Abstract
Among Old World monkeys, pig-tailed macaques (Pt) are uniquely susceptible to human immunodeficiency virus type 1 (HIV-1), although the infection does not persist. We demonstrate that the susceptibility of Pt T cells to HIV-1 infection is due to the absence of postentry inhibition by a TRIM5 isoform. Notably, substitution of the viral infectivity factor protein, Vif, with that from pathogenic SIVmne enabled replication of HIV-1 in Pt T cells in vitro. When inoculated into juvenile pig-tailed macaques, the Pt-tropic HIV-1 persistently replicated for more than 1.5 to 2 years, producing low but measurable plasma viral loads and persistent proviral DNA in peripheral blood mononuclear cells. It also elicited strong antibody responses. However, there was no decline in CD4+ T cells or evidence of disease. Surprisingly, the Pt-tropic HIV-1 was rapidly controlled when inoculated into newborn Pt macaques, although it transiently rebounded after 6 months. We identified two notable differences between the Pt-tropic HIV-1 and SIVmne. First, SIV Vif does not associate with Pt-tropic HIV-1 viral particles. Second, while Pt-tropic HIV-1 degrades both Pt APOBEC3G and APOBEC3F, it prevents their inclusion in virions to a lesser extent than pathogenic SIVmne. Thus, while SIV Vif is necessary for persistent infection by Pt-tropic HIV-1, improved expression and inhibition of APOBEC3 proteins may be required for robust viral replication in vivo. Additional adaptation of the virus may also be necessary to enhance viral replication. Nevertheless, our data suggest the potential for the pig-tailed macaque to be developed as an animal model of HIV-1 infection and disease.
INTRODUCTION
The AIDS epidemic is primarily caused by human immunodeficiency virus type 1 (HIV-1), a virus highly specific and pathogenic for humans that evolved after serial human-to-human passage of ancestral simian immunodeficiency virus (SIV) from chimpanzees (SIVcpz) (30). This species-specific tropism presents a major problem for AIDS research because it complicates the development of an animal model of HIV-1 infection and disease that can facilitate preclinical testing of novel antiviral therapies, vaccines, and treatment strategies. Indeed, HIV-1 does not infect most nonhuman primate species, including rhesus macaques (Rh), which are commonly used for AIDS research. Consequently, SIV infection of Asian macaques or SIV/HIV-1 chimeric viruses (SHIVs) containing the HIV-1 envelope or reverse transcriptase are the most commonly used models of HIV-1 infection for the study of disease, proof-of-concept vaccine strategies, and antiretroviral therapy (5). However, while these models have been informative, they have shortcomings that result from the genetic differences between HIV-1 and SIV. The absence of other HIV-1 genes in SHIVs prevents (i) the evaluation of the virally encoded proteins in replication or as antiviral targets in vivo; (ii) efficacy testing of some drugs, which are inactive against SIV enzymes; and (iii) vaccine testing against epitopes found only in HIV-1.
The resistance of nonhuman primates to HIV-1 also provides a useful model for understanding mechanisms of innate immunity to retroviral infection. Recent studies with rhesus macaque cells have shown that two key cellular factors influence cross-species infection by inhibiting replication of the transmitted virus. The first is mediated by Rh TRIM5α, which engages the HIV-1 capsid protein and interferes with its postentry activities in replication (83). The second is caused by the Rh cytidine deaminase, APOBEC3G (A3G), which the HIV-1 Vif protein is incapable of recognizing and inhibiting (29, 57, 78). Overcoming these blocks by functional replacement of the HIV-1 capsid and vif sequences with those from SIV enable robust replication in rhesus macaque T cells, but only after repeated in vitro passage (29, 39). An alternative macaque species, Macaca nemestrina (pig-tailed macaque [Pt]), may be a more suitable host for HIV-1 animal model development. In contrast to other macaque species, pig-tails have been shown to support an acute, but limited, infection by HIV-1 (2, 3, 23, 41). This greater susceptibility of Pt cells may be due to the absence of a TRIM5 isoform restricting postentry events in HIV-1 replication (13, 14, 63, 89, 91). These data support earlier in vitro and in vivo experiments, which suggest there may be fewer restrictions to HIV-1 replication in pig-tails (12, 23, 24, 47). Thus, a minimally modified HIV-1 may potentially be more easily engineered to replicate in this species.
In this study, we generated a Pt-tropic HIV-1 clone by substitution of the vif gene with that from a pathogenic SIVmne clone. We show that this chimeric virus (HSIV-vif) replicates in Pt peripheral blood mononuclear cells (PBMC) and that it persistently replicates in the pig-tail host for up to 2 years. Containment was in part associated with loss of recovery of replication-competent virus. Furthermore, incomplete degradation of A3 proteins by the Pt-tropic HIV-1 may underlie its eventual control by the host.
MATERIALS AND METHODS
Cell lines.
The 293T and CrFK cell lines were cultured in Dulbecco modified Eagle medium (DMEM) supplemented with 10% heat-inactivated fetal bovine serum (HI-FBS), 2 mM glutamine, 100 U of penicillin/ml, and 100 μg of streptomycin/ml (DMEM complete). TZM-bl cells were obtained from the NIH AIDS Reagent Program and grown in DMEM complete (20, 90). MAGI-CCR5 cells were maintained as described previously (72).
Construction of HIV-1/SIV vif chimeras.
To generate the HIV-1/SIV chimera (HSIV-vif) that replaces the HIV-1 Vif with that from SIV, we used overlap extension PCR to substitute the vif gene in the proviral sequence of NL4-3 (1) with that from SIVmne027 (44, 45). The HIV-1 Vif start codon and two additional Met codons upstream of the SIV Vif start codon were mutated by QuikChange mutagenesis (Stratagene). Additional HIV-1/SIV vif chimeras based on the HIV-1 molecular clones Bru-Yu2 (60) and NL-AD8 (22) were constructed in a similar manner.
Cloning, expression, and assay of activity of pig-tail TRIM5 isoforms.
The predominantly expressed cDNA of pig-tail TRIM5 isoform, TRIM5η, was cloned by two-step reverse transcription-PCR (RT-PCR). First-strand cDNA was generated from total RNA of pig-tailed macaque PBMC using the PCR primer complementary to the 3′ end of the TRIM5 coding sequence and Superscript III (Invitrogen, Carlsbad, CA). PCR amplification was performed using a Taq polymerase master mix (Invitrogen) and PCR primers based on the rhesus TRIM5α sequence (TRIM5.51, 5′-GTGGAGAAGCTGCTATGGCTTCTGGAATCCTGCTTAATG-3′; TRIM5.31, 5′-GGGGCTGAGTGTGTAAGAAGGTTCAAGAGCTTGGTGAG-3′). Amplification was initiated at 94°C for 5 min and followed by 35 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 90 s. Amplified products were inserted into a TOPO cloning vector (Invitrogen), and the sequences were determined.
Pt TRIM5η activity was examined in CCR5-MAGI cells (72) because it was initially demonstrated that the postentry inhibitory effects of rhesus TRIM5α could be observed in HeLa-derived HIV-1 indicator cells (83). Stable cell lines expressing TRIM5α from humans or rhesus macaques or TRIM5η from pig-tailed macaques were generated by transducing CCR5-MAGI with the pBabe-blasti vector encoding the TRIM5 cDNAs epitope tagged on the 5′ end with 3×-FLAG and hemagglutinin (HA) on the 3′ end. Western blotting with an antibody against FLAG was used to confirm expression of the TRIM5 proteins. Antibody directed against β-actin served as a protein loading control. Initially, infections of the TRIM5 expressing and control cells were performed with increasing amounts (2 to 100 ng) of HIV-GFP/VSV pseudotyped virus (51). Two days after infection, the cells were analyzed by flow cytometry for green fluorescent protein (GFP) expression.
