell types, including fibroblasts, epithelial cells, and conventional dendritic cells (40, 49, 54, 60, 61). RLRs respond to virus infection by signaling through the mitochondrial antiviral signaling adaptor (MAVS, also known as IPS-1/VISA/Cardif), which results in activation of host transcription factors, including interferon regulatory factor 3 (IRF3) and NF-κB, that orchestrate an early IFN-independent transcriptional program (40, 54, 61). Transcripts synthesized during this phase, including those encoding IFN-β and other antiviral proteins such as p54, p56, and p15, characterize the early antiviral state (49). Interferons, synthesized and secreted from infected cells, in turn direct amplification of the response and transcription of a broader range of antiviral genes in both an autocrine and paracrine manner, leading to establishment of the antiviral state (49). The two best characterized RLRs—RIG-I (retinoic acid-inducible gene I) and MDA-5 (melanoma differentiation-associated gene-5)—are cytoplasmic RNA helicases belonging to the DEXD/H box family, and each responds to distinct viral ligands (7, 31, 37–39, 44, 47, 51, 63). RNA viruses can be sensed by RIG-I (hepatitis C virus, Sendai virus, influenza virus, vesicular stomatitis virus, Newcastle disease virus, rabies virus, reovirus, and Japanese encephalitis virus), by MDA-5 (picornaviruses), or by both RIG-I and MDA-5 (dengue virus and West Nile virus) (7, 60). Other studies using purified RNA ligands, including viral genomic dsRNA (38), have demonstrated that RIG-I primarily recognizes short dsRNA and phosphate present as 5′-triphosphate moieties or monophosphates on the 5′ or 3′ ends of short dsRNA, whereas MDA-5 mainly recognizes longer dsRNA and higher-order structures formed by RNA (46).
With the exception of strains that contain rearranged genes resulting in truncated NSP1 proteins (3–5, 24, 29), nondefective rotavirus strains can generally negate the IFN response. At least two distinct mechanisms by which rotaviruses inhibit the host IFN response have been reported: (i) NSP1-mediated proteasomal degradation of host transcription factors IRF3 and IRF7, resulting in diminished IFN-β gene transcription and antiviral activity (4, 5), and (ii) proteasomal degradation of β-TrCP, leading to inhibition of NF-κB activity and subsequent IFN-β gene transcription (24). In contrast to these examples, we reported that the UK bovine rotavirus strain was unable to block IFN-β secretion in murine fibroblasts (17), and this correlated with the lack of murine IRF3 degradation by UK NSP1 (53). However, the defect in UK NSP1 function was specific to murine IRF3 and not to human or simian IRF3 proteins. UK was thus identified as a rotavirus strain that was unable to block IFN secretion via the activation of the early innate immune response in murine but not simian cells (17, 53). Although studies have addressed several mechanisms by which rotaviruses inhibit the IFN response, it is still not clear how rotaviruses that are host restricted are able to activate the host early innate immune response.
In this study, we found that rotaviruses activate the early innate transcriptional response and IFN-β secretion in an MAVS/IPS-1-dependent manner. Using MEFs derived from either RIG-I or MDA-5 knockout mice as well as double knockdowns induced by transient expression of small interfering RNA (siRNA), we found that neither RIG-I nor MDA-5 is essential for rotavirus-mediated activation but that both PRRs contributed to the IFN response. Using siRNA-induced inhibition, we found that TLR3 was dispensable for mediating the host response to UK rotavirus. In addition, our studies demonstrated that the double-stranded RNA-dependent protein kinase (PKR) is essential for IFN-β secretion from rotavirus-infected cells. However, PKR is not required for the early antiviral transcriptional response to the virus or for the transcription of IFN-α and IFN-β genes. In contrast, IRF3 is essential for both transcriptional and secretory aspects of the antiviral response. These findings reveal that rotaviruses trigger the host innate immune response by both MDA-5- and RIG-I-dependent signaling to MAVS/IPS-1 and IRF3, resulting in transcription of several early antiviral genes, including the IFN-β gene, and that PKR is essential for secretion of IFN-β from activated cells.
MATERIALS AND METHODS
Cells and viruses.
African green monkey kidney MA104 cells were purchased from the American Type Culture Collection (ATCC) and were maintained in Dulbecco's modified Eagle medium (DMEM; Cellgro) or M199 medium for MA104 cells containing 10% fetal calf serum (Invitrogen) supplemented with penicillin and streptomycin (complete DMEM). Mutant mouse embryonic fibroblasts used were described earlier: MAVS/IPS-1−/− (58), RIG-I−/− (Katherine Fitzgerald, University of Massachusetts) (37), MDA-5−/− (23), IRF3−/− (30), and PKR−/− (8). Control wild-type (WT) MEFs derived from an identical genetic background (harboring MAVS/IPS-1, MDA-5, and IRF3) or identical mouse strain (RIG-I and PKR) were used. Rotavirus strains RRV and UK were propagated in MA104 cells, and titers were determined plaque assay as described previously (32). Inactivated virus was prepared by incubation with 80 μg/ml psoralen (Sigma) and exposure to a UV light source (GBL-100C; G. B. Gate and Co.) on ice at a distance of 7.5 cm for 40 min. Inactivated virus preparations were not infectious as analyzed by plaque assay (data not shown).
Reagents and antibodies.
Poly(I:C) was purchased as a sodium salt (Sigma) and dissolved in water to obtain a 2.5 mg/ml stock solution. Short (0.2 to 1.0 kb) and long (1.5 to 8.0 kb) poly(I:C) preparations were purchased as formulations precomplexed with a transfection reagent [LyoVec-poly(I:C); Invitrogen]. Purified RRV dsRNA was prepared from concentrated virus stocks obtained by cesium chloride density gradient ultracentrifugation as described previously (15) using Trizol (Invitrogen) per the manufacturer's recommendations. Following chloroform extraction, RNA was precipitated with 4 M LiCl at 4°C, washed twice in 70% ethanol, and resuspended in RNase-free water. Commercial antibodies were obtained from the following suppliers: anti-IRF3 (FL-325 rabbit antibody from Santa Cruz Biotechnology), anti-tubulin (mouse monoclonal from Sigma), anti-p56 and anti-p54 (rabbit polyclonals from Thermo Scientific), anti-IRF7 (H-246 rabbit polyclonal from Santa Cruz Biotechnology), anti-VP6 (2B4 mouse monoclonal from Santa Cruz Biotechnology), and secondary horseradish peroxidase (HRP)-conjugated anti-mouse and anti-rabbit polyclonal antibodies (Amersham).
Virus infections and transfections.
Cells were plated in 6-well or 24-well cluster plates and infected when confluent after 18 to 36 h. Cells were washed three times with DMEM without additives, and virus was added at the multiplicity of infection (MOI) specified in the figure legends and adsorbed at 37°C for 1 h. Cells were then washed three times, and infection was allowed to proceed in DMEM lacking serum for the times indicated on the figures or in the legends. For quantifying infectious virus, 0.5 μg/ml trypsin (Sigma) was included in the DMEM, infected MEFs were collected 48 h later, and virus was released by three freeze-thaw cycles. Clarified supernatants were used for infection of MA104 cells following trypsin activation at 5 μg/ml for 1 h at 37°C. Virus titration was performed using a fluorescent focus-forming assay in MA104 cells as described previously (17). For transfection of poly(I:C) or purified rotavirus dsRNA, Lipofectamine 2000 (Invitrogen) was used at a ratio of 3:1 (vol/wt, Lipofectamine-RNA) as per the manufacturer's instructions. The final concentration of poly(I:C) or rotavirus dsRNA was 1 μg/ml in the cell culture supernatants. In experiments using short or long poly(I:C), LyoVec-poly(I:C) was directly added to culture medium at a final concentration of 1 μg/ml.
siRNA-mediated silencing.
MEFs were plated in 24-well cluster plates and 16 h later were transfected with siRNA at a final concentration of 50 nM using Lipofectamine 2000 (Invitrogen) following the manufacturer's instructions. Twenty-four hours later, cells were either infected with rotavirus or transfected with dsRNA as described above and then lysed and analyzed at the times indicated on the figures. Predesigned siRNA constructs targeting mouse TLR3, RIG-I, MDA-5, and a nonspecific control siRNA were purchased from Qiagen.
IFN measurement.
Cell culture supernatants were collected, and secreted mouse IFN-β was measured in duplicate using a Verikine mouse IFN-β enzyme-linked immunosorbent assay (ELISA) kit (PBL Interferon Source). Amounts of secreted IFN-β were calculated from a standard curve created using a mouse IFN-β standard as per the manufacturer's instructions.
qRT-PCR.
For quantitative reverse transcription-PCR (qRT-PCR) analyses, cells were lysed, and RNA was prepared using Trizol (Invitrogen) or the iScript RT-qPCR sample preparation reagent (Bio-Rad). Reverse transcription was carried out at 50°C for 1 h using Superscript III (Invitrogen), and the cDNA was analyzed using TaqMan assays in a real-time PCR instrument (Mx3005P; Agilent). Data were collected and analyzed by the comparative quantification method using the instrument software, Microsoft Excel spreadsheet software, and Prism statistical analysis software (GraphPad). TaqMan assays were obtained from Applied Biosystems and were as follows: IFN-stimulated gene 54 (ISG54; Mm00492606_m1), ISG56 (Mm00515153_m1), ISG15 (Mm01705338_s1), IRF7 (Mm00516788_m1), IFN-β (Mm00439546_s1), RIG-I (Mm00554529_m1), MDA-5 (Mm00459183_m1), A20 (Mm00437121_m1), IFN-α4 (Mm00833969_s1), IFN-α5 (Mm00833976_s1), GBP-2 (Mm00494575_m1), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Mm99999915_g1). The TaqMan assay for rotavirus gene 11 plus strand consists of the following primers and a probe labeled at the 5′ end with Cy5 and at the 3′ end with Iowa Black RQ-Sp from Integrated DNA Technologies: forward oligonucleotide, 5′-CTG CTT CAA ACG ATC CACTCA C-3′; reverse oligonucleotide, 5′-TGA ATC CAT AGA CAC GCC-3′; probe, 5′-TCA AAT GCA GTT AAG ACA AAT GCA GAC GCT-3′.
