Abstract
Human cytomegalovirus (HCMV) encodes multiple G protein-coupled receptor (GPCR) homologues, including pUS27, pUS28, pUL33, and pUL78. To explore the function of pUS27, we constructed pUS27-deficient derivates of two clinical isolates of HCMV. BFX-GFPstopUS27 is a FIX variant with a single base pair change in the US27 open reading frame, generating a stop codon that ablates accumulation of the GPCR homologue, and TB40/E-mCherrydlUS27 lacks the entire US27 coding region. BFX-GFPstopUS27 generated 10-fold less extracellular progeny in fibroblasts, and TB40/E-mCherrydlUS27 exhibited a similar defect in endothelial cells. The pUS27-deficient FIX derivative produced normal quantities of viral DNA and viral proteins tested, and a late virion protein was appropriately localized to the cytoplasmic assembly zone. After infection at a low multiplicity with wild-type FIX virus, neutralizing antibody reduced the accumulation of intracellular viral DNA and intracellular virions, as would be expected if the virus is limited to direct cell-to-cell spread by neutralization of extracellular virus. In contrast, the antibody had little effect on the spread of the BFX-GFPstopUS27 virus. Further, after infection at a low multiplicity, the pUS27-deficient TB40/E virus exhibited a growth defect in endothelial cells, where the clinical isolate normally generates extracellular virus, but the TB40/E derivative exhibited little defect in epithelial cells, where the wild-type virus does not produce extracellular virus. Thus, mutants lacking pUS27 rely primarily on direct cell-to-cell spread, and we conclude that the viral GCPR homologue acts at a late stage of the HCMV replication cycle to support spread of virus by the extracellular route.
INTRODUCTION
Human cytomegalovirus (HCMV) is a ubiquitous pathogen belonging to the betaherpesvirus subfamily, which establishes a life-long infection within its human host (5). Infections in immunocompetent children and adults are generally asymptomatic, but the virus can cause life-threatening disease in immunologically immature and immunocompromised individuals. Sequence analysis of the viral genome (8) revealed four open reading frames (ORFs) encoding proteins with strong homology to G protein-coupled receptors (GPCRs): US27, US28, UL33, and UL78 (1, 8). UL33 and UL78 homologues are found in primate as well as rodent cytomegaloviruses, whereas US27 and US28 homologues are restricted to viruses infecting primates (reviewed in reference 2).
GPCRs comprise a large family of seven transmembrane receptor proteins. When activated, GPCRs initiate signaling cascades by undergoing a conformational change that leads to the recruitment and activation of heterotrimeric G proteins, followed by the production of second messengers such as cyclic AMP (cAMP), calcium, or phosphoinositides (reviewed in reference 12). The G protein-coupling specificity of each GPCR determines the nature of its downstream signaling targets. Activated GPCRs regulate a wide variety of cellular processes including adhesion and migration, proliferation, differentiation, apoptosis, cytoskeletal rearrangement, chemotaxis, and cell survival (reviewed in reference 26). GPCRs include sensory receptors and receptors for neurotransmitters, hormones, and chemokines. The subset of GPCRs responding to chemokines is characterized by well-defined sequence and structural motifs. The HCMV US27, US28, and UL33 ORFs encode GPCR homologues with hallmarks of chemokine receptors (reviewed in reference 2).
pUS28 is the best-characterized HCMV chemokine receptor (reviewed in reference 30). It signals constitutively, inducing phospholipase C to generate inositol triphosphate and activating NF-κB (7). pUS28 also signals in response to multiple CC chemokines and the CX3C-chemokine, fractalkine, mobilizing calcium (10, 14) and activating mitogen-activated protein (MAP) kinase signaling (3). pUS28 can induce chemotaxis of smooth muscle cells (27) and macrophages (30), and ectopic expression of pUS28 can induce a transformed phenotype in murine 3T3 cells with elevated secretion of vascular endothelial growth factor (18).
Since the US27 and US28 ORFs encode proteins with similar sequences and since they reside beside each other on the viral genome, it is likely that the gene pair evolved by gene duplication followed by divergence. Given its similarity to pUS28, pUS27 might also function as a chemokine receptor (9), but this has not yet been proven to be the case. In contrast to pUS28, pUS27 expression failed to significantly induce inositol phosphate turnover or activate NF-κB or CREB (31). It is possible that pUS27 does not signal constitutively, and, to date, no activating ligand has been reported.
pUS27 is endocytosed rapidly from the cell surface, and it accumulates in vesicles with markers of late endosomes and lysosomes (9). As has been proposed for pUS28 (4, 7), pUS27 might serve as a sink that clears immunomodulatory chemokines from the microenvironment of infected cells. As a constituent of the virion (17, 29), pUS27 could interact with a cell surface receptor to facilitate virus binding to a cell, or it might activate a signaling cascade upon fusion of the virion envelope with a cellular membrane, perhaps making the cellular environment more amenable to viral replication. However, the role for pUS27 during infection has remained uncertain.
To decipher the role of pUS27 in the HCMV replication cycle, we generated mutant viruses in which expression of the protein was ablated. At a low multiplicity of infection, the pUS27-deficient mutants display a growth defect compared to wild-type virus. We determined that although the mutant can spread directly from cell to cell, it is deficient for spread via the production of extracellular virus.
MATERIALS AND METHODS
Cells and viruses.
Primary human lung fibroblasts (MRC5 [ATCC]; passages 23 to 31) and human foreskin fibroblasts (HFF; passages 10 to 17) were maintained in Dulbecco's modified Eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 1 mM sodium pyruvate, 10 mM HEPES, 2 mM l-glutamine, 0.1 mM nonessential amino acids, and 100 U/ml each of penicillin and streptomycin. Primary human retinal pigment epithelial cells (ARPE-19 [ATCC]; passages 27 to 35) were maintained in 1:1 DMEM-HAM's F12 containing 10% FBS, 2.5 mM l-glutamine, 0.5 mM sodium pyruvate, 15 mM HEPES, 1.2g/liter NaHCO3, and 100 U/ml each of penicillin and streptomycin. Human umbilical vascular endothelial cells (HUVECs) were isolated from umbilical cords as previously described (32). The cells were cultured in Primaria tissue culture plates (BD Falcon) and maintained in EBM-2 medium containing 10% FBS with EGM-2 supplements (Lonza). For infection, HUVECs were used between passages 3 and 5. All cells were propagated at 37°C in 5%CO2.
Two clinical HCMV isolates that had been passaged to a limited extent in fibroblasts were used in these studies. BFXwtGFP (where wt is wild type) (19) expresses an enhanced green fluorescent protein (eGFP) marker from the simian virus 40 (SV40) early promoter, and it is derived from the HCMV bacterial artificial chromosome (BAC) clone termed FIX (11). TB40/Ewt-mCherry expresses an mCherry marker gene (construction described below), and it is derived from an endothelial cell-tropic BAC-cloned isolate of TB40/E termed TB40-BAC4 (25).