CrFK cells expressing TRIM-Cyp from pig-tailed macaques and owl monkeys were obtained from Michael Emerman (Fred Hutchinson Cancer Research Center) and Shiu-Lok Hu (University of Washington) (13). To examine restriction activity against HIV-1NL4-3, CrFK cells expressing either the pig-tail or owl monkey TRIM-Cyp or control cells were transduced with an HIV-1NL4-3-luciferase vector pseudotyped with VSV-G. At 2 days postinfection, the cells were lysed, and the luciferase activity was measured as an indicator of infection using a Promega luciferase assay system (Promega, Madison, WI) and a tube luminometer (Berthold). Infectivity was reported relative to the control CrFK cells.
Viral infection and replication assays.
PBMC were isolated from heparinized rhesus, pig-tail, or human peripheral blood by Ficoll gradient centrifugation and activated by costimulation with anti-CD3/anti-CD28 monoclonal antibodies and interleukin-2 as previously described (7, 9, 46). At 3 days after costimulation the cells were >98% CD3+ T cells, as indicated by flow cytometry.
The susceptibility of macaque T cells to HIV-1 was examined by transducing peripheral blood-derived T cells from rhesus and pig-tailed macaques with HIV-GFP pseudotyped with vesicular stomatitis virus-glycoprotein (VSV-G). Flow cytometry was used to determine the percentage of infected cells by measuring the number of GFP positive and negative cells.
For viral replication assays, infectious stocks of SIVmne027, HIV-1NL4-3, HIV-1Bru-Yu2, HIV-1NL-AD8, and HSIV-vif clones based on NL4-3, Bru-Yu2, or NL-AD8 were produced by transfection of 293T cells. Infectious titers were determined by limiting dilution infection analysis using TZM-bl indicator cells (20, 90). To compare viral replication in costimulated human or pig-tailed macaque PBMC, 106 cells were infected at a multiplicity of infection (MOI) of 0.01. After a 3-h incubation, the cells were washed twice with phosphate-buffered saline to remove unbound virus and then resuspended in fresh medium. To monitor viral replication, supernatants were harvested every 3 to 4 days for measurement of infectious virus by limiting dilution using TZM-bl cells. Alternatively, HIV-1 p24gag antigen or SIV p27gag antigen were quantified by enzyme-linked immunosorbent assay (ELISA; Zeptometrix, Buffalo, NY, or Advanced Bioscience Laboratories, Kensington, MD, respectively).
APOBECE3 protein cloning, expression, and activity.
Total RNA was isolated from the Pt PBMC by using an RNeasy minikit (Qiagen) according to the manufacturer's instructions. First-strand cDNA was then synthesized from total RNA with the SuperScript III first-strand synthesis system for an RT-PCR kit (Invitrogen) and used to amplify Pt A3G and Pt A3F cDNAs and add 3′ HA-epitope tags. The primers for Pt A3G amplification were PtA3G-F (5′-GAAACAATGGTGGAGCCAATGGATC-3′) and CEM15-HA (5′-TAGAAGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATAGTTTTCCTGATTCTGGAG-3′) (57). Primers for APOBEC3F amplification were A3F-ST (5′-CAAGGATGAAGCCTCACTTCAGAAAC-3′) and A3F-HA (5′-TAGAAGCTCGAGTCAAGCGTAATCTGGAACATCGTATGGATACTCGAGAATCTCCTGCAG-3′). PCR amplification was carried out using AmpliTaq Gold DNA polymerase (Applied Biosystems) under the following conditions: denaturation at 94°C for 2 min, followed by 30 cycles of 94°C for 30 s, 60°C for 30 s, and 72°C for 90 s. The amplified products were cloned into pCR2.1 TOPO vector (Invitrogen) for sequencing and transferred into mammalian expression vectors pcDNA3 or pCDNA3.1(−) (Invitrogen).
293T cells were cotransfected with HIV-1NL4-3, SIVmne027, or HIV-1/SIV vif chimeras and HA-tagged Pt A3G or Pt A3F expression vectors. At 48 h posttransfection, culture supernatants were spun at low speed to remove cellular debris and used to measure viral infectivity or concentrate viral particles by centrifugation at 23,600 × g for 1 h at 4°C. Protein lysates were prepared from the cells and virions. Proteins from virions or cells were separated by SDS-PAGE using (12 or 15%) Tris-HCl ready gels (Bio-Rad) and transferred to nitrocellulose membranes for Western blotting. Blots were probed overnight at 4°C with antibodies to HA (Roche), HIV-1 p24gag and SIV p27gag (monoclonal antibody 183-H12-5C) (19, 86), SIV Vif (custom rabbit antisera to the Vif N-terminal peptide EEKRWIAVPTWRIPERLER; Open Biosystems), or anti-β actin (Sigma). The infectivity of viral supernatants was determined by using TZM-bl reporter cells and luciferase assays. The relative infectivity was calculated by dividing the relative light units generated by infection of virions produced in the presence of Pt A3 proteins by those determined for the viruses produced in the absence of Pt A3 proteins.
Inoculation of pig-tailed macaques.
Two juvenile and two newborn pig-tailed macaques were inoculated intravenously with 105 TCID50 of HSIV-vif based on NL4-3. Animals were specific pathogen free for simian T lymphotropic virus type 1, SIV, simian retrovirus type D, and herpes B virus. At several time points postinoculation, peripheral blood was drawn for CD4+ T-cell count determinations and isolation of plasma, sera, and PBMC. All animals were housed and cared for in accordance with the guidelines of the American Association for Accreditation of Laboratory Animal Care and the Animal Care and Use Committee of the University of Washington.
Plasma viral loads, CD4+ T-cell counts, and antibody determinations.
Plasma viral load measurements were determined by using the Roche Amplicor HIV-1 monitor test, version 1.5 according to the manufacturer's protocol. CD4+ T-cell counts were determined as previously described (68). HIV-1-specific antibodies were measured by ELISA as previously described, except that gradient-purified and disrupted whole HIV-1 virions were used as the capture antigen (34, 68).
PCR cloning of HSIV-vif sequences from pig-tailed macaques.
The sequences of HSIV-vif gag, vif, env, and nef were amplified by nested PCR from DNA isolated from PBMC by using a QIAamp DNA minikit (Qiagen). Gag sequences were amplified with the primers HIV-gag-762 (5′-TTGACTAGCGGAGGCTAGAA-3′) and HIV-gag-1658 (5′-TTTGGTCCTTGTCTTATGTCCAGAATGC-3′) in round 1 and HIV-gag-836 (5′-GGGAAAAAATTCGGTTAAGGCC-3′) and HIV-gag-1610 in round 2. The same amplification conditions were used in both rounds: 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, 56°C for 30 s, and 72°C for 90 s. A final extension at 72°C for 10 min completed each round. Vif sequences were amplified with primers pol-4933 (5′-CAGGGACAGCAGAGATCCAGTTTG-3′) and vpr-5842 (5′-CTACTGGCTCCATTTCTTGCTCTC-3′) in round 1 and pol-4986 (5′-GAAAGGTGAAGGGGCAGTAGTAATAC-3′) and vpr-5666 (5′-GTTGTCCTAAGTTATGGAGCCATATC-3′) in round 2. PCR amplification for both rounds was initiated at 95°C for 5 min; this was followed with 35 cycles of 95°C for 1 min, 60°C for 1 min, and 72°C for 1 min, with a final extension at 72°C for 10 min. For both gag and vif reactions, PCR was carried out using a 250 nM concentration of each primer and AmpliTaq Gold DNA polymerase and buffer (Applied Biosystems). env sequences were amplified with previously described primers (65), but with AmpliTaq Gold DNA polymerase and buffer (Applied Biosystems). Each round of amplification was initiated at 94°C for 5 min, followed by 35 cycles of 94°C for 30 s, annealing for 30 s, and 72°C for 3 min. Annealing was performed at 45°C in the first round and 50°C in the second. A final extension for 5 min at 72°C completed each round. nef sequences were amplified using previously described primers and conditions (7, 66). For each primer set ∼1 μg of DNA was used per reaction. All PCR products were cloned into pCR2.1 TOPO and sequenced.