Cell lysis and immunoblotting.
Cells were washed twice in phosphate-buffered saline (pH 7.0) and were lysed in Laemmli buffer containing 2% SDS and 5% β-mercaptoethanol. Cell lysis was performed at room temperature for 20 min, and lysates were passed through a 25-gauge needle to reduce sample viscosity. Cell lysates were boiled for 5 min and briefly centrifuged, and approximately 10% of the total lysate was loaded on 12% SDS-PAGE gels. Following electrophoresis, proteins were transferred onto nitrocellulose membranes (Amersham Biosciences), and blots were probed using the antibodies indicated on the figures. Blots were exposed to autoradiography film (Amersham) and developed using an enhanced chemiluminescence kit from GE Healthcare. Blots were subsequently stripped and reprobed using a commercially available kit (Re-blot; Millipore).
RESULTS
Rotaviruses induce early antiviral transcriptional responses regardless of IFN-β secretion later in infection.
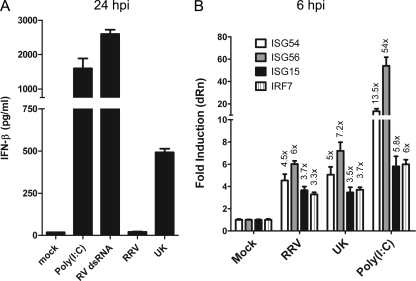
We previously showed that the bovine rotavirus UK induces IRF3 activation and subsequent secretion of IFN-β in primary MEFs, and this property was mapped to a host cell-specific dysfunction in the viral NSP1 protein (17, 53). In contrast, infection with a simian rotavirus RRV resulted in degradation of endogenous IRF3 and an effective blockage of IFN-β secretion. In agreement with these findings and as shown in Fig. 1A, we found that infection with UK led to significant IFN-β secretion at 24 h postinoculation (hpi) compared to RRV. Transfection of either of two RNA ligands—a commercial preparation of poly(I:C) or purified RRV dsRNA containing dsRNA segments ranging in length from ∼650 to 3,300 bp—was used as a positive control, and both controls induced a robust IFN response. Although we previously noted that infection with RRV does not lead to accumulation of detectable phospho-serine 396 IRF3 early in infection (6 hpi), as detected by immunoblot analysis (17), activation of the IFN response by UK suggested that RRV might trigger virus recognition pathways before significant levels of NSP1 are present in infected cells. In order to examine this possibility, we measured levels of antiviral transcripts, including ISG54, ISG56, ISG15, and IRF7, which are typically synthesized following virus recognition by cytosolic PRRs, using a sensitive TaqMan qRT-PCR method. As shown in Fig. 1B, infection with UK virus led to upregulation of each transcript examined (ISG54, ∼5.0-fold over mock; ISG15, ∼3.5-fold; IRF7, ∼3.7-fold; ISG56, ∼7.2-fold). Interestingly, infection with RRV also resulted in induction of these transcripts (ISG54, ∼4.5-fold; ISG15, ∼3.7-fold; IRF7, ∼3.3-fold; ISG56, ∼6.0-fold) at 6 hpi. The positive control, transfected poly(I:C), induced all measured transcripts. In other experiments (see Fig. 5E and 8B; also data not shown), IFN-β and ISG transcript levels in RRV-infected MEFs at 24 hpi were significantly downregulated compared to levels in UK-infected MEFs, supporting the conclusion that RRV blocks amplification of IFN responses despite its capacity to induce an early transcriptional response. Thus, although RRV is very efficient at blocking the secretion of IFN-β from MEFs, it activates early virus recognition pathways in MEFs similar to UK.
Fig. 1.
Rotavirus induces an early antiviral transcriptional response. (A) WT MEFs were infected with RRV or UK at an MOI of 3.0 or were transfected with 1 μg/ml of poly(I:C) or purified rotavirus dsRNA (RV dsRNA). Twenty-four hours later, culture supernatants were assayed for the presence of IFN-β by ELISA. Data are shown as average of duplicate measurements with standard error bars. (B) WT MEFs were infected with RRV or UK or transfected with poly(I:C) (long or short, indicated as L-pIC or S-pIC, respectively), and total RNA was purified from cells at 6 hpi for qRT-PCR using TaqMan assays for the transcripts indicated. Data are presented after normalization to internal GAPDH levels as fold increase over mock controls, and bars represent the standard errors among triplicate measurements. Data are representative of two independent experiments. dRn, delta normalized reporter.
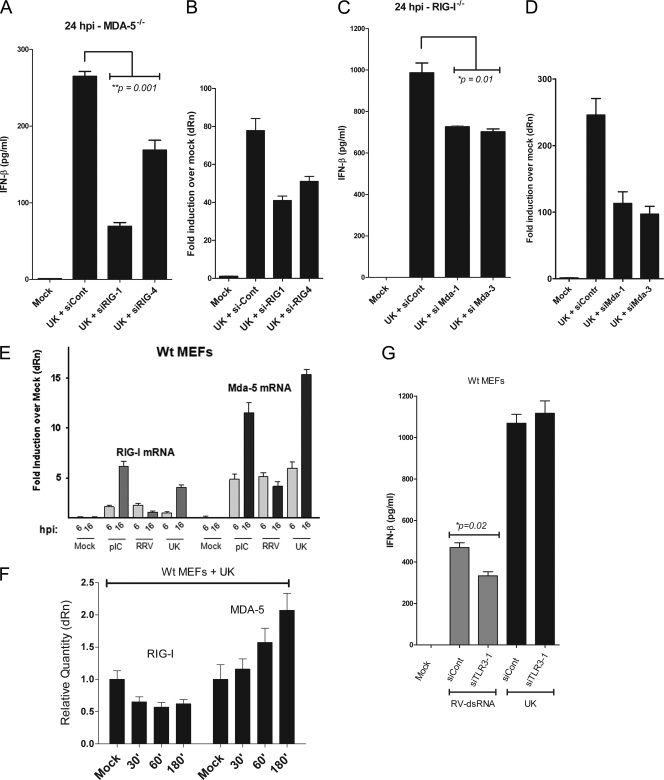
Fig. 5.
RIG-I and MDA-5 both respond to rotavirus infection. (A) MDA-5−/− MEFs were infected with UK virus 24 h after transfection with siRNA to RIG-1 (siRIG-1 and siRIG-4) or a control siRNA (siCont). Secreted IFN-β was measured at 16 hpi. (B) Cells were lysed, and qRT-PCR was used to determine the levels of RIG-I transcripts. (C) RIG-I−/− cells were transfected with siRNA targeting MDA-5 (siMDA-1 and siMDA-3). Secreted IFN-β was measured at 16 hpi. (D) Cells were lysed, and qRT-PCR was used to determine the levels of MDA-5 transcripts. (E) WT MEFs were infected with RRV and UK virus or were transfected with poly(I:C), and cells were lysed at 6 hpi and 16 hpi, and levels of RIG-I and MDA-5 transcripts were determined by qRT-PCR. (F) Levels of RIG-I and MDA-5 transcripts were measured as a function of time (in minutes; x axis) in UK-infected WT MEFs. (G) WT MEFs were transfected with siRNA targeting TLR3 (siTLR3-1) or a control siRNA (siCont) and 24 h later were infected with UK or transfected with purified rotavirus dsRNA. Levels of secreted IFN-β were measured. Statistical significance was calculated using a Student's t test. Data are representative of two independent experiments.
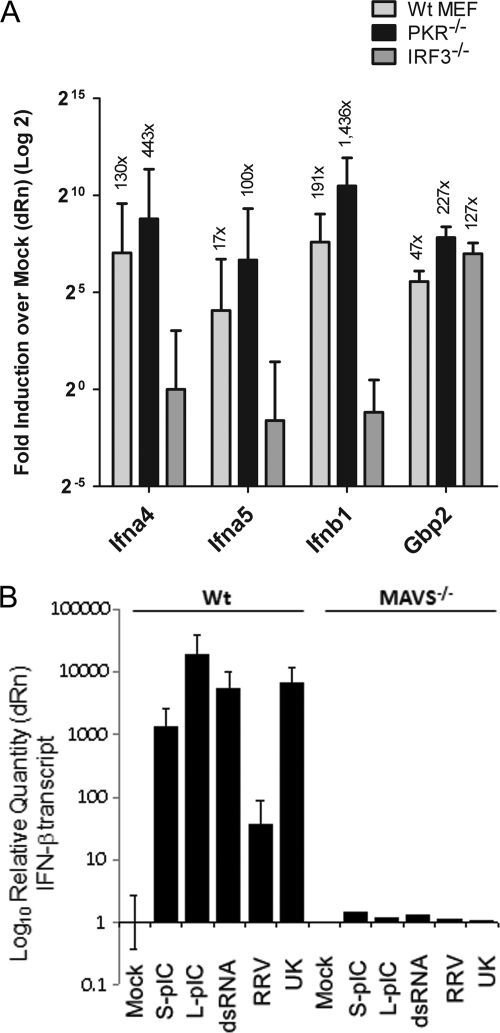
Fig. 8.
Levels of ISG transcripts in the absence of PKR, IRF3, and MAVS/IPS-1 in response to rotavirus infection. (A) WT, PKR−/−, or IRF3−/− MEFs were infected with UK, and 16 h later RNA was purified for qRT-PCR analysis of IFN-α4, IFN-α5, IFN-β1, and GBP-2 gene transcripts. Data are presented on a 2-log scale as fold induction over levels of mock-infected controls, and actual fold values are indicated above the respective bars. (B) WT or MAVS/IPS-1−/− MEFs were transfected with the ligands indicated or infected with RRV and UK, and IFN-β transcript levels were measured at 24 hpi by qRT-PCR. Data are representative of two independent experiments.
Rotavirus-induced IFN-β secretory response and early antiviral transcription are both MAVS/IPS-1 dependent.