BFXwtGFP was used to generate a virus expressing FLAG-tagged pUS27 (BFXwt-US27-3×F) (where 3×F indicates three copies of the FLAG tag) and two identical mutants carrying stop codons within the US27 ORF (BFX- GFPstopUS27-1 and BFX-GFPstopUS27-2). Two independent mutants were pre- pared and characterized to ensure that their phenotypes did not result from an off-target mutation. To construct BFXwt-US27-3×F, three tandem FLAG sequences (3×FLAG) were cloned upstream of a kanamycin resistance cassette (Kan) flanked by a Flp recombination target (FRT) site in pGEM-T-Easy (Promega), to generate pGTE-3×FLAG-Kan-FRT. This template was used to PCR amplify the epitope and cassette using the following primers: FOR 5′-TATGACAGAAAACATGCACCTATGGAGTCCGGGGAGGAGGAATTTCTGTTGTTAGATTATAAAGATGATGATGATAAA-3′ and REV 5′-CGTGCAATTAGCAAAAATAGATGTGCGGCGGACGCGTGAGAGAGGATCGAAGGCCGCGGGAATTCGAAGTT-3′, where the underlined sequences bind to the 3×FLAG-Kan-FRT cassette. The resulting amplification product was used to transform recombination-competent Escherichia coli SW105 cells containing BFXwtGFP and generate the desired alterations by linear recombination as previously described (15). The Kan-FRT cassette was then excised, by the arabinose-inducible flp. To generate the mutants containing stop codons, BFXwt-US27-3×F was modified by using the galK recombineering system previously described (19, 33). Briefly, the galK gene was amplified by PCR using the following primers: FOR 5′-CCTGCAAGGTAATGACTACATCTACTACAACTACCACTAATATCATGCTACCTGTTGACAATTAATCATCGGCA-3′ and REV 5′-ACTGATAAATTTCGGTGCTATTCAAGGTGTGATTCGTTACGTTGCTCACCTTCAGCACTGTCCTGCTCCTT-3′, where the underlined sequences correspond to galK. The resulting product was used to transform recombination-competent E. coli SW105 cells containing BFXwt-US27-3×F. GalK-expressing clones were subsequently selected and electroporated with 5′-CTGCAAGGTAATGACTACATCTACTACAACTACCACTAATATCATGCTATAGGTGAGCAACGTAACGAATCACACCTTGAATAGCACCGAAATTTATCA-3′ annealed to its complementary oligonucleotide. The underlined sequence shows the stop codon in the US27 gene, generated by altering the first amino acid in the codon from C to T. The resultant mutants were counter-selected against galK and sequenced to ensure incorporation of the stop codon.
The TB40/E BAC (25) was engineered to express mCherry (TB40/Ewt-mCherry). Using galK recombineering methodology as described above, galK was inserted between US34 and TRS1 using FOR 5′-TGTATTTGTGACTATACTATGTGCAGTCGTGTGTCGATGTTCCTATTGGGCCTGTTGACAATTAATCATCGGCA-3′ and REV 5′-GATGTCTTCCTGCGTCCCACCATTCTTTATACCTCCTACATTCACACCCTTTCAGCACTGTCCTGCTCCTT-3′, where the underlined sequences correspond to galK. A cassette containing mCherry controlled by the SV40 promoter and containing the bovine growth hormone poly(A) motif was then PCR amplified using FOR 5′-TTGTATTTGTGACTATACTATGTGCAGTCGTGTGTCGATGTTCCTATTGGGATCTGCGCAGCACCATGGCCTGAAATAACCTCTGAAAG-3′ and REV 5′-GATGTCTTCCTGCGTCCCACCATTCTTTATACCTCCTACATTCACACCCTTCTGCCCCAGCTGGTTCTTTCCGCCTCAGAAGCCATAGA-3′, where the underlined sequences are complementary to the intergenic region between the HCMV US34 and TRS-1 ORFs. The resulting PCR product was electroporated into competent E. coli SW105 cells and counter-selected against galK to generate TB40/Ewt-mCherry. An epitope-tagged variant was constructed from TB40/E as described above, resulting in TB40/Ewt-US27-3×F. A virus lacking the complete US27 ORF, TB40/E-mCherrydlUS27, was constructed from TB40/Ewt-US27-3×F using the galK recombineering system described above, with the following primers for galK insertion: FOR 5′-GTGTAATGCTTTTTACAGGACCGTTCAACAGGTGATACTACCTGCAAGGTACCTGTTGACAATTAATCATCGGCA-3′ and REV 5′-CGTGCAATTAGCAAAAATAGATGTGCGGCGGACGCGTGAGAGAGGATCGAATCAGCACTGTCCTGCTCCTT-3′, where the underlined sequences correspond to galK. The following oligonucleotide, annealed to its complementary sequence, was used to delete the US27 ORF: 5′-TTTACAGGACCGTTCAACAGGTGATACTACCTGCAAGGTATTCGATCCTCTCTCACGCGTCCGCCGCACATCTATTTTTG-3′. The mutation was confirmed by sequence analysis and by demonstrating by immunofluorescence that pUS27-3×F no longer accumulated after infection (data not shown).
Wild-type and mutant virus stocks were propagated by transfecting fibroblasts with BAC DNA, cultures were harvested when full cytopathic effect was evident, and virus was then partially purified by centrifugation through a 20% sorbitol cushion. Virus stocks were stored at −80°C in DMEM containing 10% FBS plus 1.5% bovine serum albumin (BSA). Virus stock titers were determined by 50% tissue culture doses (TCID50) on fibroblasts.
Analysis of virus growth and spread.
Multistep growth kinetics were analyzed by infecting 2.5 × 105 fibroblasts at a multiplicity of 0.01 PFU/ml. Viral growth was also analyzed at a multiplicity of infection of 0.5 PFU/ml, where indicated in the figure legends. In each case, medium was collected at various times after infection and used to infect fresh cultures of fibroblasts. Virus yield was determined by quantifying the number of IE1-positive cells by using an IE1-specific monoclonal antibody (clone 1B12) (34) and a fluorescently labeled secondary antibody in three randomly chosen fields, as previously described (28). For the analysis of cell-associated virus yield, 7.0 × 104 infected cells were harvested by scraping into fresh medium and lysed by subjecting them to three freeze-thaw cycles. Cellular debris was pelleted by centrifugation, and the amount of infectious virus in the resulting supernatant was determined by TCID50 assay. To assess viral growth in ARPE cells, approximately 5.0 × 105 cells per time point were infected at a low multiplicity (0.1 PFU/ml). Cells and medium were collected at various times after infection, and virus titers were determined on fibroblasts by either quantifying IE1-positive cells, as described above, or TCID50 assay.