Proviral DNA sequence analysis.
For hypermutation analysis of env surface gene (env su) proviral sequences, we used Hypermut 2.0 (http://www.hiv.lanl.gov/content/sequence/HYPERMUT/hypermut.html) (71). The highlighter tool (http://www.hiv.lanl.gov/content/sequence/HIGHLIGHT/highlighter.html) was used to indicate mutations in aligned env su sequences.
GenBank accession numbers.
Sequences were deposited under numbers EU047926 to EU047928 (Pt TRIM5η), HQ007903 to HQ007913 (HSIV-vif partial env proviral sequences), HM991859 (Pt A3G), and HM991860 (Pt A3F).
RESULTS
Pt T cells are more susceptible than Rh T cells to HIV-1.
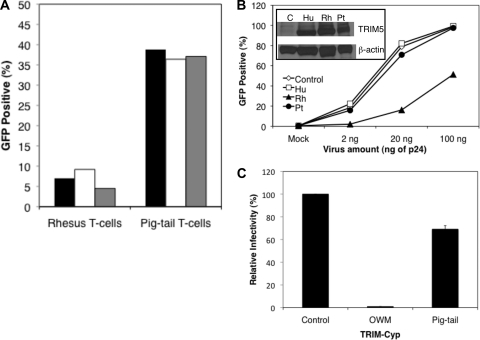
A previous study suggested that there was no early postentry restriction to HIV-1 infection of Pt T cells, which is in contrast to what has been reported for rhesus macaque cells (17, 31, 47, 79). We therefore initially compared the susceptibility of Pt and Rh cells to HIV-1 infection by transducing anti-CD3/anti-CD28 costimulated PBMC from each species with an HIV-1NL4-3 vector encoding GFP (HIV-GFP) pseudotyped with VSV-G. Interestingly, we observed a higher level of HIV-1 transduction in Pt T cells than in Rh T cells. The percentage of transduced cells was 36.4 to 39.8% in Pt PBMC versus 4.5 to 9.2% in Rh PBMC (Fig. 1A). In addition, the mean fluorescence intensity of GFP expression was ∼10-fold higher in Pt PBMC compared to Rh PBMC, suggesting a higher level of transduction and greater permissiveness of Pt cells than Rh cells to HIV-1 infection. These data demonstrate greater susceptibility of Pt T cells than Rh T cells to HIV-1 infection.
Fig. 1.
Infection of pig-tail cells with HIV-1 and restriction by TRIM5 isoforms. (A) PBMC costimulated with anti-CD3 and anti-CD28 from rhesus and pig-tailed macaques were transduced with HIV-GFP virus pseudotyped with VSV-G. The cells were obtained from three experimental donor animals of each species, as indicated by three different columns (black, white, and gray). After costimulation, >98% of the cells were T cells. (B) Transduction of MAGI indicator cells expressing human (Hu) or rhesus (Rh) TRIM5α or pig-tail (Pt) TRIM5η or control cells with increasing amounts of HIV-GFP pseudotyped with VSV-G. Infections were performed in duplicate and the average is shown. One of three representative experiments is shown. The inset shows a Western blot for flag-tagged TRIM5 proteins. (C) Transduction of control cells or cells expressing owl monkey (OWM) or pig-tail TRIM-Cyp with HIV-1NL4-3-luc/VSV-G. Infections were performed in triplicate. The average level of infection relative to the level of infection of the control cells is shown.
Recent studies demonstrated that pig-tailed macaques, in contrast to rhesus macaques, do not express the alpha isoform of TRIM5. Instead, two novel TRIM5 isoforms (η and θ), as well as a novel TRIM5-cyclophilin A (TRIM-Cyp) fusion protein, are synthesized (13, 14, 63, 89). TRIM5η lacks only exon 7 due to disruption of the intron 6 splice acceptor and alternative splicing, while TRIM5θ is prematurely truncated because of a frameshift in exon 7 (14). We confirmed the presence of the single nucleotide polymorphism in the intron 6 splice acceptor and the generation of the TRIM5 isoforms in pig-tails, and we tested whether the predominant novel TRIM5η or TRIM-CYP proteins from pig-tailed macaques would inhibit HIV-1 infection if overexpressed in the HeLa cell-derived HIV-1 indicator line, CCR5-MAGI. We found that in comparison to Rh TRIM5α, which inhibited HIV-1 infection as previously described (83), Pt TRIM5η was inefficient at blocking transduction by an HIV-GFP vector pseudotyped with VSV-G (Fig. 1B). The percentage of GFP positive Pt TRIM5η expressing cells was 2 to 8-fold greater than that of cells expressing Rh TRIM5α, depending on the amount of virus used for infection. However, it was similar to those percentages observed for the negative control and human TRIM5α-expressing cells. Furthermore, Pt TRIM-Cyp was limited in blocking transduction by HIV-1, unlike owl monkey TRIM-Cyp, which almost completely inhibited infection, as previously shown (76) (Fig. 1C). These data demonstrate that the greater susceptibility of pig-tailed macaque cells to HIV-1 infection may be in part due to the lack of postentry inhibition by TRIM5η and TRIM-Cyp.
SIV vif substitution is sufficient for HIV-1 replication in Pt T cells.
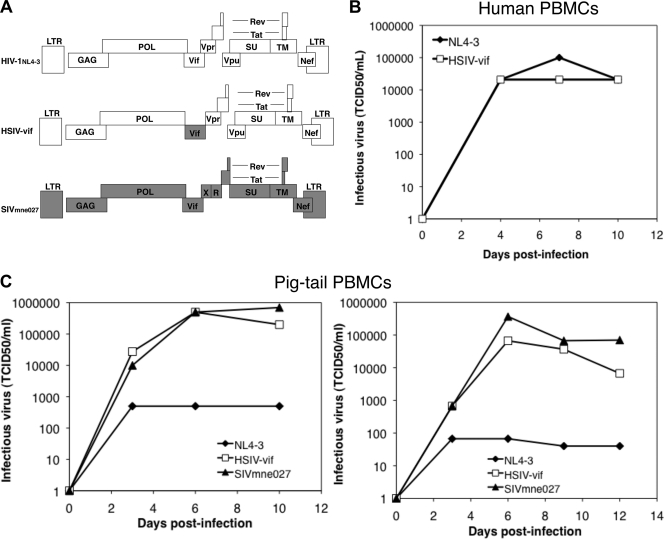
Given the absence of a postentry block to HIV-1 infection in Pt T cells, we hypothesized that the primary reason that HIV-1 fails to replicate in Pt macaque cells may be the inability of the HIV-1 Vif protein to counteract the antiviral activity of A3 proteins. If the hypothesis is correct, incorporation of the SIV Vif protein into HIV-1 should overcome the block and significantly improve viral replication. Thus, we generated an HIV-1/SIV vif chimera (HSIV-vif) by PCR-mediated overlap extension that replaces the HIV-1 vif of NL4-3 with that from SIVmne027 (Fig. 2A), a highly pathogenic SIVmne variant that evolved after serial passage of SIVmne in Pt macaques (44, 45). The SIVmne027 vif sequences were inserted in frame with the HIV-1 vif open reading frame but 3′ to the end of pol and just 5′ to vpr. The HIV-1 Vif start codon and two additional Met codons upstream of the SIV Vif start codon were mutated without altering the coding sequence for pol. Initially, to determine whether HSIV-vif could replicate, we infected human PBMC costimulated with anti-CD3 and anti-CD28 and assayed conditioned supernatants at 3- to 4-day intervals postinfection for infectious virus using TZM-bl indicator cells (20). We found that replication kinetics of HSIV-vif were similar to wild-type HIV-1NL4-3, indicating that incorporation of the SIV sequences did not adversely affect HIV-1 replication (Fig. 2B).