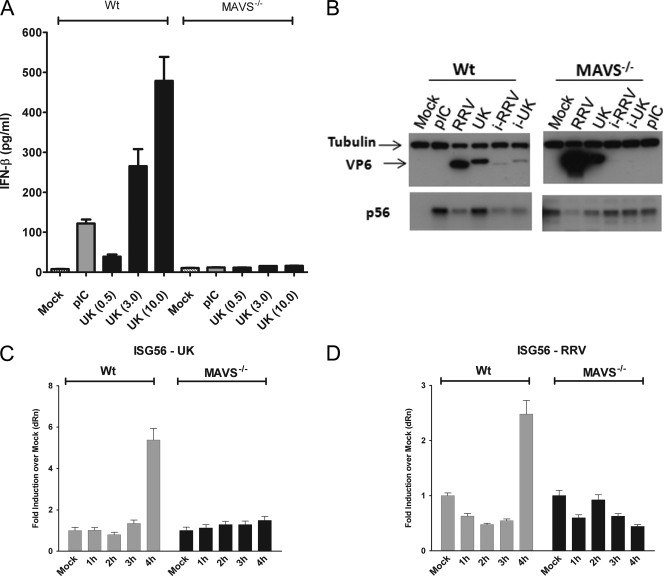
Most natural (i.e., nondefective) rotavirus strains studied thus far successfully negate the IFN response in selected cell types by mechanisms involving proteasomal degradation of IRFs and β-TrCP (5, 24). In contrast, UK robustly induces the IFN response in a host cell-specific manner in MEFs (17), providing us with a tractable system to determine the mechanisms involved in activation of early innate immune responses by rotavirus. We first examined the role of MAVS/IPS-1, a critical adaptor for host recognition of cytosolic RNA viruses, using MEFs lacking MAVS/IPS-1. Wild-type (WT) or MAVS/IPS-1−/− MEFs were infected with UK virus at MOIs of 0.5, 3.0, or 10.0, and IFN-β levels were measured in the culture supernatants at 16 hpi (Fig. 2A). In contrast to dose-dependent induction of IFN-β secretion by UK in WT MEFs, there was a profound loss of IFN-β secretion from MAVS/IPS-1−/− MEFs in response to UK infection. These results reveal an essential role for MAVS/IPS-1 in IFN secretion in response to UK rotavirus infection. In order to further confirm the loss of IFN response in the absence of MAVS/IPS-1, we infected WT or MAVS/IPS-1−/− MEFs with UK, RRV, or psoralen/UV-inactivated UK and RRV or transfected cells with 1 μg/ml of poly(I:C). Cells were lysed at 24 hpi and analyzed by immunoblotting for the presence of p56 and the VP6 viral antigen. As shown in Fig. 2B, p56 was not detected in uninfected WT fibroblasts, but its expression was strongly induced following infection with UK virus or with the positive control, poly(I:C). In contrast, RRV, and to an even lesser extent inactivated UK virus and RRV, only weakly induced p56 expression. The lack of p56 expression by inactivated UK demonstrates that induction of IFN response is replication dependent, as observed by us earlier (17). In the absence of MAVS, we detected low levels of basal ISG expression similar to previous studies (44) although to our knowledge the mechanism of elevated basal p56 expression is not understood. However, in this case, neither UK virus infection nor poly(I:C) treatment resulted in further increase in p56 expression. These data demonstrate a critical role for MAVS/IPS-1 in an effective IFN response to rotavirus in MEFs. In order to examine whether MAVS/IPS-1 is important for the induction of early antiviral transcripts in response to rotavirus, we infected WT or MAVS/IPS-1−/− MEFs with UK virus or RRV, purified total RNA from cells at different times after infection (1 to 4 hpi), and measured levels of ISG56 transcripts using qRT-PCR. Both UK and RRV induced ISG56 transcription between 3 and 4 hpi in WT fibroblasts, and this activation was dependent on MAVS (Fig. 2C and D). Similar results were also obtained at later time points and for the ISG54 transcript (data not shown), demonstrating that in the absence of MAVS/IPS-1, there is a loss of the early transcriptional response to rotavirus.
Fig. 2.
Innate immune response to rotavirus is MAVS/IPS-1 dependent. (A) WT or MAVS/IPS-1−/− MEFs were infected with UK at MOIs of 0.5, 3.0, or 10.0, and IFN-β secretion was measured by ELISA at 16 hpi. (B) WT or MAVS/IPS-1−/− MEFs were transfected with poly(I:C) or infected with RRV, UK, or psoralen/UV-inactivated viruses (i-RRV and i-UK). Cell lysates were examined at 24 hpi for levels of tubulin, VP6 antigen, or p56 expression by immunoblotting. (C) WT or MAVS/IPS-1−/− cells were infected with UK at an MOI of 3.0, and ISG56 transcript levels were determined by qRT-PCR. The data are presented as fold increase over levels of mock-infected 1-hpi cells after normalization to GAPDH for triplicate measurements. (D) WT or MAVS/IPS-1−/− cells were infected with RRV, and data were collected as described in panel C. Data are representative of two or more independent experiments. pIC, poly(I:C).
Rotavirus replication is restricted in a MAVS/IPS-1-dependent manner.
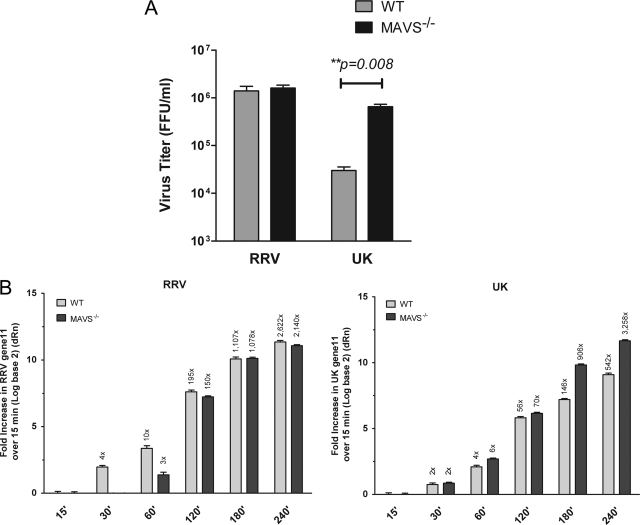
We previously reported that in MEFs the replication of most heterologous rotaviruses, including the bovine UK strain, is restricted compared to simian RRV and that this restriction can be significantly rescued in the absence of a functional type I IFN response (17). Since our data revealed that MAVS/IPS-1 is critical for the IFN response to UK, we next examined whether UK replication could be rescued in the MAVS/IPS-1 knockout cells. Wild-type or MAVS-deficient MEFs were infected with RRV or UK virus at an MOI of 1.0, and at 48 hpi, infectious virus produced was quantified by a fluorescent focus-forming assay on fresh monolayers of MA104 cells. As shown in Fig. 3A, UK, but not RRV, displayed more than a 10-fold increase in titer in MAVS/IPS-1−/− compared to WT fibroblasts (3.0 × 104 versus 6.5 × 105 FFU per ml, respectively). Thus, UK replication in heterologous murine cells in the absence of MAVS is similar to the previously observed effect of ablation of the type I IFN response (17).
Fig. 3.
Replication of UK virus is rescued in MAVS/IPS-1−/− MEFs early during infection. (A) WT or MAVS/IPS-1−/− MEFs were infected with RRV or UK at an MOI of 1.0. After 48 h, cells were freeze-thawed, and titers of the infectious virus released were determined on MA104 cells using a fluorescent focus-forming assay. Errors are from triplicate measurements. Statistical significance was determined using a two-way analysis of variance. (B) WT or MAVS/IPS-1−/− MEFs were infected with RRV or UK, and the viral gene 11 transcript was quantified by qRT-PCR at the times indicated. Data are given as fold increase compared to a 15-min postinfection control. The actual mean fold increase is indicated above each bar. Data are representative of two independent experiments. FFU, focus-forming units.
In order to measure differences in virus replication at early times in infection that more accurately corresponded to MAVS/IPS-1 dependent signaling, we used a TaqMan assay capable of detecting the gene 11 mRNA from both RRV and UK virus. Wild-type or MAVS/IPS-1−/− cells were infected with identical amounts of RRV or UK, and cells were lysed at various times following 1 h of virus adsorption. Compared to the signal detected at 15 min following virus adsorption (presumably mostly input viral genome and used as a calibrator across all samples), there was a time-dependent increase in gene 11 levels for both RRV and UK virus in WT MEFs (Fig. 3B). The increases observed upon UK virus infection were significantly higher in the MAVS/IPS-1−/− than WT MEFs (P < 0.0001 at 180 and 240 min). For example, the increases at 180 and 240 min postinfection were 146- and 542-fold in WT MEFs, respectively, and 906- and 3,258-fold in MAVS/IPS-1−/− MEFs, respectively. In contrast, there was no significant increase at any time point in gene 11 RNA levels between MAVS/IPS-1−/− and WT MEFs infected with RRV (Fig. 3B). These results are in agreement with the infectious virus titer data (Fig. 3A) and provide us a quantitative measure of replication for early times following rotavirus infection. Of note, the results demonstrate that the MAVS-dependent restriction of UK occurs during early virus replication and is likely related to MAVS-dependent virus recognition.
The cytosolic sensors MDA-5 and RIG-I are individually dispensable for the IFN response to rotavirus.