To monitor spread mediated by extracellular virus versus direct cell-to-cell spread, HCMV neutralizing antibody, CytoGam (National Hospital Specialties), was used as described previously (22). Fibroblasts (7.0 × 104 cells/condition) were infected for 1 h at 37°C and washed twice with phosphate-buffered saline (PBS), and fresh medium was added with or without a sufficient quantity of CytoGam to neutralize infectious virus produced in the cultures (3%, vol/vol). Medium and CytoGam were replaced daily. At 15 days postinfection (dpi), cells were harvested for cell-associated DNA analysis by quantitative PCR (qPCR) and to determine the yield of cell-associated virus by TCID50 assay.
Assay of viral DNA and proteins.
Quantification of DNA in virus stocks and in infected cells was performed as previously described (28). To determine the particle-to-PFU ratio, genomes were used as a metric for virus particles. The genome number, determined by qPCR, was divided by the infectious yield, assayed by TCID50. Cell-associated viral DNA levels were normalized using primers specific for actin: FOR 5′-TCCTCCTGAGCGCAAGTACTC-3′ and REV 5′-CGGACTCGTCATACTCCTGCTT-3′. In each case, viral DNA was quantified using primers to UL123: FOR 5′-GCCTTCCCTAAGACCACCAAT-3′ and REV 5′-ATTTTCTGGGCATAAGCCATAATC-3′. Samples were analyzed in triplicate by SYBR green assay using an ABI 7600 real-time PCR instrument.
To label viral proteins exposed on the cell surface, biotinylation was carried out using a Cell Surface Protein Isolation Kit (Thermo Scientific) according to the manufacturer's instructions. In brief, virus was absorbed to 1.8 × 106 fibroblasts at a multiplicity of infection of 0.5 PFU/cell for 1 h at 37°C; cells were washed twice with PBS, and the cultures were reconstituted with fresh medium. At 96 hpi, medium was removed, and cells were biotinylated; then the reaction was quenched, and cells were harvested for lysis according to the manufacturer's instructions. One-tenth of each lysate was reserved as a control. Biotinylated proteins were isolated by binding to NeutrAvidin beads and denatured for SDS-PAGE according to the manufacturer's instructions. Proteins were transferred to Protran (Whatman), and blots were probed with anti-FLAGm2 antibody (Sigma), diluted 1:7,500, and anti-insulin receptor antibody (Millipore), diluted 1:500, followed by horseradish peroxidase (HRP)-conjugated goat-anti-mouse secondary antibody (Jackson Laboratories), diluted 1:10,000.
To assess viral protein expression, 2.5 × 105 cells per time point were infected with either mutant or wild-type virus at a multiplicity of 0.5 PFU/ml. Virus was adsorbed for 1 h at 37°C and washed twice with PBS, and cultures were replenished with fresh medium. Cells were harvested at various times after infection and lysed in radioimmunoprecipitation assay (RIPA) buffer, and equal amounts of protein (determined by Bradford assay) were subjected to immunoblot assay. Primary antibodies used were anti-IE1 (clone 1B-12) (34) and anti-pUL99 (clone 10B4-29) (24), both diluted 1:100, anti-UL44 (Virusys), diluted 1:2,500, and anti-tubulin (Sigma), diluted 1:5,000, followed by HRP-conjugated goat anti-mouse secondary antibody.
For analysis of protein localization by immunofluorescence, approximately 2.0 × 105 cells were grown on coverslips and infected at a multiplicity of 0.5 PFU/ml. Cells were washed with PBS, fixed for 20 min at 37°C with 2% paraformaldehyde, washed again with PBS, and then permeabilized with 0.1% Triton X-100 plus 0.2% Tween in PBS for 15 min at room temperature. Slides were stored at 4°C with blocking buffer (2% BSA in PBS with 0.2% Tween) until all time points were harvested and then incubated with anti-FLAGm2 (Sigma) diluted 1:500, anti-pUL55 (glycoprotein B [gB]) (Abcam) diluted 1:100, or anti-pUL99 (24) diluted 1:10 in blocking buffer for 1 h at room temperature. Cells were then treated with Alexa Fluor 546 anti-mouse secondary antibody (Molecular Probes) and 4′,6′-diamidino-2-phenylindole (DAPI) and washed as above. Slides were mounted with SlowFade antifade reagent (Molecular Probes). Images were collected by using a Zeiss LSM 510 confocal microscope.
RESULTS
pUS27 is expressed on the surface and in the assembly zone of infected cells.
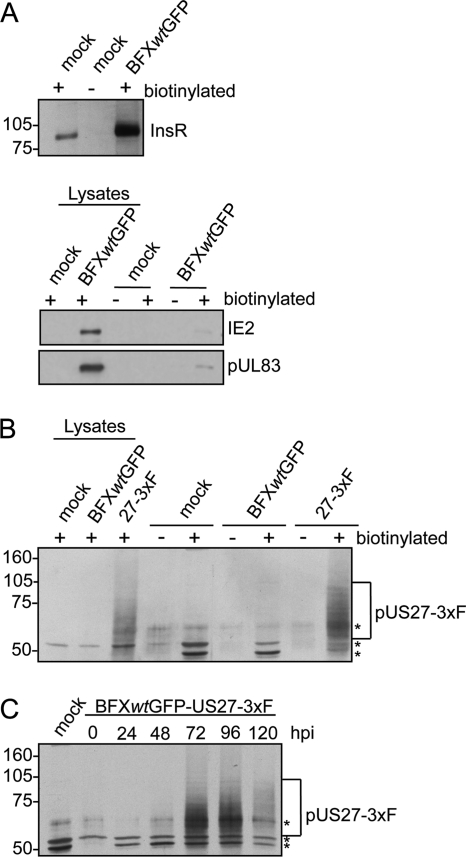
To extend the earlier studies on pUS27 localization in transfected cells and cells infected with a laboratory strain (9, 31), we generated a derivative of the clinical isolate, BFXwtGFP. The variant, termed BFXwt-US27-3×F, expresses a pUS27 fusion protein with three tandem repeats of a FLAG epitope at its carboxy terminus. We initially used the variant to determine if pUS27 is expressed on the surface of infected fibroblasts, where it could potentially respond to environmental signals. To this end, we biotinylated intact cells, prepared extracts, and captured proteins with avidin beads for analysis. As a control, we assayed our ability to detect a known cell surface protein, the insulin receptor (Fig. 1A, top panel). The receptor was detected, and it was captured only when samples were biotinylated. Interestingly, the control experiment indicated that the insulin receptor is elevated after infection, consistent with the earlier observation that insulin receptor RNA is elevated after HCMV infection (6). We also assayed the proteins captured with avidin beads for two viral proteins that are not known to be exposed on the cell surface, IE2 (nuclear) and pUL83 (nuclear and cytoplasmic), and they were present at very low levels (Fig. 1A, bottom panel). A large portion of input insulin receptor was captured, and minimal portions of total cellular IE2 and pUL83 were captured, which supports the conclusion that biotinylation was substantially limited to proteins exposed on the cell surface. We next tested for the presence of pUS27-3×F on the cell surface at 96 hpi with BFXwt-US27-3×F (Fig. 1B). The protein was detected both in whole-cell lysates and after capture of biotinylated surface proteins. It was not detected in mock-infected cells or cells infected with a virus expressing pUS27 without a FLAG tag, confirming specificity of the antibody. pUS27-3×F migrated as a smear, and this was expected because other investigators have reported that pUS27 is glycosylated (17). Finally, we performed a time course analysis (Fig. 1C), which indicated that the amount of pUS27 exposed on the cell surface is maximal during the late phase of infection.