Fig. 2.
Replication of the HSIV-vif chimera. (A) Schematic diagram comparing the genomic structures of HIV-1NL4-3, HSIV-vif, and SIVmne027. (B) Replication of HIV-1NL4-3 and HSIV-vif in anti-CD3/anti-CD28-costimulated human PBMC. (C) Replication of HSIV-vif in comparison to SIVmne027 and HIV-1NL4-3 (NL4-3) in anti-CD3/anti-CD28-costimulated Pt PBMC. Duplicate infections were carried out at an MOI of 0.01. Replication was monitored by determining the amount of infectious virus produced per ml at 3- to 4-day intervals postinfection using TZM-bl reporter cells. Infections of two different macaque donors are shown and were performed using independently derived stocks of virus.
To examine whether the HSIV-vif chimera would replicate in primary Pt macaque cells, we infected PBMC from different macaque donors after costimulation with anti-CD3/anti-CD28 monoclonal antibodies. The amount of infectious virus produced from the HSIV-vif-infected cells increased to levels nearly as high as those observed in cells infected with the pathogenic, SIVmne027, and to a level almost 1,000-fold higher than wild-type HIV-1NL4-3, which remained low, suggesting that the inclusion of the SIV Vif protein enabled HIV-1NL4-3 to replicate in Pt PBMC and continuously produce infectious virus (Fig. 2C). To confirm that the virus produced in the HSIV-vif was HIV-1 in origin, we tested the supernatants for HIV-1 p24gag by antigen ELISA, which detects the HIV-1 but not SIV capsid protein. Concomitant with the accumulation of infectious virus, HIV-1 p24 antigen increased in HSIV-vif-infected but not in HIV-1NL4-3-infected Pt PBMC, verifying that replicating virus was HSIV-vif (data not shown). SIV p27gag was not detected in the HSIV-vif cultures, indicating that SIVmne027 contamination did not account for the increase in infectious virus. By comparison, SIV p27gag increased in association with the amount of infectious virus in the SIVmne027-infected cells (data not shown). Similar results were observed in experiments using PBMC derived from four additional donors. Notably, the high level of replication of HSIV-vif was achieved without passaging and recloning any part of the viral genome, suggesting that extensive adaptation of HIV-1 beyond the inclusion of the SIV Vif protein is unnecessary for robust viral replication in Pt T cells in vitro.
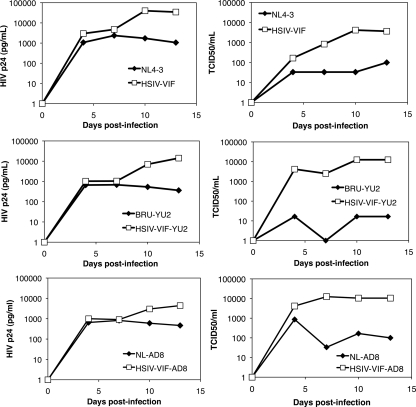
To verify that insertion of the SIV vif gene was necessary and sufficient for replication of HIV-1 in Pt T cells, we generated two additional HIV-1 vif chimeras based on the CCR5-tropic clones, HIV-1NL-AD8 and HIV-1Bru-Yu2, and tested their replication. Both clones (HSIV-vifAD8 and HSIV-vifYu2) were similar to the prototype HSIV-vif based on HIV-1NL4-3 and showed increasing production of Gag p24 protein and infectious virus from costimulated Pt T cells (Fig. 3), suggesting productive infection by these viruses. In contrast, the wild-type HIV-1 clones showed low p24 accumulation and decreasing amounts of infectious virus, suggesting loss of infectivity. Together, these data suggest that A3 proteins are the major impediment to HIV-1 replication in Pt cells and that a minimal SIV vif substitution is sufficient for high-level replication.
Fig. 3.
Replication of different HIV-1 vif chimeras. The replication kinetics of two additional HIV-1 chimeras with minimal vif (HSIV-vif-Yu2 and HSIV-vif-AD8) substitutions are shown in comparison to the HSIV-vif clone based on HIV-1NL4-3. The experiments were performed as described for Fig. 2D.
Replication of a HIV-1/SIV vif chimera in pig-tailed macaques.
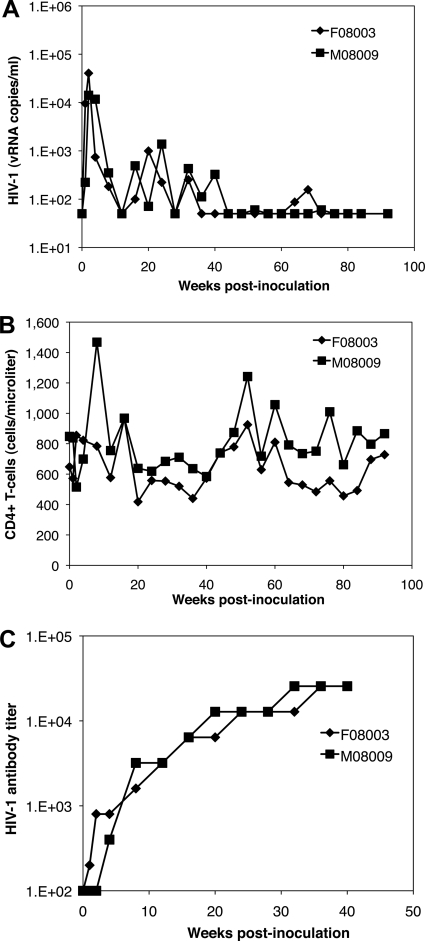
Previous attempts to infect pig-tailed macaques with HIV-1 showed that the molecular clone HIV-1NL4-3 could establish acute transient infections (2). Recent studies also demonstrated that capsid and vif modifications extended replication for up to 6 months (28, 35). We therefore determined if the SIV vif substitution was sufficient for persistent HIV-1 replication in vivo by intravenously inoculating two juvenile pig-tailed macaques with 105 50% tissue culture infectious doses (TCID50) of the prototype HSIV-vif based on HIV-1NL4-3 (Fig. 4). Plasma viral loads peaked at (1.4 to 4.04) × 104 viral RNA (vRNA) copies per ml at 2 weeks postinoculation (wpi) and decreased through 12 wpi to 50 vRNA copies/ml (Fig. 4A). The plasma viral loads then rebounded to levels between 100 and 1,400 vRNA copies/ml until 32 to 40 wpi, at which point they decreased again to levels below detection in plasma, except for the small peak at 72 wpi in animal M08009 and the increase between 64 and 72 wpi in animal F08003. CD4+ T-cell counts in peripheral blood demonstrated modest fluctuations, although there was no significant and prolonged decrease resulting from infection (Fig. 4B). To confirm infection, we measured antibody responses to HIV-1 by ELISA. Figure 4C shows a robust response against whole virus antigens and an increase in antibody titer to over 25,000. In addition, PBMC were PCR positive for gag, vif, env, and nef sequences through 92 wpi, verifying persistent infection (Table 1). However, plasma viral RNA was below the limit of detection at the late time points, some viral genes were difficult to detect by PCR at 92 wpi, and infectious virus could not be recovered after 64 wpi by cocultivation of infected macaque PBMC with PBMC from uninfected donor pig-tails (data not shown). Together, these data indicate that HSIV-vif replication is persistent in pig-tailed macaques for up to 2 years, but the level of infection is low.