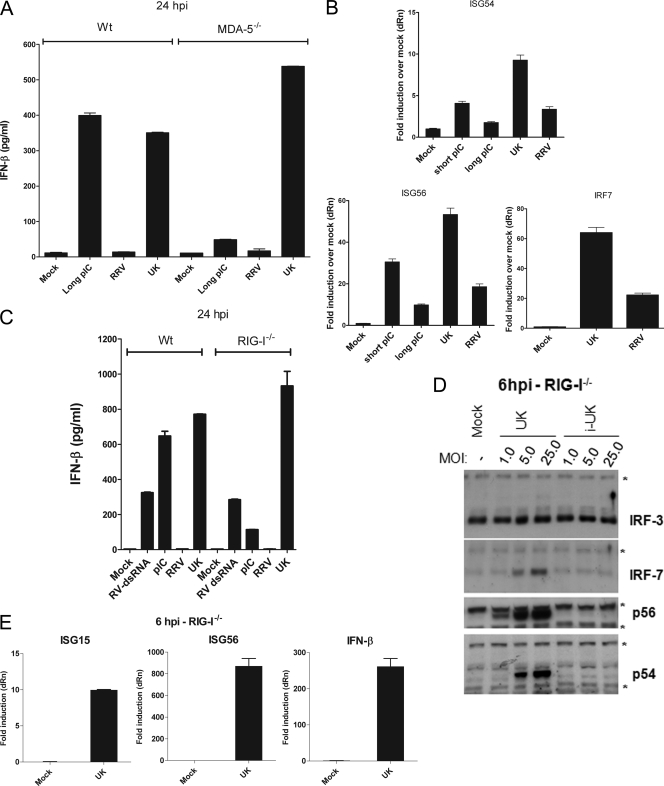
The MAVS/IPS-1-mediated IFN response to RNA viruses in the cytoplasm occurs through two upstream PRRs, MDA-5 and RIG-I. In order to determine the role of MDA-5 in the UK virus-mediated IFN response, WT or MDA-5−/− MEFs were infected with UK or transfected with a commercial preparation of high-molecular-weight poly(I:C), an MDA-5 ligand, as a control, and IFN-β secretion was measured at 24 hpi. As shown in Fig. 4A, infection of either WT or MDA-5−/− MEFs with UK virus resulted in comparable amounts of IFN-β secretion at 24 hpi. In contrast, secretion of IFN-β in response to long poly(I:C) was intact in WT cells but severely attenuated in the absence of MDA-5. Thus, the absence of MDA-5 does not mimic the complete lack of IFN-β secretion in UK virus-infected cells observed in the absence of MAVS/IPS-1 (Fig. 2). The presence of an intact antiviral response was further confirmed by measuring levels of ISG54, ISG56, and IRF7 transcripts in MDA-5−/− MEFs at 6 h following UK infection or stimulation with long- and short-poly(I:C) preparations (Fig. 4B). The transcription of ISG54 and ISG56 was significantly enhanced by UK infection at 6 hpi as well as by the short poly(IC) positive control. In these experiments, long poly(I:C) was a less effective trigger than short poly(I:C), supporting data from earlier reports that these ligands preferentially activate MDA-5 and RIG-I, respectively. UK infection also led to a significant increase in IRF7 transcript levels, indicating that intact IRF3-dependent signaling occurs in UK-infected MDA-5−/− MEFs. From these findings, we conclude that in the absence of MDA-5, UK likely triggers IRF3 activation, resulting in ISG transcription and the subsequent secretion of IFN-β.
Fig. 4.
The IFN response to rotavirus proceeds in the absence of RIG-I or MDA-5. (A) WT or MDA-5−/− MEFs were infected with RRV or UK or transfected with long poly(I:C), and secreted IFN-β was measured 24 h later by ELISA. (B) MDA-5−/− MEFs were infected with RRV or UK or were transfected with the short or long poly(I:C). RNA was prepared at 6 hpi, and levels of ISG54, ISG56, and IRF7 were quantified by qRT-PCR. Data are presented as fold increase compared to levels of mock controls, and errors are for triplicate measurements after GAPDH normalization. (C) WT or RIG-I−/− MEFs were infected with RRV or UK or transfected with rotavirus dsRNA or poly(I:C). Secreted IFN-β was measured 24 h later by ELISA. (D) WT or RIG-I−/− MEFs were infected with UK or inactivated UK (i-UK) at the MOIs indicated. At 6 hpi, cells were lysed for immunoblot analysis of IRF3, IRF7, p56, and p54. The asterisk indicates nonspecific background bands. (E) RIG-I−/− MEFs were infected with UK, and levels of ISG15, ISG56, and IFN-β were determined at 6 hpi by qRT-PCR. Data are representative of two independent experiments.
Next, we examined the role of RIG-I by infecting RIG-I−/− MEFs with UK or RRV virus or transfecting cells with either short poly(I:C) or purified rotavirus dsRNA containing a mixture of dsRNAs of different lengths. After 24 h, levels of IFN-β were measured. We found that in the absence of RIG-I, UK virus induces the secretion of IFN-β to levels comparable to those of WT MEFs (Fig. 4C). In contrast, the response of RIG-I−/− MEFs to short poly(I:C) was significantly reduced (P = 0.002). There was no significant difference in secretion of IFN-β between RIG-I−/− MEFs and WT MEFs treated with rotavirus dsRNA; presumably rotavirus dsRNA activates non-RIG-I pathways (38). These results demonstrate that UK is able to trigger IFN responses in the absence of RIG-I and in cells lacking MDA-5.
We next examined IRF3, IRF7, p56, and p54 protein levels in RIG-I−/− MEFs infected with UK. Cells were infected with UK at MOIs of 1, 5, and 25 or with a control inactivated UK preparation. Cell lysates prepared at 6 hpi had dose-dependent and replication-dependent increases in levels of IRF7, p56, and p54 proteins (Fig. 4D). There was no IRF3 degradation, similar to what was reported previously (53). Thus, the replication-dependent induction of early antiviral responses by UK is intact in RIG-I−/− MEFs. We then measured levels of ISG15, ISG56, and IFN-β transcripts 6 h after UK infection (Fig. 4E). Mirroring the protein expression data, there was a substantial increase in the abundance of transcripts dependent on IRF3 (ISG15 and IFN-β) and on both IRF3 and NF-κB (ISG56). Collectively, the data suggest that RIG-I is dispensable for UK-mediated early IRF3 activity, transcriptional responses, and IFN-β secretion in MEFs. From these experiments, we conclude that, unlike the profound effect of the MAVS knockout, neither MDA-5 nor RIG-I is essential for the early innate antiviral transcriptional response or IFN-β secretion following rotavirus infection.
Combined deficiency of RIG-I and MDA-5 results in attenuation of the rotavirus-induced IFN response.
Although neither RIG-I nor MDA-5 is essential for mounting an effective IFN response to rotavirus infection, these RLRs could act cooperatively or redundantly to induce a response to rotavirus, a phenomenon observed during infection with other viruses (19, 35, 58). In order to address this possibility, we used an RNA interference approach to inhibit expression of one RLR in the reciprocal knockout cell. First, RIG-I was depleted using siRNA in MDA-5−/− MEFs. Cells were transfected with one of two different siRNAs targeted to the RIG-I mRNA or a control scrambled siRNA. Cells were infected after 24 h with UK, and IFN-β was measured 24 h later. We found that compared to the control siRNA, both RIG-I siRNA constructs resulted in significantly decreased IFN-β secretion (36 to 74% decrease; P = 0.001) in the supernatants in response to UK infection (Fig. 5A, lanes 3 to 4). The cells were lysed and examined by qRT-PCR for transcripts encoding RIG-I in order to assess the efficiency of mRNA silencing (Fig. 5B). By this approach there was an approximately 30 to 50% reduction in RIG-I mRNA levels compared to control siRNA-treated cells. Thus, in the absence of MDA-5, the depletion of RIG-I led to a significant decrease in the IFN response to UK infection. This indicated that, although not essential, RIG-I is involved in the recognition of rotavirus during infection.
In order to determine the role of MDA-5 in a RIG-I-deficient background, we used siRNA to inhibit MDA-5 expression in RIG-I-deficient cells. As shown in Fig. 5C, UK infection of RIG-I−/− MEFs transfected with MDA-5-specific siRNA, but not control siRNA, resulted in a modest but significant decrease (∼26 to 30% decrease; P = 0.01) in IFN-β secretion relative to untreated cells at 24 hpi. We confirmed that the siRNA treatment resulted in a significant decrease (greater than 50%) in relative MDA-5 mRNA levels by qRT-PCR (Fig. 5D).
MDA-5 siRNA treatment of RIG-I−/− MEFs did not lead to as substantial a reduction in IFN-β levels as RIG-I siRNA treatment of MDA-5−/− MEFs. One possible explanation is that following rotavirus infection, MDA-5 transcription is amplified more efficiently than RIG-I transcription as part of a positive feedback loop, and consequently the efficiency of MDA-5 mRNA depletion was lower. In order to examine this possibility, we infected WT MEFs with RRV or UK virus or transfected the cells with poly(I:C). Cells were lysed at 6 hpi and 16 hpi, and qRT-PCR was used to determine levels of RIG-I and MDA-5 transcripts. Following infection with UK, there was a significant increase in MDA-5 transcripts at 6 hpi, and levels increased further by 16 hpi relative to levels in uninfected cells (Fig. 5E). Interestingly, although RRV triggered increased MDA-5 transcription comparable to that induced by UK at 6 hpi, there was no further increase at 16 hpi. A similar pattern of transcriptional activation was observed for RIG-I transcripts following UK and RRV infections although the increase was much less than that observed for MDA-5. These results suggest that MDA-5 is more efficiently amplified following UK infection than RIG-I. This conclusion was confirmed using WT MEFs infected with UK and lysed at time points ranging from 30 min to 3 h postinfection (Fig. 5F). During this early time frame, UK induced MDA-5, but not RIG-I, transcription in a time-dependent manner, confirming that MDA-5 is induced earlier and more effectively by UK than RIG-I. This robust amplification of MDA-5 transcription provides a likely explanation for the weaker effect of the siRNA-mediated inhibition of MDA-5 (Fig. 5C) in comparison to the siRNA-mediated RIG-I inhibition (Fig. 5A).
Of note, our experiments do not exclude the existence of additional sensors proximal to MAVS/IPS-1 in the rotavirus-induced IFN response. In order to partially address this possibility, we treated MEFs with a TLR3-specific siRNA. Wild-type MEFs were transfected with siRNA targeting TLR3 or a control and were subsequently either transfected with purified rotavirus dsRNA or infected with UK virus. Twenty-four hours later, culture supernatants were assayed for levels of IFN-β. Treatment with siRNA resulted in a slight but significant (∼30%; P = 0.02) decrease in IFN-β levels in response to purified dsRNA but no significant decrease in response to UK infection (Fig. 5G). Thus, although preliminary and requiring further study, these results are consistent with the conclusion that TLR3 is unlikely to play a major role in mediating IFN-β secretion following infection of MEFs with UK. This conclusion is also supported by the complete abolition of IFN-β secretion in the MAVS KO mice. Taken together, the data obtained from these experiments demonstrate that depletion of both MDA-5 and RIG-I leads to decreased IFN responses to rotavirus and establish redundant roles for these PRRs during rotavirus infection.