Fig. 1.
pUS27 is expressed on the surface of infected cells. Fibroblasts were mock infected or infected with BFXwtGFP or BFXwtGFP-US27-3×F at a multiplicity of 0.5 PFU/cell. Cell surface proteins were biotinylated 96 h later and captured with avidin-conjugated beads. Each lysate sample assayed received protein from 1.4 × 105 cells per lane, and each biotinylated sample assayed was the material captured from 7.9 × 105 cells. (A) Detection of cell surface insulin receptor InsR in biotinylated (+) samples but not in untreated cells (−) by immunoblot assay using InsR-specific antibody (top) and minimal detection of IE2 (nuclear localization) and pUL83 (nuclear plus cytoplasmic localization) in biotinylated samples (bottom). (B) Detection of FLAG-tagged pUS27-3×F in whole-cell lysates and in biotinylated samples by immunoblot assay using antibody to the FLAG epitope. The bracket marks glycosylated pUS27-3×F, and the asterisks denote nonspecific bands due to biotinylation. (C) Peak expression of pUS27-3×F at 72 and 96 hpi. Fibroblasts were biotinylated at various times after infection and then assayed as in panel B.
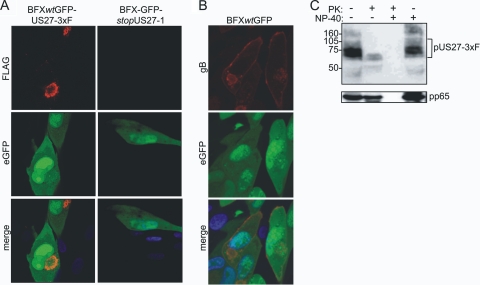
Although we detected pUS27 on the cell surface in the sensitive biotinylation assay, it has been reported that the protein's localization is dynamic and that it is rapidly internalized. In transient transfection assays, very little of the GPCR homologue is localized to the cell surface (9, 31). We therefore assessed the localization of pUS27 by immunofluorescence in BFXwtGFP-US27-3×F-infected fibroblasts. At 96 hpi, pUS27-3×F localized almost exclusively to the perinuclear assembly zone (Fig. 2A, upper left panel), and none was evident on the cell surface. In a control experiment the pUL55-coded gB was readily detected on the cell surface with an intensity similar to that in the assembly zone (Fig. 2B). The presence of pUS27 in the assembly zone is not surprising because it is a virion component (17, 29), and we readily detected pUS27-3×F exposed on the surface of partially purified virions (Fig. 2C). Apparently, only a small portion of the total pUS27 resides at the surface of infected cells. The epitope tag very likely did not alter the localization of pUS27 because the virus expressing the tagged protein grew normally, in contrast to pUS27-deficient viruses, which displayed a growth defect (Fig. 3B).
Fig. 2.
pUS27 is localized to the assembly zone of infected cells, and it is present in virions. (A) Immunofluorescence detection of pUS27-3×F in the assembly zone. Fibroblasts were infected with BFXwtGFP-US27-3×F or BFX-GFPstopUS27 at a multiplicity of 0.5 PFU/ml. Cells were assayed at 96 hpi by using anti-FLAGm2 antibody (red) and DAPI (blue). eGFP (green) is expressed from the viral genome and serves as a marker of infection. (B) Immunofluorescent detection of gB on the cell surface. Fibroblasts were infected with BFXwtGFP at a multiplicity of 0.5 PFU/ml and assayed at 96 hpi by using anti-gB antibody (red) and DAPI (blue). (C) pUS27 is a constituent of the viral envelope. BFXwtGFP-US27-3×F virus particles were partially purified by centrifugation through a 20% sorbitol cushion and, where indicated, treated with proteinase K (PK) in the presence or absence of NP-40. pUS27-3×F was assayed by immunoblotting using antibody to the FLAG epitope. The bracket marks glycosylated pUS27-3×F. As a control, the internal virion protein, pUL83, was detected using a monoclonal antibody to the viral protein.
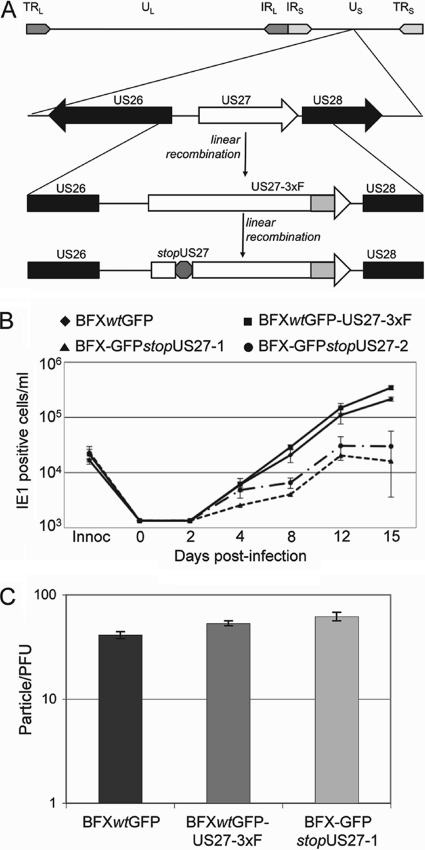
Fig. 3.
pUS27-deficient virus generates reduced yields of extracellular virus. (A) Diagram of the BFXwtGFP-US27-3×F and BFX-GFPstopUS27 genomes. TR, terminal repeat; IR, inverted repeat; U, unique sequence; L and S, long and short, respectively. (B) Growth kinetics of wild-type and mutant viruses. Fibroblasts were infected at a multiplicity of 0.01 PFU/ml with the indicated viruses, samples of medium were collected at the indicated time points, and viral progeny was assayed by infecting fibroblasts and quantifying IE1-positive cells 24 h later by immunofluorescence. Each sample was measured in triplicate, and the dashed line represents the level of detection for the assay. (C) pUS27-deficient virions exhibit normal infectivity. The number of genomes in a virus stock of known titer was determined by qPCR. The ratio of particles per PFU was determined by dividing the number of genomes by the titer for each stock. Each sample was measured in triplicate.
pUS27-deficient viruses produce reduced yields of extracellular virus.