Fig. 4.
Infection of juvenile pig-tailed macaques. Two juvenile pig-tails were inoculated intravenously with HSIV-vif based on HIV-1NL4-3. (A) Plasma viral load measurements; (B) CD3+ CD4+ T-cell counts in peripheral blood; (C) HIV-1 antibody titer.
Table 1.
Detection of HSIV-vif in PBMC by nested PCR
| Animal | wpia | Virus target sequencesb |
|||
|---|---|---|---|---|---|
| Gag | Vif | Env | Nef | ||
| F08003 | 2 | + | NT | +/− | NT |
| 80 | + | + | + | + | |
| 92 | + | − | + | − | |
| M08009 | 2 | + | NT | +/− | NT |
| 32 | + | + | + | + | |
| 80 | + | + | + | + | |
| 92 | +/− | + | + | + | |
| K09186 | 24 | + | − | + | + |
| 40 | + | + | + | + | |
| T09187 | 24 | + | + | + | + |
| 40 | + | − | + | + | |
wpi, weeks postinoculation.
Proviral sequences in PBMC from HSIV-vif-infected pigtails were detected by nested-PCR amplification. +, PCR amplification of viral sequence; −, no amplification of viral sequence; +/−, weak amplification of the targeted sequence. NT, not tested.
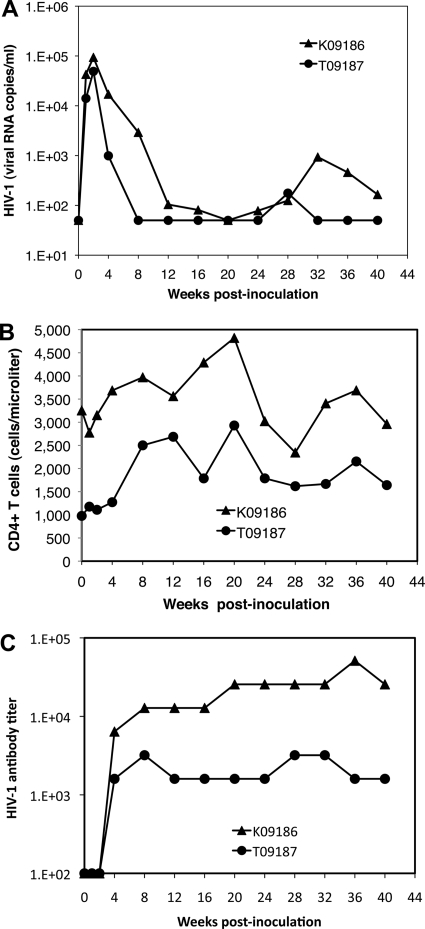
Accessory genes of HIV-1 and SIV play critical roles for enhancing viral replication by increasing viral infectivity and counteracting the host innate and adaptive immune responses (reviewed in references 6 and 54). For example, SIVmac239 with deletions in nef and vpr was shown to be highly attenuated in adult rhesus macaques (92). However, this virus caused rapid disease in newborn rhesus macaques (8), indicating that these accessory genes are not essential for robust replication and disease in newborns. We therefore hypothesized that even if HIV-1 accessory genes such as vpr and nef lack critical functional activities, leading to the control of HSIV-vif replication in juvenile pig-tailed macaques, newborns may be more susceptible to infection and disease. To test this hypothesis, we intravenously inoculated two newborn pig-tails with HSIV-vif and monitored the infections. Interestingly, while the peak plasma viral loads at 2 wpi were slightly higher than in the juveniles (0.5 × 105 to 1.0 × 105 vRNA copies/ml), they rapidly declined and were below the limit of detection within 8 to 20 wpi, indicating faster control of the infection by the host (Fig. 5A). Plasma viral loads modestly rebounded to levels between 100 and 1,000 vRNA copies/ml after 24 wpi. However, there were no significant declines in CD4+ T cells in either of the animals (Fig. 5B). Anti-HIV-1 antibody levels were induced to high-levels by HSIV-vif infection in animal K09186, but they were >10-fold lower in animal T09187, which demonstrated greater control of HSIV-vif (Fig. 5C). These data suggest that HSIV-vif does not disrupt host resistance enough for persistent high-level replication in the host, as predicted by the initial cell culture experiments. Importantly, this may be distinct from the possible absence of HIV-1 Nef, Vpu, and Vpr functions that could contribute to control of viral replication in the juvenile pig-tailed macaques.
Fig. 5.
Infection of newborn pig-tailed macaques. Two newborn pig-tails were inoculated intravenously with HSIV-vif based on HIV-1NL4-3. (A) Plasma viral loads; (B) CD3+ CD4+ T-cell counts in peripheral blood; (C) HIV-1 antibody titer.
Inhibition of APOBEC3 proteins by Pt-tropic HIV-1 may be suboptimal for replication in the host.
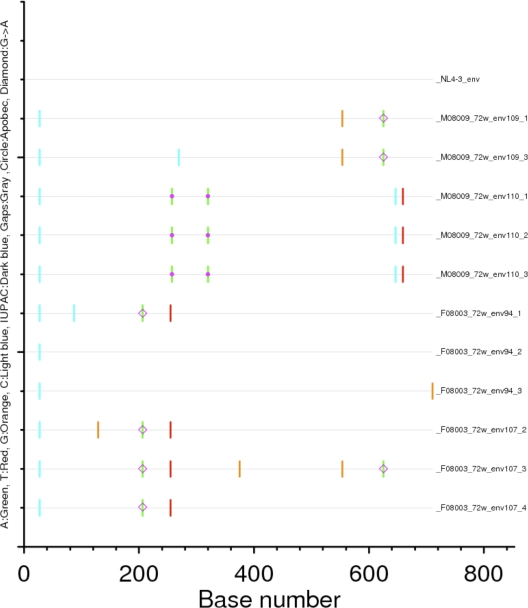
Because plasma viral loads rapidly decreased in both juvenile and newborn pig-tailed macaques, we hypothesized that HSIV-vif may not be as effective as SIVmne027 at inducing degradation of Pt A3 proteins. To address whether control of viral replication may be due Pt A3-mediated restriction, we initially examined envelope surface gene (env su) sequences cloned from PBMC of the infected juvenile pig-tails at 72 wpi for evidence of hypermutation. While G-to-A mutations were observed in most of the env genes, there were relatively few mutations overall, and those potentially attributable to Pt A3 proteins were found in only three clones (Fig. 6). In addition, relatively few mutations were also observed in gag, vif, and nef alleles at 72 wpi. The limited number of mutations likely reflects the low level of viral replication during the later stages of infection. However, even at 2 wpi when the plasma viral loads peaked, gag remained highly conserved with no indication of hypermutation (data not shown).
Fig. 6.