The dsRNA-dependent protein kinase, PKR, is essential for IFN secretion but does not regulate the early antiviral transcriptional response to UK.
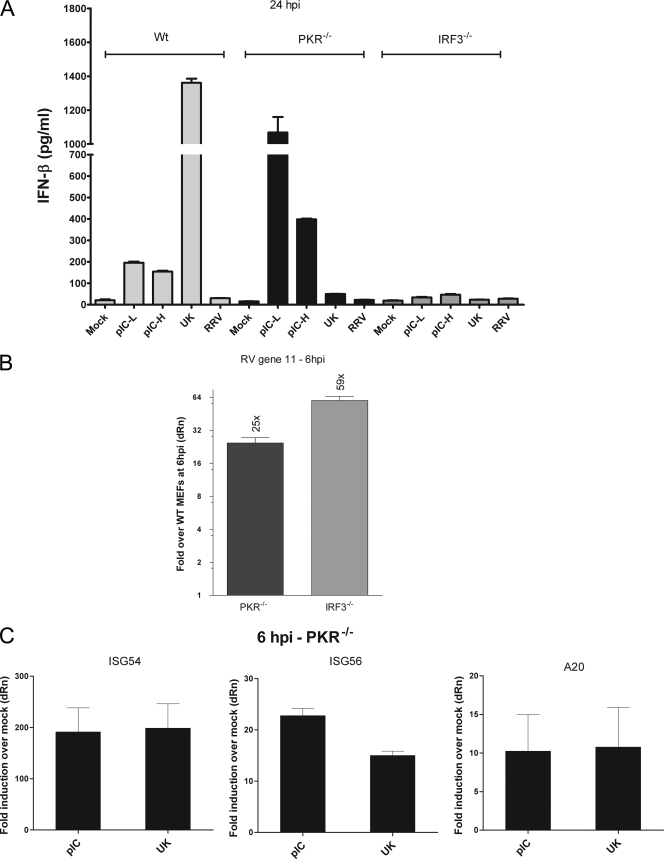
Recently, several studies have demonstrated a critical role for PKR in the IFN response to RNA viruses that are recognized by MDA-5 but not by RIG-I (6, 11, 22, 52). The role of PKR in the response to viruses that activate both MDA-5 and RIG-I and whether PKR acts as a sensor upstream of IRF3 have not been determined. The roles of PKR and IRF3 in the early innate immune response to rotavirus have not been directly studied. Since our results revealed a role for both RIG-I and MDA-5 in rotavirus recognition, we compared the ability of PKR−/− and IRF3−/− MEFs to respond to infection with UK. MEFs were infected with UK virus or transfected with short or long poly(I:C). IFN-β secretion was measured by ELISA 24 h later (Fig. 6A). Unexpectedly, we found that IFN-β secretion in response to UK was severely abrogated in PKR−/− as well as IRF3−/− MEFs (Fig. 6A, lanes 4 and 9). In contrast, poly(I:C)-induced IFN-β secretion was dependent on IRF3 but not PKR. These results indicate a novel essential role for PKR in the innate immune response to UK rotavirus and provide experimental evidence for an essential role of IRF3 in this process. In order to confirm these results, we compared the replication of UK in PKR−/− and IRF3−/− MEFs by evaluation of gene 11 levels. At 6 hpi, gene 11 RNA levels were significantly higher in PKR−/− MEFs (∼25-fold) than in WT MEFs although there was a more substantial increase (∼60-fold) in IRF3−/− MEFs (Fig. 6B). These data confirm the roles for both PKR and IRF3 in the UK virus antiviral response and replication restriction.
Fig. 6.
Role of PKR and IRF3 in the rotavirus-mediated IFN response. (A) WT, PKR−/−, or IRF3−/− MEFs were infected with UK and RRV or were transfected with short or long poly(I:C). Levels of secreted IFN-β were measured at 24 hpi. pIC-L and pIC-H refer to the low- and high-molecular-weight forms of poly (I:C), respectively. (B) WT, PKR−/−, or IRF3−/− MEFs were infected with UK. At 6 hpi, total RNA was analyzed by qRT-PCR to quantitate gene 11 transcription. Data are presented after GAPDH normalization as fold increase over levels in WT MEFs on a 2-log scale, and errors are for triplicate measurements. (C) PKR−/− MEFs were infected or transfected as indicated, and levels of ISG56 were quantified by qRT-PCR at 6 hpi. Data are presented as fold increase over levels of mock-infected controls. Data are representative of three independent experiments with similar results.
In order to determine whether PKR acts as an initial PRR for rotavirus leading to the transcription of IRF3- or NF-κB-dependent genes, we examined levels of ISG54, ISG56, and A20 transcripts in UK virus-infected PKR-deficient fibroblasts. In contrast to our earlier results obtained using MAVS/IPS-1−/− MEFs (Fig. 2C), infected PKR−/− cells responded effectively to UK infection, as evidenced by significant upregulation of the ISGs examined (ISG54, ∼200-fold relative to uninfected cells; ISG56, ∼15-fold; A20, ∼11-fold) (Fig. 6C); this upregulation was similar in magnitude to the response of PKR−/− MEFs to poly(I:C). Despite the critical role of PKR in IFN-β secretion, transcription activated by IRF3 (ISG54 and ISG56) and NF-κB (ISG56 and A20) in response to rotavirus was not significantly different from that of WT cells, indicating that PKR is unlikely to act as an initial virus sensor directing IRF3/NF-κB activation.
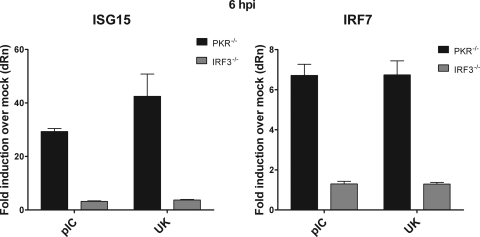
To confirm these findings, we directly compared the ability of UK to induce the transcription of the IRF3-dependent genes ISG15 and IRF7 in IRF3−/− and PKR−/− MEFs. At 6 hpi, transcription of ISG15 and IRF7 in response to UK was not observed in IRF3−/− cells (Fig. 7). In contrast, UK-infected PKR−/− MEFs increased ISG15 mRNA levels by ∼42-fold and IRF7 levels by ∼7-fold over mock-infected controls (Fig. 7). Thus, the severe loss of an interferon response to rotavirus in PKR−/− MEFs is unlikely to be a result of IRF3 dysfunction. Instead, our data indicate that PKR functions independently of IRF3-mediated early transcription to promote IFN secretion from infected cells.
Fig. 7.
Comparison of ISG15 and IRF7 transcriptional activation in PKR−/− and IRF3−/− MEFs. PKR- or IRF3-deficient MEFs were infected with UK or transfected with poly(I:C). At 6 hpi, RNA was prepared, and levels of ISG15 and IRF7 transcripts were quantitated by qRT-PCR. Data are presented as fold induction over levels of mock-infected controls, and error bars are from triplicate measurements. Results are representative of two independent experiments with similar results.
PKR acts downstream of the transcription of type I IFN genes during rotavirus infection.
In order to examine whether the defect in IFN-β secretion in PKR-deficient MEFs was specifically due to a lack of transcription of a subset of type I IFN genes, we compared the accumulation of transcripts of IFN-α4, IFN-α5, and IFN-β genes in WT, IRF3-deficient, and PKR-deficient MEFs at 16 hpi. As shown in Fig. 8A, infection of WT fibroblasts with UK resulted in significant induction of IFN transcripts compared to mock-infected cells (IFN-α4, ∼130-fold; IFN-α5, ∼17-fold; IFN-β, ∼191-fold). As expected, this induction was dependent on IRF3 and was abrogated in IRF3−/− MEFs. Interestingly, UK-infected PKR−/− MEFs also expressed high levels of the IFN transcripts (IFN-α4, ∼443-fold relative to mock-infected controls; IFN-α5, ∼100-fold; IFN-β, ∼1,436-fold). GBP-2, which is transcriptionally upregulated during rotavirus infection (33), was induced by UK virus in all three types of MEFs (∼47-fold, ∼227-fold, and ∼127-fold in WT, PKR−/−, and IRF3−/− MEFs, respectively), as shown in Fig. 8A. A similar analysis for the presence of IFN-β transcripts in WT and MAVS/IPS-1-deficient MEFs (Fig. 8B) revealed that there was a profound defect in transcription of IFN-β in MAVS/IPS-1−/− MEFs compared to WT cells during infection. These findings demonstrate that PKR acts at a stage following transcription of IFN genes during the IFN response to rotavirus.
DISCUSSION
In most mammalian nonimmune cell lineages, the IFN response to RNA viruses depends on detection of viral RNA by the cytosolic host PRRs, RIG-I and MDA-5. The host PRRs recognize specific viral PAMPs and propagate virus-induced signaling through MAVS/IPS-1 (also called Cardif or VISA), an adaptor found on the outer mitochondrial membrane and in peroxisomes (37, 40, 45, 54, 61, 63, 65). Mechanisms by which rotavirus initially activates host innate immune responses are unclear since most WT RV strains reported to date efficiently suppress IFN responses. Previous studies examining rotavirus activation of innate immunity have employed purified viral genomic dsRNA (38) or defective rotavirus strains encoding truncated NSP1 proteins (29). We found that the RV strains UK and RRV exert opposing effects on IFN-β secretion in primary MEFs (Fig. 1A) (17) as well as in murine alveolar epithelial LA4 and murine cholangiocyte cells (data not shown). Infection of these murine cells with UK resulted in robust IFN-β secretion, providing us with a WT rotavirus strain that is unable to negate the host IFN response. Interestingly, despite differences in their ability to induce IFN-β secretion late in infection, both RRV and UK virus triggered expression of antiviral transcripts early in infection (6 hpi) that are characteristic of PRR activation by virus, including genes requiring functional IRF3 (ISG54, ISG15, and IRF7) and/or NF-κB (ISG56) for their transcription. This activation of transcriptional responses indicated that both RRV and UK triggered host cell sensors similarly leading to early activation of IRF3 and NF-κB signaling. Presumably, synthesis of NSP1 following this initial activation influences subsequent amplification of the response and IFN-β secretion. RRV NSP1 targets murine IRF3 for proteasomal degradation much more efficiently when IRF3 is activated extrinsically with poly(I:C), and a phospho-mimetic mutant resembling activated IRF3 (IRF3-5D) is functionally inhibited by RRV NSP1 (53). Thus, virus-induced early IRF3 activation observed here may, in fact, confer an advantage to RRV by leading to more efficient blockade by NSP1 during later stages of infection. Since UK virus infection activates both the early (transcriptional) and later (IFN-β secretion) phases of the antiviral response, we examined the role of various host factors in UK-directed IFN responses using MEFs derived from various strains of knockout mice.