We next generated two identical, but independently constructed, stop mutants in the US27-3×F ORF present in BFXwtGFP-US27-3×F, which are termed BFX-GFPstopUS27-1 and BFX-GFPstopUS27-2 (Fig. 3A), in order to test for a requirement for pUS27 within infected fibroblasts. To generate the stop variants, we changed one base pair in the US27 coding sequence to create a stop codon. By making the mutation in BFXwtGFP-US27-3×F, we could easily confirm by immunofluorescence that expression of the epitope-tagged pUS27 was abrogated (Fig. 2A, right panels). It has been reported that pUS27 is dispensable for growth in cultured fibroblasts, with mutant virus showing no defect compared to wild-type virus (4). However, this earlier work employed a laboratory strain of HCMV at a relatively high multiplicity of infection. To determine the growth properties of our mutant derivatives of a clinical strain, we infected fibroblasts at a low multiplicity (0.01 PFU/cell) and measured the production of infectious extracellular progeny in the media of cultures infected with each of the two identical stop mutant viruses (BFX-GFPstopUS27-1 and BFX-GFPstopUS27-2) compared to growth of wild-type viruses (BFXwtGFP and BFXwtGFP-US27-3×F). At a low multiplicity of infection, the mutant viruses displayed an approximately 10-fold growth defect in extracellular virus accumulation on days 8 to 15 after infection (Fig. 3B). The observation that the two independently isolated stop mutant viruses displayed similar growth defects argues that neither contains off-target mutations contributing to the mutant phenotype.
Since pUS27 is a constituent of virions, we asked if there was a difference in the specific infectivity of the mutant progeny by determining the particle-to-PFU ratio for BFXwtGFP, BFXwt-US27-3×F, and BFX-GFPstopUS27. There is no statistically significant difference in this ratio for parental compared to mutant virus (Fig. 3C), and we conclude that BFX- GFPstopUS27 produces virions of normal infectivity.
pUS27 is not needed for the accumulation of viral DNA or representative viral proteins.
We next asked if the loss of pUS27 expression affected the accumulation of viral DNA, viral proteins, or intracellular infectivity. To this end, we infected fibroblasts at a multiplicity of 0.5 PFU/cell and harvested DNA and protein for further analyses. Initially, we assayed production of extracellular virus, confirming the growth defect of the pUS27 mutant compared to growth of its wild-type parent after infection at this input multiplicity (Fig. 4A). Importantly, the mutant virus accumulated somewhat reduced levels of intracellular infectivity as well. The phenotype of the mutant appears to be multiplicity dependent because the defect in the production of infectious progeny is less at an input of 0.5 PFU/cell (Fig. 4A) than at 0.01 PFU/cell (Fig. 3B).
Fig. 4.
Viral DNA and protein accumulate normally in BFX- GFPstopUS27-infected cells. Fibroblasts were infected at a multiplicity of 0.5 PFU/cell with BFXwtGFP or BFX-GFPstopUS27, and either medium or cells were harvested for analysis after various time intervals. (A) Growth of wild-type and mutant viruses. The accumulation of cell-associated and extracellular virus progeny at 96 and 120 hpi was assayed by infecting fibroblasts and quantifying IE1-positive cells by immunofluorescence. Each sample was measured in triplicate, and differences between wild-type and mutant were statistically significant (extracellular virus, P of 0.005 at 96 hpi and of 0.025 at 120 hpi; cell-associated virus, P of 0.003 at 96 hpi and of 0.015 at 120 hpi). Innoc, innoculum. (B) Normal DNA accumulation in mutant virus-infected cells. Viral DNA was quantified by qPCR and normalized to cellular β-actin DNA, and the results are displayed as arbitrary values. Each sample was measured in triplicate. (C) Normal protein accumulation in mutant virus-infected cells. Representative viral proteins were assayed by immunoblotting, and cellular tubulin was monitored as a loading control.
To test whether earlier events in the virus replication cycle occurred normally, we quantified DNA accumulation by qPCR (Fig. 4B), and protein lysates were assayed by immunoblotting for the accumulation of representatives of the three kinetic classes of viral proteins, i.e., IE1 (immediate early), pUL44 (early), and pUL99 (late) (Fig. 4C). Compared to wild-type virus, BFX-GFPstopUS27 displayed normal DNA accumulation. Production of the immediate-early protein was also normal, whereas the early and late proteins accumulated to modestly elevated levels (about 2-fold) in the mutant compared to accumulation in wild-type virus-infected cells at 48 hpi; levels were normal at 72 and 96 hpi. Although it is possible that the lack of pUS27 influences the expression of a protein that was not assayed, it appears likely that the GPCR homologue is not required for substantially normal viral gene expression.
Although viral protein synthesis is not impaired, we asked if the lack of pUS27 affected the proper localization of proteins necessary for virion assembly by comparing the localization of pUL99 in mutant and wild-type virus-infected cells (Fig. 5). During the late phase of the replication cycle, pUL99 localizes to the assembly zone of infected cells (21), and it is required for envelopment of virus particles (24). pUL99 was properly localized to the assembly zone in the BFX-GFPstopUS27-infected cells.
Fig. 5.
pUL99 is localized normally in BFX-GFPstopUS27-infected cells. Infected fibroblasts were assayed by immunofluorescence for pUL99 (red) at 120 hpi. Nuclei were stained with DAPI (blue), and eGFP (green) is expressed from the viral genome, serving as a second marker of infection.
pUS27 is required for virus spread by the extracellular route.
Our results argue that in the absence of pUS27, the virus replication cycle proceeds normally up to the assembly of infectious virus. However, the amount of intracellular as well as extracellular virus is reduced in populations of cells infected with BFX-GFPstopUS27 compared to those infected with the parental virus. Therefore, we tested the hypothesis that, as a viral envelope constituent (Fig. 2B), pUS27 plays a role in the spread of virus by the extracellular route but does not influence direct cell-to-cell spread. To this end, we infected fibroblasts at a multiplicity of 0.01 PFU/cell with either wild-type or BFX-GFPstopUS27 virus, and following virus adsorption we propagated the infected cultures in the presence or absence of HCMV neutralizing antibody (CytoGam). The addition of CytoGam to infected cultures at a concentration sufficient to neutralize extracellular virus (22) allowed us to differentiate between extracellular spread and cell-to-cell spread of the virus because any virus released into the medium is neutralized and unable to infect a new cell. As a consequence, infected cultures that received this treatment are restricted to cell-to-cell spread of the virus. Following 15 days in culture, infected cells were harvested for analysis of intracellular viral DNA and cell-associated virus. In the absence of neutralizing antibody, BFX-GFPstopUS27 displayed a defect, relative to wild-type virus, in the accumulation of cell-associated viral DNA (Fig. 6A, no CytoGam) and cell-associated infectious virus (Fig. 6B, no CytoGam). However, when the infection was restricted to direct cell-to-cell spread in the presence of neutralizing antibody, BFXwtGFP accumulated less intracellular viral DNA (Fig. 6A, 3% CytoGam) and fewer intracellular infectious virions (Fig. 6B, 3% CytoGam), whereas there was little change in intracellular DNA and no change in intracellular virus for BFX- GFPstopUS27. These results argue that pUS27 plays a role in an event after the production of virion constituents needed for direct cell-to-cell spread and before the release of virions from the infected cell to allow spread mediated by extracellular virus.