Summary of mutations in the env su gene. env su sequences were cloned from PBMC DNA isolated 72 weeks postinoculation from both juvenile pig-tailed macaques. Mutations relative to the HIV-1NL4-3 env su sequence are shown. Base number 0 is position 178 in the NL4-3 env sequence. The color code for nucleotide base changes and mutation symbols are provided on the vertical axis. Alignment and highlighted mutations were generated by using Highlighter (http://www.hiv.lanl.gov/content/sequence/highlight/highlighter.html).
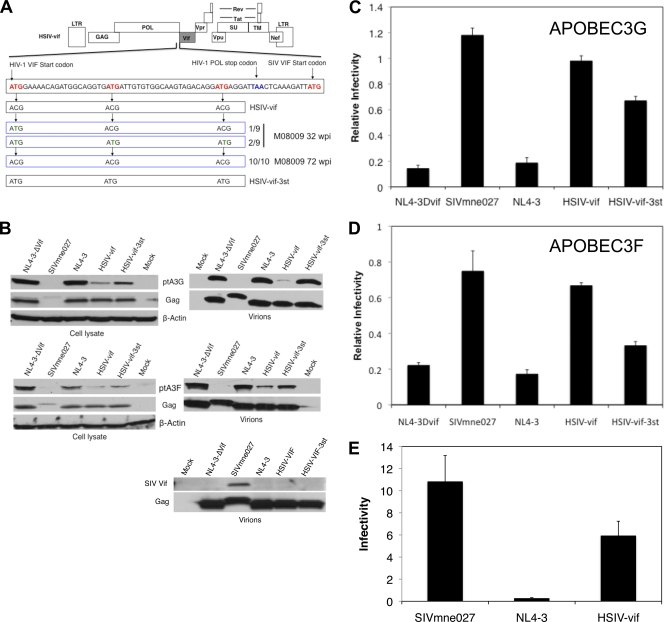
We further investigated whether SIV Vif expression from HSIV-vif may be sufficient by examining the SIV vif gene and sequences surrounding it for mutations that might alter expression. Notably, at 32 wpi in juvenile pig-tail M08009, we found reversion of the HIV-1 vif translational start site and the downstream ATG sites in a minority of clones (3 of 9) isolated from PBMC (Fig. 7A), suggesting insufficient expression of the SIV Vif protein from the parental HSIV-vif chimera. However, by 72 wpi, the reversions had disappeared. There was also no evidence for reversion of the codons in proviral sequences cloned from animal F08003 (data not shown). Together, the data suggest that the HIV-1 Vif start codon may not improve SIV Vif expression.
Fig. 7.
Vif start site mutations and degradation of Pt APOBEC3 proteins. (A) Diagram of the sequences between the HIV-1 vif gene start site and SIV vif gene start site. In HSIV-vif, each ATG within the HIV-1 vif sequence was changed to ACG. Reversion mutations observed in the HIV-1 vif met codons located 5′ to the SIV vif start site at 32 and 72 weeks postinoculation are shown. A mutant of HSIV-vif with the HIV-1 vif met codons (HSIV-vif-3st) is also shown. (B) Vif protein expression and degradation of Pt APOBEC3 proteins by SIVmne027, HIV-1NL4-3, NL4-3ΔVif, HSIV-vif, and HSIV-vif-3st. Each provirus was cotransfected into 293T cells with HA-tagged Pt A3G or Pt A3F. Protein expression in cell lysates or concentrated virions prepared from supernatants was determined by immunoblotting with anti-HA, anti-Gag p24 antibody, or a control antibody against β-actin. (C and D) The relative infectivity of each clone was determined by comparing infectivity in the presence or absence of Pt A3G (C) or Pt A3F (D). Infections were performed in quadruplicate, and means ± the standard errors of the mean are shown. (E) Infectivity of viruses produced from Pt PBMC. The infectivity of each virus was determined using supernatants harvested at 3 to 7 days postinfection with HSIV-vif, HIV-1NL4-3, or SIVmne027. Infectivity is defined as the average TCID50 per pg of p24 (HIV-1NL4-3 or HSIV-vif) or p27 (SIVmne027) ± the standard deviations of five independent experiments.
To examine the effect of SIV Vif on Pt A3 protein degradation and incorporation into virions, we compared Pt A3 protein inhibition by HSIV-vif with SIVmne027, HIV-1NL4-3 (NL4-3), a vif gene deletion mutant (NL4-3Dvif or NL4-3-ΔVif), and a mutant containing reversions of the HIV-1 Vif start codon and two downstream met codons (HSIV-vif-3st; Fig. 7A). Interestingly, HSIV-vif degraded both Pt A3G and Pt A3F but not to the extent of SIVmne027, leading to greater incorporation of these Pt A3 proteins in virions (Fig. 7B). In addition, there was a notable absence of SIV Vif associated with HSIV-vif viral particles compared to SIVmne027 particles. Whether this results from a failure of SIV Vif packaging by HIV-1 core particles or low Vif expression is unclear because the anti-Vif antiserum did not detect the SIV Vif protein in lysates of cells transfected with the proviral clones (data not shown). These data suggest that either an incomplete inhibition of Pt A3 proteins or a lack of Vif incorporation, or both, may contribute to the poor replication fitness of HSIV-vif in pig-tailed macaques.
To examine the effect of Pt A3 protein incorporation, we compared the relative infectivity of the viruses produced in the presence or absence of Pt A3G or Pt A3F in transient-transfection assays. Surprisingly, we did not observe significant decreases in relative viral infectivity when comparing SIVmne027 and HSIV-vif by this method (Fig. 7C and D). On the other hand, HSIV-vif-3st was less able to inhibit Pt A3G and Pt A3F than either wild-type HSIV-vif or SIVmne027. This was also reflected in virions, which showed greater amounts of both Pt A3G and Pt A3F and a significant decrease in infectivity (Fig. 7B to D).
To further investigate whether there are differences in infectivity of HSIV-vif and SIVmne027, we measured the infectivity of virus produced from costimulated Pt PBMC. Unlike the transfection experiments, HSIV-vif produced from infected Pt PBMC appeared to be 50% less infectious in comparison to SIVmne027 (Fig. 7E). Both viruses were also more infectious than wild-type HIV-1NL4-3 produced from Pt PBMC. Together, these data suggest that while including the SIV vif in HIV-1 greatly improves replication in pig-tail cells in vitro and persistent replication in vivo, the level of expression may not be sufficient for persistent high-level viral replication.
DISCUSSION
Earlier studies showed that pig-tailed macaques are partly susceptible to HIV-1 infection and that the greater relative susceptibility may be due to the absence of a postentry block to infection (2, 23, 41, 47). We demonstrate that Pt T cells are indeed more susceptible to infection by HIV-1 than Rh T cells, and we found that this greater susceptibility is in part due to the absence of a TRIM5 isoform capable of restricting infection, confirming the previous work of others (13, 14, 89, 91). Importantly, we show that a minimal substitution of vif with that from a pathogenic SIVmne clone is sufficient for HIV-1 to replicate in Pt PBMC, indicating that APOBEC3 proteins are a major impediment to HIV-1 replication in this species. Finally, in contrast to the rapid clearance of HIV-1 previously observed in pig-tailed macaques, we show for the first time that the Pt-tropic HIV-1 clone, HSIV-vif, is capable of persistently replicating in vivo for up to 2 years, although the plasma viral load is very low and more similar to what may be observed in HIV-1 long-term nonprogressors than progressors.