We observed that MAVS/IPS-1-deficient MEFs do not secrete IFN-β or induce p56 protein following UK infection and that UK virus titers are higher in MAVS/IPS-1−/− MEFs than in WT MEFs. These data reveal an essential role for MAVS-dependent signaling in the IFN response to rotavirus. Using a sensitive approach to quantify viral RNA at early times during infection, we found that the MAVS/IPS-1-dependent restriction of UK virus replication is evident as early as 2 to 3 h after infection. In addition, the ISG56 transcriptional response that occurs at 3 to 4 h after infection in WT cells was not observed in MAVS-deficient MEFs. The timing of host transcriptional activation of ISG54 and ISG56 genes in infected WT MEFs and the finding that the early activation of IRF3 by UK requires viral replication (17, 53) suggest that the MAVS/IPS-1-dependent rotavirus recognition is mediated by an early product of viral replication.
The critical function of MAVS in the IFN-β induction pathway for rotaviruses prompted us to examine the role of the cytosolic RNA sensors RIG-I and MDA-5 using MEFs derived from knockout mice. UK infection of either RIG-I−/− or MDA-5−/− MEFs resulted in the secretion of IFN-β despite reduced IFN-β responses in these knockout cells to control ligands. In addition, the expression levels of IRF3- and NF-κB-dependent genes in RIG-I−/− or MDA-5−/− MEFs following UK virus infection were similar to levels observed in WT cells, suggesting a potential redundancy in the functions of RIG-I and MDA-5 during rotavirus infection.
Similar findings have been obtained for other viruses, including dengue virus and West Nile virus, which evoke MAVS/IPS-1 signaling but lack an absolute requirement for either RIG-I or MDA-5 to potentiate an IFN response (19, 22, 35, 44, 52). Notably, for such viruses, RIG-I and MDA-5 function redundantly to generate a response. Using RNA interference to deplete RIG-I in MDA-5−/− MEFs, we found that the combined loss of both PRRs resulted in significantly reduced IFN-β secretion, demonstrating that both RIG-I and MDA-5 are involved in the IFN response to UK. In comparison to the suppression of IFN secretion in MDA-5−/− cells by RIG-I siRNAs, the complementary approach of siRNA-mediated depletion of MDA-5 in RIG-I−/− MEFs resulted in a smaller reduction in virus-induced IFN-β secretion. The different efficiencies of RIG-I and MDA-5 knockdowns are likely attributable to differences in UK-mediated feedback amplification of RIG-I and MDA-5 transcripts since in WT MEFs UK infection amplified MDA-5 transcripts more efficiently than RIG-I transcripts. Interestingly, both RRV and UK induced comparable increases in MDA-5 transcript at 6 hpi. RRV was unique in blocking subsequent amplification of this process between 6 and 16 hpi, and this blockade correlated with its capacity to suppress the IFN response. The induction of MDA-5 transcript by UK was more efficient than induction of RIG-I and occurred earlier, indicating that MDA-5 and RIG-I may be activated by rotavirus in a temporally distinct manner.
A previous study using human intestinal HT29 cells showed that RIG-I knockdown by siRNA results in reduction of IFN-β secretion and increases in levels of replication after infection with simian rotavirus strain SA11-5S, a strain that encodes a truncated NSP1 protein due to a genetic lesion (13). Although the role of MDA-5 in rotavirus recognition was not examined in that study, our results support the involvement of both MDA-5 and RIG-I in rotavirus recognition in a MAVS/IPS-1-dependent pathway. Given the critical nature of MAVS/IPS-1 and the lack of a detectable decrease in IFN-β secretion following treatment with TLR3 siRNA, rotavirus sensors in fibroblasts are likely to be MAVS/IPS-1 dependent rather than TLR mediated. Despite the clear role of RIG-I and MDA-5 in the IFN response to UK, our experiments do not exclude involvement of additional sensors dependent on MAVS/IPS-1.
The rotavirus NSP1 protein interacts with IRF3 and directs its proteasomal degradation by an unknown mechanism (4, 5, 25). We previously found that IRF3 is the minimal host factor involved in the species-specific degradation of IRF3 by NSP1 since UK NSP1 effectively degrades transiently expressed human IRF3 in murine cells (53). Inhibition of IRF3 has been documented for several strains of rotavirus (4, 5, 25, 53). In this study, we obtained direct experimental evidence that IRF3 is required for IFN-β secretion in response to rotavirus since IFN-β secretion was completely abolished in UK-infected IRF3−/− MEFs. As expected, IRF3 is essential for the induction of transcripts immediately after virus infection, confirming its importance in both the early and later phases of the antiviral response to UK. Thus, rotavirus likely triggers MDA-5 and RIG-I signaling through MAVS/IPS-1, resulting in IRF3 activation, transcription of early ISGs, and subsequent secretion of IFN-β. Certain strains of rotavirus, including RRV, trigger the early transcriptional response but degrade IRF3 subsequently by an NSP1-mediated process (53) and thus suppress IFN-β secretion and amplification of the host innate immune response. Although IRF3 is essential for the IFN response to rotavirus, other host factors also are likely to be important regulators of this process and present potential targets for viral inhibition. For example, the porcine OSU strain targets the IκBα-associated E3 ligase SCFβ-TrCP for proteasomal degradation and inhibits IFN-stimulated response element (ISRE) activity and transcription of the IFN-β gene (24).
An important finding of this study is that the dsRNA-dependent kinase PKR regulates IFN-β secretion during rotavirus infection. Fibroblasts lacking PKR were unable to secrete IFN-β in response to rotavirus although the induction of several antiviral genes, including those regulated by transcriptionally active IRF3 and NF-κB, appeared to be intact. Thus, in the context of rotavirus activation of the innate immune response, PKR is critical for the secretory phase but not for the early PRR-mediated transcriptional phase, which is dependent instead on MAVS, RIG-I/MDA-5, and IRF3. Mammalian PKR is an IFN-inducible Ser/Thr kinase though it is also basally expressed in MEFs and is associated with ribosomes in an unphosphorylated state (12, 59). In the context of PKR as a protein synthesis inhibitor, the activation of PKR by dsRNA leads to inhibitory Ser51 phosphorylation of the translation initiation factor eIF2α (14). In contrast, our findings as well as other recent studies (52) implicate PKR as a critical positive regulator of IFN protein synthesis/secretion; i.e., the presence of PKR is apparently required for IFN-β secretion to occur. The phosphorylation of eIF2α by PKR does not seem to be required for IFN production in response to virus (52), indicating that PKR promotes IFN-β secretion by a different mechanism. Although PKR can activate NF-κB by phosphorylation of IκB (20, 21, 41, 64) and can activate IRF1 (42), we found that transcription of ISG and IFN-α/β was intact in PKR−/− MEFs, indicating that PKR is unlikely a major determinant of early virus-induced signaling events.
Unlike PKR, we found that IRF3 is required for both ISG transcription and IFN-β secretion, suggesting that it occupies a relatively more critical functional role in the innate immune response to rotavirus. In support of this notion, UK rotavirus RNA synthesis increased significantly in IRF3−/− MEFs compared to PKR−/− MEFs although both knockout cells supported much higher levels of viral RNA synthesis than WT MEFs. Interestingly in certain situations, rotavirus infection can lead to phosphorylation of eIF2α in a PKR-dependent manner (50), and it has been noted that triple-layered (i.e., intact) rotavirus particles replicate more efficiently in PKR−/− MEFs than WT MEFs (50). An earlier study found that PKR is critical for reovirus-induced IFN-β responses and influences virulence in a mouse model of myocarditis (56). Our findings thus reveal PKR as an additional host factor that restricts UK virus replication in an IFN-related manner similar to the functions of type I IFN receptors (16), STAT1, MAVS, and IRF3, and delineate PKR function at a step downstream of IFN and ISG transcriptional responses. Although we have not addressed the mechanism of PKR function in promoting the secretion of IFN-β in this study, recent reports have demonstrated that PKR is critical for the synthesis of IFN-β protein in response to infection with viruses that primarily engage MDA-5 (encephalomyocarditis virus, Theiler's murine encephalomyelitis virus, Semliki Forest virus, and West Nile virus) (6, 22, 52) but does not apparently regulate this process for RIG-I-dependent viruses (Newcastle disease virus, influenza virus, and Sendai virus) (55, 62). We found that PKR is required for the secretion of IFN-β in response to rotavirus, which activated both RIG-I and MDA-5, and similar to the recent findings above, uses a mechanism operant following IFN-β gene transcription. Although the exact mechanism involved is currently not clear, given this new role for PKR in the IFN response to rotavirus, it is possible that rotaviruses employ strategies to specifically target PKR functions related to IFN-β secretion.