Fig. 6.
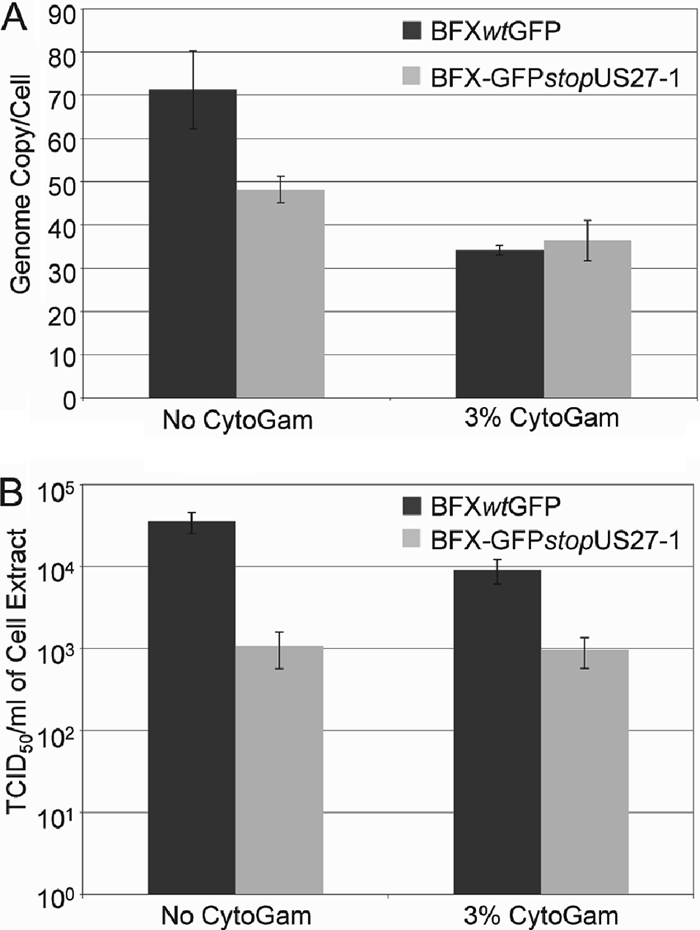
BFX-GFPstopUS27 depends on direct cell-to-cell spread. Fibroblasts were infected with either the wild type or the US27 mutant at a multiplicity of 0.01 PFU/cell and maintained for 15 days in the presence (+) or absence (−) of the neutralizing antibody CytoGam. (A) Intracellular DNA was assayed to monitor virus spread. DNA was quantified by qPCR and normalized to β-actin. Assays were performed in triplicate. (B) Cell-associated virus was assayed to monitor virus spread. Infected cells were lysed, and virus yields were measured in triplicate by TCID50 assay.
To further test for a role of pUS27 in extracellular spread of the virus, we infected epithelial and endothelial cells with wild-type, TB40/Ewt-mCherry, and a pUS27-deficient mutant derivative, TB40/E-mCherrydlUS27, of an endothelial cell-tropic, clinical isolate, TB40/E (25). While HCMV spreads both extracellularly and by cell to cell in cultured fibroblasts, infection of primary retinal pigmented epithelial cells (ARPE-19 cells) with TB40/Ewt-mCherry resulted in exclusively cell-to-cell spread (Fig. 7A). We detected no virus released into the medium of infected ARPE-19 cells over a 30-day time course. We therefore hypothesized that infecting epithelial cells with TB40/E-mCherrydlUS27 would result in no defect in virus replication compared to infection with wild-type TB40/Ewt-mCherry since virus production in a culture infected at a low input multiplicity would be dependent entirely on cell-to-cell spread. We collected infected cells at each time point and determined the relative titer of cell-associated virus. The TB40/E-mCherrydlUS27 mutant virus grew similarly to TB40/Ewt-mCherry in ARPE-19 cells (Fig. 7B). To assess the growth of this mutant in human umbilical vascular endothelial cells (HUVECs), where the extracellular virus is normally produced (25), we quantified infectious virus in the medium at each time point. Similar to results with fibroblast infections, we observed significantly reduced titers of extracellular virus in HUVECs infected with the TB40/E-mCherrydlUS27 mutant virus compared to infection with its wild-type counterpart (Fig. 7C).
Fig. 7.