Recently, other studies have shown that HIV-1 strains with modified gag and/or vif sequences achieved transient but longer replication in pig-tailed macaques than reported for wild-type HIV-1 (28, 35). However, curtailment of viremia occurred within 12 to 25 wpi. The HIV-1 construct used in our experiments had extended replication through 44 wpi and transient small rebounds after 60 wpi in juvenile animals. Plasma viral loads were also measurable in newborn pigtails through at least 40 wpi. The differences in results from the previous studies are likely due to differences in the Pt-tropic HIV-1 clones. The virus constructed by Kamada et al. had both gag and vif modifications (39). Even after in vitro passage, this virus had relatively limited replication capacity compared to pathogenic SIVmac239, perhaps owing to the introduction of changes in gag that were nonessential for replication in pig-tail cells (39). The relatively poor replication capacity may explain its transient nature in the pig-tail host. Additional in vitro attempts to adapt the virus to cynomolgus monkeys did not improve replication in this alternative macaque species (73). On the other hand, the macaque-tropic HIV-1 clones of Hatziioannou et al. (28) contained only minimal vif substitutions similar to our construct used in the present study. However, although we used the same backbone clone (HIV-1NL4-3), their clones included the rhesus macaque adapted HIV-1 env from SHIVKB9, while our clone included the NL4-3 env. In addition, we used a vif allele from pig-tailed adapted SIVmne027, instead of the rhesus adapted SIVmac239 or HIV-2 alleles. Significantly, neither the SIVmac239 nor HIV-2 Vif efficiently downregulated rhesus A3F in in vitro experiments (88). However, we found that SIVmne027 Vif effectively reduces both Pt A3G and Pt A3F in cells and virions. That our clone replicated for a longer period of time suggests that inclusion of the NL4-3 env and a pig-tail adapted SIVmne vif gene may have contributed to longer persistent replication in the pig-tailed macaque host.
Previous studies with SIVmac showed that deletion of vpr and nef attenuate viral replication and disease in adult macaques (25, 32, 42), but that these genes are not required for high-level replication and disease in newborns (8). Furthermore, the HIV-1 accessory gene vpu also contributes to pathogenicity of SHIVs in the macaque host, although it is not essential for SHIV induced AIDS (59, 81). We therefore examined whether HSIV-vif would replicate more robustly and cause disease in newborn pig-tails, hypothesizing that the lack of activity of the HIV-1 accessory genes may account for the attenuated phenotype in the juvenile animals. Surprisingly, we found that the Pt-tropic HIV-1 is more quickly controlled in newborn than in juvenile pig-tailed macaques. If the pathogenesis of HIV-1 is akin to SIVsm/SIVmac in macaques, our data suggest that there may be a potential defect in the Pt-tropic HIV-1 clone, HSIV-vif, that is unrelated to the functionality or expression of accessory proteins other than Vif.
Our data suggest that SIV Vif expression from the Pt-tropic HIV-1 clone may not be optimal for high-level replication in the pig-tail host. We demonstrated that the Pt-tropic HIV-1 clone, HSIV-vif, does not direct degradation of Pt A3G and Pt A3F proteins as well as SIVmne027. Consequently, Pt A3 proteins are incorporated into HSIV-vif virions at amounts that are higher than observed in SIVmne027 virions. This may impact viral replication because although HSIV-vif's ability to inhibit Pt A3 proteins is clearly sufficient for it to replicate in activated Pt T cells in vitro, its infectivity was lower than that of SIVmne027. Thus, it is possible that HSIV-vif is more sensitive than SIVmne027 to changes in Pt A3G and A3F expression levels that may occur with the host immune response in vivo. Its less efficient inhibition of Pt A3G and A3F proteins could result in reduced infectivity and replication in the host. Support for this hypothesis comes from studies showing that cytokines, chemokines, and cellular activation events can upregulate human A3G and A3F expression (18, 50, 67, 69, 70, 75, 82, 93), and differences in A3G expression in human primary CD4+ T cells modulate HIV-1 infectivity (87). Recent vaccine experiments also indicate that a protective effect induced by Toll-like receptor agonists and interleukin-15 as mucosal adjuvants correlates with upregulation of A3G expression (84). In addition, greater virus-specific cellular immune responses are induced by virions harboring A3G (16), which could lead to greater control of infection. Finally, the appearance of reversion mutations of the HIV-1 Vif start codon and other Met codons upstream from and in-frame with the SIV Vif start codon in HSIV-vif further suggest the possibility that the level of Vif expression was not sufficient for high-level replication in the host. However, these mutations did not persist, suggesting a lack of an advantage for replication over time. Testing of reversion mutants confirmed their poor ability to inhibit A3 proteins. The data demonstrate the potency of A3 proteins in controlling HIV-1 replication in pig-tails and imply that Vif expression must be improved for robust viral replication in the host.
Hypermutation is one proposed mechanism by which A3G can inactivate HIV-1 in the absence of Vif (27, 56, 95). Although we did not find evidence for A3G hypermutation of the HSIV-vif proviral genome from infected macaques, it is possible that virion incorporation of A3 proteins inhibits HSIV-vif replication via an alternative mechanism. For example, A3G has been shown to inhibit reverse transcription and reduce the accumulation of viral cDNA in newly infected cells independent of hypermutation (10, 11, 26, 33, 36, 58). There is also evidence indicating that catalytically inactive A3G still blocks HIV-1 replication (33, 62), although other studies show that deaminase activity is essential (61, 77). Finally, A3G editing activity in T cells and G-to-A mutations in viral cDNAs from vif-deleted HIV-1-infected PBMC are lower than expected (49, 85), further suggesting hypermutation may have a limited role in inhibiting viral replication.
We observed that inclusion of A3 proteins by the pathogenic SIVmne027 clone and HSIV-vif clones was inversely associated with Vif detection in viral particles. This may suggest that Vif incorporation into HSIV-vif is important for functionality. However, the significance of this observation is unclear because we could not detect Vif in lysates of cells transfected with either the proviral clone of HSIV-vif or SIVmne027 and because Vif packaging into HIV-1 virions does not appear to be necessary for A3 protein inhibition or viral infectivity in culture (21, 40, 43, 52, 53, 80). Furthermore, it is unknown whether Vif inclusion in virions is in fact required for pathogenesis of HIV-1 or SIV in vivo or activities other than A3 protein inhibition that enhance viral replication such as G2 cell cycle arrest, modulation of reverse transcription, or inhibition of IRF-3 (15, 37, 64, 74). The importance of virion association could potentially be tested with the present model if the SIV Vif can be adapted to HIV-1 core particles. Regardless of the functional significance of Vif incorporation, the lack of SIV Vif detection in HSIV-vif particles may indicate a lower level of gene expression from the context of the chimeric provirus, resulting in insufficient inhibition of A3 proteins, prevention of their inclusion in virions, and suboptimal infectivity.
Overcoming innate resistance by Pt A3 proteins may require the development of a new macaque-tropic HIV-1 with greater SIV Vif expression for more complete degradation of Pt A3 proteins and exclusion from virions, as well as intravenous serial in vivo passage of the virus to select for variants with increased replication capacity and pathogenicity. In fact, it may be important to consider using a different HIV-1 clone as the backbone in future experiments. Our study and both previous ones by Igarashi et al. (35) and Hatziioannou et al. (28) modified HIV-1NL4-3. While each subsequent experiment showed improved replication and persistence in vivo, the viruses were eventually controlled. In this regard, it is possible that a macaque-tropic HIV-1 based on NL4-3 is not very fit for the pig-tail host. It is conceivable that even if all of the innate blocks to virus replication in pig-tailed macaques are overcome, a macaque-tropic HIV-1 may still not replicate due to susceptibility of the virus to adaptive immunity. Indeed, Hatziioannou et al. showed that anti-CD8 depletion increased virus production and that neutralizing antibodies to the infecting macaque-tropic HIV-1 clone were induced during infection (28). Potentially, HIV-1 will require adapting to the pig-tailed macaque via extensive passaging in order to evolve escape mutants that avoid recognition by cytotoxic T cells and neutralizing antibodies. Alternatively, escape from other intrinsic restriction factors or counteraction of innate and adaptive immunity may be equally important.