While this study was under review, Broquet and coworkers also reported on a role for MAVS/IPS-1 in rotavirus recognition in intestinal cells (10). In general, their findings are similar to ours and indicate that both RIG-I and MDA-5 play roles in sensing rotavirus infection. However, there are some interesting and currently unexplained differences between the two studies. First, findings in the Broquet et al. study indicate that RIG-I and MDA-5 are additive in their function while we find them to be redundant. The reason for this difference is not clear at present but might relate to differences in both the virus strains and cell lines studied. An additional potential difference relates to RRV and its effects on IRF3 activation in HT-29 cells. Broquet et al. provide data to indicate that RRV does not inhibit IRF3 and IFN responses in HT-29 cells while our previous (18, 53) and current studies involving RRV demonstrate substantial IRF3 and IFN antagonism in both murine and simian fibroblasts as well as in murine biliary and alveolar epithelial cells. In addition, in a recent publication, Arnold and Patton demonstrated efficient IRF3 antagonism by RRV in HT-29 cells (2). The basis for these differences awaits further study. Finally, Broquet and coworkers studied RRV and SA11 replication in vivo using PKR knockout mice and concluded that neither serum IFN levels nor virus replication was significantly different from that in WT mice following infection with simian rotavirus. We have not yet performed similar studies. However, we (15) along with others (48) have been unable to employ RRV or SA11 replication in adult mice as a useful model because the replication and shedding of these heterologous rotaviruses in adult mice are highly restricted and of short duration in our hands. The basis for the more robust simian rotavirus replication and sustained in vivo shedding in adult mice in the Broquet study will require further analysis. Of note, several murine strains of rotavirus are shed efficiently over a number of days in adult mice (27), and we plan to examine these strains in appropriate PKR KO mice in future studies.
The exact nature of rotavirus-encoded PAMPs that trigger host PRRs is not completely clear, especially in the context of virus infection. Prior studies have reported that purified rotavirus dsRNA genome segments can stimulate IFN responses by both RIG-I- and MDA-5-dependent processes (38). Short dsRNA was an effective ligand for RIG-I, whereas longer dsRNA segments potently triggered MDA-5-dependent responses, indicating a role for both PRRs in recognizing the rotavirus genomic RNA (38). Both the viral PAMPs and host PRRs for rotavirus are likely to differ in plasmacytoid dendritic cells (pDCs) from those identified here in fibroblasts as pDCs secrete IFN independently of RIG-I and MDA-5 (57). We recently found that in human pDCs, rotaviral (genomic) dsRNA encapsidated by intact virus particles is the likely signal for IFN activation although the nature of the PRR involved is unknown (13). In pDCs, unlike fibroblasts or epithelial cells, the type I IFN response is independent of viral replication (13). On the other hand, in fibroblasts we found that inactivated UK virus does not trigger IRF3 phosphorylation (53) or expression of antiviral genes such as ISG54 and ISG56. Thus, unlike purified dsRNA introduced by transfection, “incoming” encapsidated viral dsRNA genome is unlikely to be available in the cytoplasm for PRR engagement, at least immediately after virus entry. Early rotavirus replication results in synthesis of single-stranded, plus-sense RNA transcripts that, unlike host RNAs, are 5′ methyl capped and nonpolyadenylated (28). These nascent ssRNA molecules are synthesized and extruded by double-layered particles within the cytoplasm from within viral cores, thus ensuring that the input rotavirus dsRNA genome is not exposed to the cytosol (43). Rotavirus genomic dsRNA contains a 5′ m7GpppGp(m)GpCp (36) and, depending on the capping efficiency of the viral VP3 protein, uncapped or partially capped mRNA is likely to exist in infected cells. Based on the evidence available, rotavirus transcripts present early in infection that are potential ligands for RIG-I and MDA-5 can be proposed as follows: (i) capped or uncapped resulting in masking or exposure of the 5′ triphosphate moiety, respectively; (ii) single-stranded or double-stranded RNA (arising from local structures and panhandles between complementary termini); or (iii) short or long RNA. We are currently examining the contribution of these rotavirus replication products to the activation of RIG-I and MDA-5.
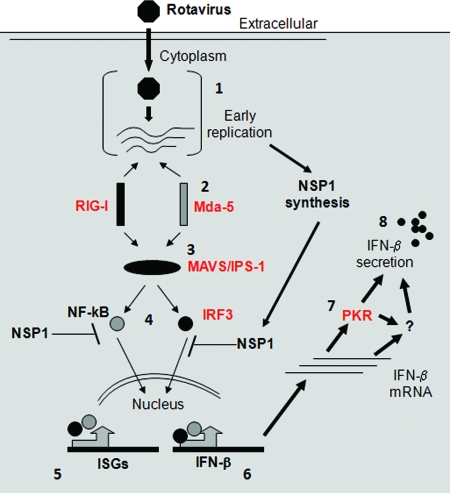
Based on the findings presented here and earlier studies by us and others (3–5, 10, 13, 15–17, 24, 25, 53), we are able to propose a model for the early events related to the rotavirus innate immune response (Fig. 9). Rotavirus cell entry and subsequent replication lead to MDA-5 and RIG-I activation; the two PPRs may be activated at different times and by different types of viral ligands. The adaptor MAVS/IPS-1 associates with activated PRRs and propagates signaling to activate IRF3 and NF-κB. This early antiviral phase is characterized by the transcription of antiviral genes including ISG15, ISG54, ISG56, and IRF7. For certain rotavirus strains, such as RRV, subsequent synthesis of the nonstructural protein NSP1 leads to IRF3 and IRF7 degradation and inhibition of the antiviral response and IFN-β secretion. When strains such as UK infect certain cells, such as murine cells, the lack of NSP1-mediated IRF inhibition means that the initial PRR activation is perpetuated downstream and ultimately results in IFN-β secretion and restriction of viral replication. PKR is critical for the phase of the antiviral response that occurs between IFN-β transcription and secretion of the protein and may act either directly or through an unidentified mediator. The mechanisms of PKR action and rotavirus regulation of PKR in its dual contexts as an IFN-induced protein synthesis inhibitor and as a primary inducer of the IFN response are not yet understood.
Fig. 9.
Model of events occurring during early innate immune recognition of rotavirus leading to IFN secretion. Numbers refer to steps in the pathway. Once inside the host cell (1), rotavirus particles synthesize nascent single-stranded RNA transcripts to direct the expression of viral proteins, including the IFN antagonist NSP1. Viral replication activates host proteins MDA-5 and RIG-I (2), leading to MAVS/IPS-1 recruitment and activation of IRF3 and NF-κB (3). These transcription factors induce expression of ISGs, including ISG54, ISG56, ISG15, and IRF7 (4 and 5). In addition, they also induce the expression of IFN-β itself (6). Rotavirus NSP1s from certain strains and in certain cells target IRF3 and/or the NF-κB regulatory factor β-TrCP for proteasomal degradation, inhibiting the interferon response. Rotavirus strains, such as UK, that trigger IFN secretion (7) lead to further induction and amplification of IFN secretion via a process that depends on PKR to promote IFN synthesis and/or secretion (8) at a posttranscriptional level. This function of PKR may be direct or indirect and is distinct from its role in preventing cellular translation. The model is based on our present study (indicated in red) as well as on several earlier reports (3–5, 17, 24, 25, 29, 53).
ACKNOWLEDGMENTS
We thank Ningguo Feng, Catherine Cruz, and Nandini Sen for helpful discussions and critical comments. We also thank Phuoc Vo for technical assistance and Joyce Troiano for administrative support. We are grateful to Shizuo Akira, Katherine Fitzgerald, Marco Colona, and Robert Finberg for generously providing RIG-I−/−, MDA5−/−, and MAVS/IPS-1−/− MEFs.
This study was supported in part by a VA Merit Award and NIH grant R01 AI021362-26 and P30DK56339 (H.B.G.). Additional support was provided by NIH grants U19AI083025 (A.G.-S.), F32 NS071986 (A.J.P.), R01 AI050080 (T.S.D.), P30 CA68485 for the Vanderbilt-Ingram Cancer Center, P60 DK20593 for the Vanderbilt Diabetes Research and Training Center, and the Elizabeth B. Lamb Center for Pediatric Research.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Angel J., Franco M. A., Greenberg H. B. 2007. Rotavirus vaccines: recent developments and future considerations. Nat. Rev. Microbiol. 5:529–539 [DOI] [PubMed] [Google Scholar]
- 2. Arnold M. M., Patton J. T. 2011. Diversity of interferon antagonist activities mediated by NSP1 proteins of different rotavirus strains. J. Virol. 85:1970–1979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bagchi P., et al. 2010. Rotavirus nonstructural protein 1 suppresses virus-induced cellular apoptosis to facilitate viral growth by activating the cell survival pathways during early stages of infection. J. Virol. 84:6834–6845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Barro M., Patton J. T. 2005. Rotavirus nonstructural protein 1 subverts innate immune response by inducing degradation of IFN regulatory factor 3. Proc. Natl. Acad. Sci. U. S. A. 102:4114–4119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Barro M., Patton J. T. 2007. Rotavirus NSP1 inhibits expression of type I interferon by antagonizing the function of interferon regulatory factors IRF3, IRF5, and IRF7. J. Virol. 81:4473–4481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Barry G., et al. 2009. PKR acts early in infection to suppress Semliki Forest virus production and strongly enhances the type I interferon response. J. Gen. Virol. 90:1382–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Baum A., Sachidanandam R., Garcia-Sastre A. 2010. Preference of RIG-I for short viral RNA molecules in infected cells revealed by next-generation sequencing. Proc. Natl. Acad. Sci. U. S. A. 107:16303–16308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Bergmann M., et al. 2000. Influenza virus NS1 protein counteracts PKR-mediated inhibition of replication. J. Virol. 74:6203–6206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Broome R. L., Vo P. T., Ward R. L., Clark H. F., Greenberg H. B. 1993. Murine rotavirus genes encoding outer capsid proteins VP4 and VP7 are not major determinants of host range restriction and virulence. J. Virol. 67:2448–2455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Broquet A. H., Hirata Y., McAllister C. S., Kagnoff M. F. 2011. RIG-I/MDA5/MAVS are required to signal a protective IFN response in rotavirus-infected intestinal epithelium. J. Immunol. 186:1618–1626 [DOI] [PubMed] [Google Scholar]
- 11. Carpentier P. A., Williams B. R., Miller S. D. 2007. Distinct roles of protein kinase R and toll-like receptor 3 in the activation of astrocytes by viral stimuli. Glia 55:239–252 [DOI] [PubMed] [Google Scholar]