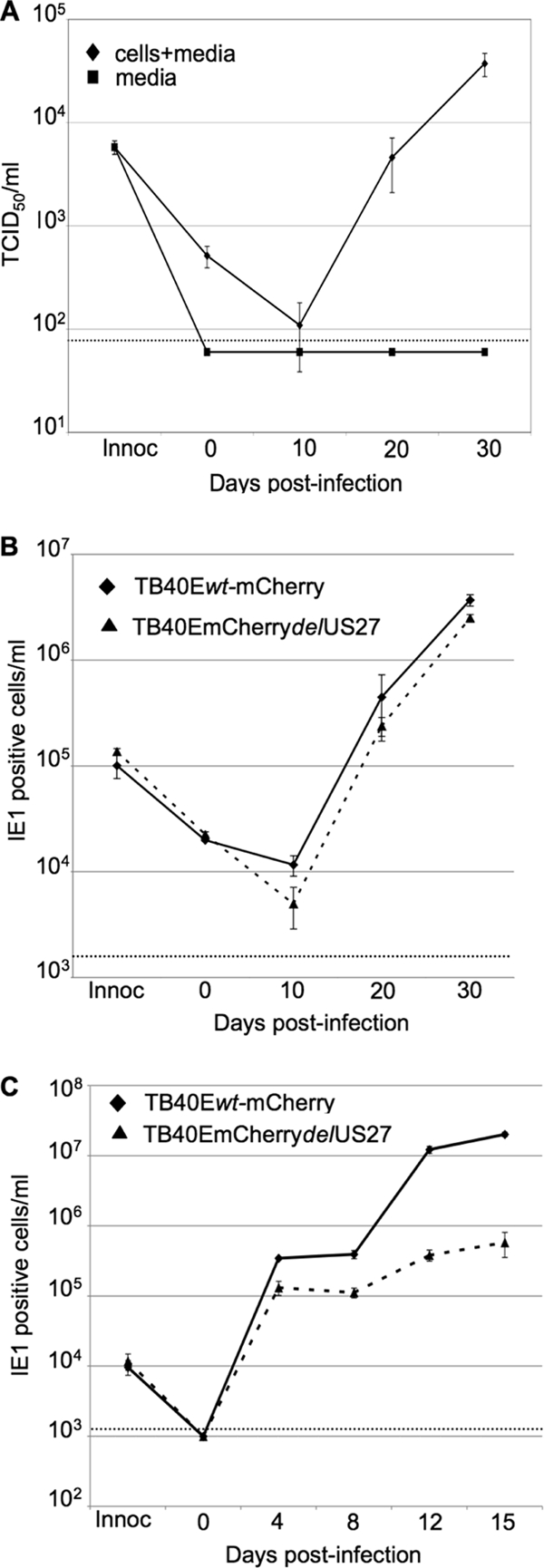
TB40/E-mCherrydlUS27 produces normal extracellular virus like wild-type virus in epithelial cells but displays a growth defect in endothelial cells. Cells were infected at a multiplicity of 0.1 PFU/cell with wild-type or mutant virus. Virus yields were assayed in triplicate, and a dashed line denotes the level of detection for each assay. (A) TB40/Ewt-mCherry remains substantially cell associated in epithelial cells. ARPE cells were infected at a multiplicity of 0.1 PFU/ml. At the indicated times after infection of ARPE-19 cells, titers were determined by TCID50 assay for virus released into the medium (squares) and virus that remained cell associated (triangles). (B) TB40/E-mCherrydlUS27 generates normal yields of cell-associated virus. At various times after infection of ARPE-19 cells, cell-associated virus was assayed by infecting fibroblasts and quantifying IE1-positive cells 24 h later by immunofluorescence. (C) TB40/E-mCherrydlUS27 exhibits a defect in the production of extracellular virus after infection of HUVECs. Cell-free virus was assayed by infecting fibroblasts and quantifying IE1-positive cells 24 h later by immunofluorescence.
Taken together, our results demonstrate that pUS27 facilitates extracellular spread but is not required for direct cell-to-cell spread of HCMV.
DISCUSSION
HCMV can spread from infected to uninfected cells either through the production of extracellular virus particles that subsequently bind and enter another cell or directly from cell to cell with little or no exposure of virus to the extracellular environment (13, 16, 22, 23). The principle conclusion of this work is that pUS27 is required for efficient spread of HCMV via the extracellular route. In fibroblasts, a pUS27-deficient derivative of the FIX clinical isolate of HCMV generated normal amounts of intracellular viral DNA and representative proteins (Fig. 4B and C), it localized a late viral protein properly to the cytoplasmic viral assembly zone (Fig. 5), and its virions exhibited normal infectivity (Fig. 3C). However, the mutant virus generated about 10-fold less extracellular progeny than its wild-type parent (Fig. 3B), and spread of the mutant within a culture of fibroblasts after infection at a low input multiplicity was inhibited to a significantly lesser extent than spread of its wild-type parent by neutralizing antibody (Fig. 6). This result argues that wild-type FIX virus spreads within fibroblast cultures by two routes: through production of extracellular virions that are sensitive to neutralizing antibody and by direct cell-to-cell spread that is resistant to the antibody. In contrast, the mutant spreads primarily by the direct cell-to-cell route. This interpretation of the mutant phenotype was reinforced by analysis of a pUS27-deficient derivative of a second HCMV clinical isolate, TB40/E. In endothelial cells, where wild-type TB40/E produced extracellular virus, the mutant exhibited a defect in the production of progeny (Fig. 7C). However, in epithelial cells, where the wild-type parent produced little extracellular virus (Fig. 7A), the mutant grew nearly as well as the wild type (Fig. 7B).
Mutations in several other HCMV genes have been shown to block the production of extracellular virus while allowing continued cell-to-cell spread. A virus unable to produce the tegument protein pUL99 failed to assemble virions and to produce detectable extracellular virus (24) but nevertheless continued to spread directly from cell to cell (23). Since enveloped particles were not observed within cells infected with the pUL99-deficient mutant, it was proposed that the tegumented capsids might spread to neighboring cells. Similarly, mutations in two virus-encoded glycoproteins, UL73 (gN) (16) and UL74 (gO) (13), have been shown to decrease the production of extracellular virus without completely blocking spread. Like the pUL99-deficient mutant, both of the mutants lacking functional glycoproteins failed to efficiently generate enveloped virions within the cytoplasm of infected cells.
What is the mechanism by which pUS27 facilitates the production of extracellular virus? It could act indirectly by influencing the accumulation of another virus-encoded protein. We did not observe changes in the levels of the small set of representative proteins studied after infection with a pUS27-deficient mutant (Fig. 4B), but it remains possible that the accumulation of a another protein was reduced, and perhaps it, in turn, functions in the production of extracellular virus. We favor the interpretation, however, that pUL27 acts directly at a late stage of the virus replication cycle, given its localization within the cytoplasmic assembly zone (Fig. 2A) and its presence in the envelope of virions (17, 29) (Fig. 2B). Intracellular as well as extracellular infectivity was reduced after infection with a pUS27-deficient mutant (Fig. 4A). So, if pUS27 acts directly at a very late stage of infection, it could facilitate the assembly or egress of virus. The modest reduction in intracellular infectivity in the absence of pUS27 might not reflect a defect in assembly because it is possible that assembled particles are degraded if they are not transported out of the cell or are transported incorrectly.
If pUS27 acts to facilitate virus assembly and/or egress, we can envision two general mechanisms. Perhaps it functions directly to support some aspect of the virion maturation/egress process. For example, it could interact with other cell- or virus-encoded proteins required for assembly of the virion, much like UL73 (gN) (16) and UL74 (gO) (13), which are needed for envelopment of particles within the assembly zone. Alternatively, since pUS27 is a GPCR homologue, it might perform an as yet unidentified signaling function that stimulates or regulates some aspect of the assembly or egress process. As noted earlier, pUS27 has not been shown to signal constitutively when expressed within transfected cells (31), and no ligand has been reported. Conceivably, its putative signaling function is activated within the viral assembly zone in response to an interaction with one or more virus-encoded proteins.
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (CA82396 and AI63335). C.M.O. was supported by fellowships from the New Jersey Commission on Cancer Research (09-1960-CCR-EO) and the American Cancer Society (PF-10-164-01-MPC).