- 12. Das S., Ward S. V., Tacke R. S., Suske G., Samuel C. E. 2006. Activation of the RNA-dependent protein kinase PKR promoter in the absence of interferon is dependent upon Sp proteins. J. Biol. Chem. 281:3244–3253 [DOI] [PubMed] [Google Scholar]
- 13. Deal E. M., Jaimes M. C., Crawford S. E., Estes M. K., Greenberg H. B. 2010. Rotavirus structural proteins and dsRNA are required for the human primary plasmacytoid dendritic cell IFNα response. PLoS Pathog. 6:e1000931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Farrell P. J., Balkow K., Hunt T., Jackson R. J., Trachsel H. 1977. Phosphorylation of initiation factor elF-2 and the control of reticulocyte protein synthesis. Cell 11:187–200 [DOI] [PubMed] [Google Scholar]
- 15. Fenaux M., Cuadras M. A., Feng N., Jaimes M., Greenberg H. B. 2006. Extraintestinal spread and replication of a homologous EC rotavirus strain and a heterologous rhesus rotavirus in BALB/c mice. J. Virol. 80:5219–5232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Feng N., et al. 2008. Role of interferon in homologous and heterologous rotavirus infection in the intestines and extraintestinal organs of suckling mice. J. Virol. 82:7578–7590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feng N., et al. 2009. Variation in antagonism of the interferon response to rotavirus NSP1 results in differential infectivity in mouse embryonic fibroblasts. J. Virol. 83:6987–6994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Feng N., et al. 29 December 2010. The roles of VP4 and NSP1 in determining the distinctive replication capacities of simian rotavirus RRV and bovine rotavirus UK in the mouse biliary tract. J. Virol. doi: 10.1128/JVI.02408-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Fredericksen B. L., Keller B. C., Fornek J., Katze M. G., Gale M., Jr 2008. Establishment and maintenance of the innate antiviral response to West Nile virus involves both RIG-I and MDA5 signaling through IPS-1. J. Virol. 82:609–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gil J., Alcami J., Esteban M. 1999. Induction of apoptosis by double-stranded-RNA-dependent protein kinase (PKR) involves the alpha subunit of eukaryotic translation initiation factor 2 and NF-κB. Mol. Cell. Biol. 19:4653–4663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Gil J., et al. 2004. TRAF family proteins link PKR with NF-κB activation. Mol. Cell. Biol. 24:4502–4512 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gilfoy F. D., Mason P. W. 2007. West Nile virus-induced interferon production is mediated by the double-stranded RNA-dependent protein kinase PKR. J. Virol. 81:11148–11158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gitlin L., et al. 2006. Essential role of Mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Graff J. W., Ettayebi K., Hardy M. E. 2009. Rotavirus NSP1 inhibits NF-κB activation by inducing proteasome-dependent degradation of beta-TrCP: a novel mechanism of IFN antagonism. PLoS Pathog. 5:e1000280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Graff J. W., Mitzel D. N., Weisend C. M., Flenniken M. L., Hardy M. E. 2002. Interferon regulatory factor 3 is a cellular partner of rotavirus NSP1. J. Virol. 76:9545–9550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Greenberg H. B., Estes M. K. 2009. Rotaviruses: from pathogenesis to vaccination. Gastroenterology 136:1939–1951 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Greenberg H. B., Vo P. T., Jones R. 1986. Cultivation and characterization of three strains of murine rotavirus. J. Virol. 57:585–590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Guglielmi K. M., McDonald S. M., Patton J. T. 2010. Mechanism of intraparticle synthesis of the rotavirus double-stranded RNA genome. J. Biol. Chem. 285:18123–18128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hirata Y., Broquet A. H., Menchen L., Kagnoff M. F. 2007. Activation of innate immune defense mechanisms by signaling through RIG-I/IPS-1 in intestinal epithelial cells. J. Immunol. 179:5425–5432 [DOI] [PubMed] [Google Scholar]
- 30. Holm G. H., et al. 2010. Interferon regulatory factor 3 attenuates reovirus myocarditis and contributes to viral clearance. J. Virol. 84:6900–6908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Holm G. H., et al. 2007. Retinoic acid-inducible gene-I and interferon-beta promoter stimulator-1 augment proapoptotic responses following mammalian reovirus infection via interferon regulatory factor-3. J. Biol. Chem. 282:21953–21961 [DOI] [PubMed] [Google Scholar]
- 32. Hoshino Y., Wyatt R. G., Greenberg H. B., Flores J., Kapikian A. Z. 1984. Serotypic similarity and diversity of rotaviruses of mammalian and avian origin as studied by plaque-reduction neutralization. J. Infect. Dis. 149:694–702 [DOI] [PubMed] [Google Scholar]
- 33. Hulst M., et al. 2008. Early transcriptional response in the jejunum of germ-free piglets after oral infection with virulent rotavirus. Arch. Virol. 153:1311–1322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hyser J. M., Estes M. K. 2009. Rotavirus vaccines and pathogenesis: 2008. Curr. Opin. Gastroenterol. 25:36–43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Ikegame S., et al. 2010. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J. Virol. 84:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Imai M., Akatani K., Ikegami N., Furuichi Y. 1983. Capped and conserved terminal structures in human rotavirus genome double-stranded RNA segments. J. Virol. 47:125–136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kato H., et al. 2005. Cell type-specific involvement of RIG-I in antiviral response. Immunity 23:19–28 [DOI] [PubMed] [Google Scholar]
- 38. Kato H., et al. 2008. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J. Exp. Med. 205:1601–1610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 40. Kawai T., et al. 2005. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat. Immunol. 6:981–988 [DOI] [PubMed] [Google Scholar]
- 41. Kumar A., Haque J., Lacoste J., Hiscott J., Williams B. R. 1994. Double-stranded RNA-dependent protein kinase activates transcription factor NF-κB by phosphorylating IκB. Proc. Natl. Acad. Sci. U. S. A. 91:6288–6292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kumar A., et al. 1997. Deficient cytokine signaling in mouse embryo fibroblasts with a targeted deletion in the PKR gene: role of IRF-1 and NF-κB. EMBO J. 16:406–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Lawton J. A., Estes M. K., Prasad B. V. 1997. Three-dimensional visualization of mRNA release from actively transcribing rotavirus particles. Nat. Struct. Biol. 4:118–121 [DOI] [PubMed] [Google Scholar]
- 44. Loo Y. M., et al. 2008. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 82:335–345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Meylan E., et al. 2005. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature 437:1167–1172 [DOI] [PubMed] [Google Scholar]
- 46. O'Neill L. A., Bowie A. G. 2010. Sensing and signaling in antiviral innate immunity. Curr. Biol. 20:R328–R333 [DOI] [PubMed] [Google Scholar]
- 47. Pichlmair A., et al. 2009. Activation of MDA5 requires higher-order RNA structures generated during virus infection. J. Virol. 83:10761–10769 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Ramig R. F. 1988. The effects of host age, virus dose, and virus strain on heterologous rotavirus infection of suckling mice. Microb. Pathog. 4:189–202 [DOI] [PubMed] [Google Scholar]
- 49. Randall R. E., Goodbourn S. 2008. Interferons and viruses: an interplay between induction, signalling, antiviral responses and virus countermeasures. J. Gen. Virol. 89:1–47 [DOI] [PubMed] [Google Scholar]
- 50. Rojas M., Arias C. F., Lopez S. 2010. Protein kinase R is responsible for the phosphorylation of eIF2α in rotavirus infection. J. Virol. 84:10457–10466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Schlee M., et al. 2009. Recognition of 5′ triphosphate by RIG-I helicase requires short blunt double-stranded RNA as contained in panhandle of negative-strand virus. Immunity 31:25–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schulz O., et al. 2010. Protein kinase R contributes to immunity against specific viruses by regulating interferon mRNA integrity. Cell Host Microbe 7:354–361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Sen A., Feng N., Ettayebi K., Hardy M. E., Greenberg H. B. 2009. IRF3 inhibition by rotavirus NSP1 is host cell and virus strain dependent but independent of NSP1 proteasomal degradation. J. Virol. 83:10322–10335 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Seth R. B., Sun L., Ea C. K., Chen Z. J. 2005. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-κB and IRF 3. Cell 122:669–682 [DOI] [PubMed] [Google Scholar]
- 55. Smith E. J., Marie I., Prakash A., Garcia-Sastre A., Levy D. E. 2001. IRF3 and IRF7 phosphorylation in virus-infected cells does not require double-stranded RNA-dependent protein kinase R or IκB kinase but is blocked by vaccinia virus E3L protein. J. Biol. Chem. 276:8951–8957 [DOI] [PubMed] [Google Scholar]
- 56. Stewart M. J., Blum M. A., Sherry B. 2003. PKR's protective role in viral myocarditis. Virology 314:92–100 [DOI] [PubMed] [Google Scholar]
- 57. Swiecki M., Colonna M. 2010. Unraveling the functions of plasmacytoid dendritic cells during viral infections, autoimmunity, and tolerance. Immunol. Rev. 234:142–162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Wang J. P., et al. 2010. MDA5 and MAVS mediate type I interferon responses to coxsackie B virus. J. Virol. 84:254–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Wang Q., Carmichael G. G. 2004. Effects of length and location on the cellular response to double-stranded RNA. Microbiol. Mol. Biol. Rev. 68:432–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Wilkins C., Gale M., Jr 2010. Recognition of viruses by cytoplasmic sensors. Curr. Opin. Immunol. 22:41–47 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Xu L. G., et al. 2005. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol. Cell 19:727–740 [DOI] [PubMed] [Google Scholar]
- 62. Yang Y. L., et al. 1995. Deficient signaling in mice devoid of double-stranded RNA-dependent protein kinase. EMBO J. 14:6095–6106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Yoneyama M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 64. Zamanian-Daryoush M., Mogensen T. H., DiDonato J. A., Williams B. R. 2000. NF-κB activation by double-stranded-RNA-activated protein kinase (PKR) is mediated through NF-κB-inducing kinase and IκB kinase. Mol. Cell Biol. 20:1278–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Zhang X., Wang C., Schook L. B., Hawken R. J., Rutherford M. S. 2000. An RNA helicase, RHIV-1, induced by porcine reproductive and respiratory syndrome virus (PRRSV) is mapped on porcine chromosome 10q13. Microb. Pathog. 28:267–278 [DOI] [PubMed] [Google Scholar]