We thank C. Sinzger (Eberhard Karls Universität, Tübingen, Germany) for the generous gift of TB40/E.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Attwood T. K., Findlay J. B. 1994. Fingerprinting G-protein-coupled receptors. Protein Eng. 7:195–203 [DOI] [PubMed] [Google Scholar]
- 2. Beisser P. S., Lavreysen H., Bruggeman C. A., Vink C. 2008. Chemokines and chemokine receptors encoded by cytomegaloviruses. Curr. Top. Microbiol. Immunol. 325:221–242 [DOI] [PubMed] [Google Scholar]
- 3. Billstrom M. A., Johnson G. L., Avdi N. J., Worthen G. S. 1998. Intracellular signaling by the chemokine receptor US28 during human cytomegalovirus infection. J. Virol. 72:5535–5544 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bodaghi B., et al. 1998. Chemokine sequestration by viral chemoreceptors as a novel viral escape strategy: withdrawal of chemokines from the environment of cytomegalovirus-infected cells. J. Exp. Med. 188:855–866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Britt W. 2008. Manifestations of human cytomegalovirus infection: proposed mechanisms of acute and chronic disease. Curr. Top. Microbiol. Immunol. 325:417–470 [DOI] [PubMed] [Google Scholar]
- 6. Browne E. P., Wing B., Coleman D., Shenk T. 2001. Altered cellular mRNA levels in human cytomegalovirus-infected fibroblasts: viral block to the accumulation of antiviral mRNAs. J. Virol. 75:12319–12330 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Casarosa P., et al. 2001. Constitutive signaling of the human cytomegalovirus-encoded chemokine receptor US28. J. Biol. Chem. 276:1133–1137 [DOI] [PubMed] [Google Scholar]
- 8. Chee M. S., et al. 1990. Analysis of the protein-coding content of the sequence of human cytomegalovirus strain AD169. Curr. Top. Microbiol. Immunol. 154:125–169 [DOI] [PubMed] [Google Scholar]
- 9. Fraile-Ramos A., et al. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218–232 [DOI] [PubMed] [Google Scholar]
- 10. Gao J. L., Murphy P. M. 1994. Human cytomegalovirus open reading frame US28 encodes a functional beta chemokine receptor. J. Biol. Chem. 269:28539–28542 [PubMed] [Google Scholar]
- 11. Hahn G., et al. 2002. The human cytomegalovirus ribonucleotide reductase homolog UL45 is dispensable for growth in endothelial cells, as determined by a BAC-cloned clinical isolate of human cytomegalovirus with preserved wild-type characteristics. J. Virol. 76:9551–9555 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Heitzler D., et al. 2009. Towards a systems biology approach of-G protein-coupled receptor signalling: challenges and expectations. C. R. Biol. 332:947–957 [DOI] [PubMed] [Google Scholar]
- 13. Jiang X. J., et al. 2008. UL74 of human cytomegalovirus contributes to virus release by promoting secondary envelopment of virions. J. Virol. 82:2802–2812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kuhn D. E., Beall C. J., Kolattukudy P. E. 1995. The cytomegalovirus US28 protein binds multiple CC chemokines with high affinity. Biochem. Biophys. Res. Commun. 211:325–330 [DOI] [PubMed] [Google Scholar]
- 15. Lee E. C., et al. 2001. A highly efficient Escherichia coli-based chromosome engineering system adapted for recombinogenic targeting and subcloning of BAC DNA. Genomics 73:56–65 [DOI] [PubMed] [Google Scholar]
- 16. Mach M., et al. 2007. The carboxy-terminal domain of glycoprotein N of human cytomegalovirus is required for virion morphogenesis. J. Virol. 81:5212–5224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Margulies B. J., Gibson W. 2007. The chemokine receptor homologue encoded by US27 of human cytomegalovirus is heavily glycosylated and is present in infected human foreskin fibroblasts and enveloped virus particles. Virus Res. 123:57–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Maussang D., et al. 2006. Human cytomegalovirus-encoded chemokine receptor US28 promotes tumorigenesis. Proc. Natl. Acad. Sci. U. S. A. 103:13068–13073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Murphy E., Vanicek J., Robins H., Shenk T., Levine A. J. 2008. Suppression of immediate-early viral gene expression by herpesvirus-coded microRNAs: implications for latency. Proc. Natl. Acad. Sci. U. S. A. 105:5453–5458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Reference deleted.
- 21. Sanchez V., Greis K. D., Sztul E., Britt W. J. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Schroer J., Shenk T. 2008. Inhibition of cyclooxygenase activity blocks cell-to-cell spread of human cytomegalovirus. Proc. Natl. Acad. Sci. U. S. A. 105:19468–19473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Silva M. C., Schroer J., Shenk T. 2005. Human cytomegalovirus cell-to-cell spread in the absence of an essential assembly protein. Proc. Natl. Acad. Sci. U. S. A. 102:2081–2086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Silva M. C., Yu Q. C., Enquist L., Shenk T. 2003. Human cytomegalovirus UL99-encoded pp28 is required for the cytoplasmic envelopment of tegument-associated capsids. J. Virol. 77:10594–10605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Sinzger C., et al. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359–368 [DOI] [PubMed] [Google Scholar]
- 26. Sodhi A., Montaner S., Gutkind J. S. 2004. Viral hijacking of G-protein-coupled-receptor signalling networks. Nat. Rev. Mol. Cell Biol. 5:998–1012 [DOI] [PubMed] [Google Scholar]
- 27. Streblow D. N., et al. 1999. The human cytomegalovirus chemokine receptor US28 mediates vascular smooth muscle cell migration. Cell 99:511–520 [DOI] [PubMed] [Google Scholar]
- 28. Terhune S., et al. 2007. Human cytomegalovirus UL38 protein blocks apoptosis. J. Virol. 81:3109–3123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Vomaske J., Nelson J. A., Streblow D. N. 2009. Human Cytomegalovirus US28: a functionally selective chemokine binding receptor. Infect. Disord. Drug Targets 9:548–556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Waldhoer M., Kledal T. N., Farrell H., Schwartz T. W. 2002. Murine cytomegalovirus (CMV) M33 and human CMV US28 receptors exhibit similar constitutive signaling activities. J. Virol. 76:8161–8168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Wang D., Shenk T. 2005. Human cytomegalovirus virion protein complex required for epithelial and endothelial cell tropism. Proc. Natl. Acad. Sci. U. S. A. 102:18153–18158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Warming S., Costantino N., Court D. L., Jenkins N. A., Copeland N. G. 2005. Simple and highly efficient BAC recombineering using galK selection. Nucleic Acids Res. 33:e36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Zhu H., Shen Y., Shenk T. 1995. Human cytomegalovirus IE1 and IE2 proteins block apoptosis. J. Virol. 69:7960–7970 [DOI] [PMC free article] [PubMed] [Google Scholar]