We recognize that a CCR5-tropic macaque-tropic HIV-1 will be desirable, since it will better reflect important aspects of natural infection and pathogenesis. However, the rationale for using the CXCR4-tropic HIV-1NL4-3 as an initial starting virus for in vivo studies seemed reasonable given that it has been previously shown to transiently infect pig-tailed macaques (2, 3) and because the initial goal was to determine whether we could improve upon those initial results and achieve persistent infection of the pig-tail host. Clearly, further development of the model will be required for it to be useful as a model of pathogenesis or vaccine studies, but the results of the current experiments, along with those of Igarashi et al. (35) and Hatziioannou et al. (28), should provide guidance for future studies.
In summary, our data show that a minimal vif substitution in HIV-1 is sufficient for persistent infection of pig-tailed macaques. Potentially, the inclusion of other SIV genes into HIV-1 will be required for high-level replication in pig-tailed macaques. For example, inclusion of the SIV Nef may be needed because it counteracts the restriction protein BST-2 (Tetherin, CD317) in macaque cells (38, 94). However, whether this will be necessary is unclear because SHIV chimeras with the HIV-1 nef are pathogenic (4, 48, 55). In other words, the HIV-1 Nef can functionally replace the SIV Nef, even though it does not interfere with macaque BST-2 restriction activity (38, 94). In addition, the lack of a SIV-encoded nef function does not explain why HSIV-vif was rapidly controlled in newborn animals, which generally progress to disease rapidly even when infected with nef deletion mutant viruses. Importantly, the construction of Pt-tropic HIV-1 chimeras with minimal SIV sequences should further the development of an improved animal model of AIDS that will facilitate studies of pathogenesis and the development of vaccines and therapies against HIV-1 and provide model system for continued investigations into host factors regulating cross-species transmission of primate lentiviruses.
ACKNOWLEDGMENTS
We thank A. Rice and J. Overbaugh for advice and discussions, B. Danielson and H. Ruan for critical reading of the manuscript, M. Emerman for CrFK cells, P. Younan for assistance with cloning pigtail TRIM5 isoforms, and the Southwest National Primate Research Center for rhesus macaque blood. NL4-3, NL-AD8, p197-1 (D-vif), anti-p24 antibody (183-H12-5C), and TZM-bl cells were obtained from M. Martin, E. Freed, R. Desrosiers, B. Cheseboro and K. Toohey, and J. Kappes, respectively, via the NIH AIDS Research and Reference Reagent Program.
This study was supported by NIH grants AI083095 and AI047725, by the Washington National Primate Research Center (RR00166), and in part by the Baylor-UT Houston CFAR (AI036211).
Footnotes
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Adachi A., et al. 1986. Production of acquired immunodeficiency syndrome-associated retrovirus in human and nonhuman cells transfected with an infectious molecular clone. J. Virol. 59:284–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Agy M. B., et al. 1992. Infection of Macaca nemestrina by human immunodeficiency virus type 1. Science 257:103–106 [DOI] [PubMed] [Google Scholar]
- 3. Agy M. B., et al. 1997. Serial in vivo passage of HIV-1 infection in Macaca nemestrina. Virology 238:336–343 [DOI] [PubMed] [Google Scholar]
- 4. Alexander L., Du Z., Howe A. Y., Czajak S., Desrosiers R. C. 1999. Induction of AIDS in rhesus monkeys by a recombinant simian immunodeficiency virus expressing nef of human immunodeficiency virus type 1. J. Virol. 73:5814–5825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Ambrose Z., KewalRamani V. N., Bieniasz P. D., Hatziioannou T. 2007. HIV/AIDS: in search of an animal model. Trends Biotechnol. 25:333–337 [DOI] [PubMed] [Google Scholar]
- 6. Arhel N. J., Kirchhoff F. 2009. Implications of Nef: host cell interactions in viral persistence and progression to AIDS. Curr. Top. Microbiol. Immunol. 339:147–175 [DOI] [PubMed] [Google Scholar]
- 7. Arora R., et al. 2010. Dendritic cell-mediated HIV-1 infection of T cells demonstrates a direct relationship to plasma viral RNA levels. J. Acquir. Immune Defic. Syndr. 54:115–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Baba T. W., et al. 1995. Pathogenicity of live, attenuated SIV after mucosal infection of neonatal macaques. Science 267:1820–1825 [DOI] [PubMed] [Google Scholar]
- 9. Biesinger T., Yu Kimata M. T., Kimata J. T. 2008. Changes in simian immunodeficiency virus reverse transcriptase alleles that appear during infection of macaques enhance infectivity and replication in CD4+ T cells. Virology 370:184–193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Bishop K. N., Holmes R. K., Malim M. H. 2006. Antiviral potency of APOBEC proteins does not correlate with cytidine deamination. J. Virol. 80:8450–8458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bishop K. N., Verma M., Kim E. Y., Wolinsky S. M., Malim M. H. 2008. APOBEC3G inhibits elongation of HIV-1 reverse transcripts. PLoS Pathog. 4:e1000231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bosch M. L., Schmidt A., Agy M. B., Kimball L. E., Morton W. R. 1997. Infection of Macaca nemestrina neonates with HIV-1 via different routes of inoculation. AIDS 11:1555–1563 [DOI] [PubMed] [Google Scholar]
- 13. Brennan G., Kozyrev Y., Hu S. L. 2008. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc. Natl. Acad. Sci. U. S. A. 105:3569–3574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Brennan G., Kozyrev Y., Kodama T., Hu S. L. 2007. Novel TRIM5 isoforms expressed by Macaca nemestrina. J. Virol. 81:12210–12217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Carr J. M., Coolen C., Davis A. J., Burrell C. J., Li P. 2008. Human immunodeficiency virus 1 (HIV-1) virion infectivity factor (Vif) is part of reverse transcription complexes and acts as an accessory factor for reverse transcription. Virology 372:147–156 [DOI] [PubMed] [Google Scholar]
- 16. Casartelli N., et al. The antiviral factor APOBEC3G improves CTL recognition of cultured HIV-infected T cells. J. Exp. Med. 207:39–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Chackerian B., Long E. M., Luciw P. A., Overbaugh J. 1997. Human immunodeficiency virus type 1 coreceptors participate in postentry stages in the virus replication cycle and function in simian immunodeficiency virus infection. J. Virol. 71:3932–3939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Chen K., et al. 2006. Alpha interferon potently enhances the anti-human immunodeficiency virus type 1 activity of APOBEC3G in resting primary CD4 T cells. J. Virol. 80:7645–7657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Chesebro B., Wehrly K., Nishio J., Perryman S. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Derdeyn C. A., et al. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Dettenhofer M., Yu X. F. 1999. Highly purified human immunodeficiency virus type 1 reveals a virtual absence of Vif in virions. J. Virol. 73:1460–1467 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Freed E. O., Englund G., Martin M. A. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J. Virol. 69:3949–3954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gartner S., et al. 1994. HIV-1 infection in pigtailed macaques. AIDS Res. Hum. Retrovir. 10(Suppl. 2):S129–S133 [PubMed] [Google Scholar]
- 24. Gartner S., et al. 1994. Adaptation of HIV-1 to pigtailed macaques. J. Med. Primatol. 23:155–163 [DOI] [PubMed] [Google Scholar]
- 25. Gibbs J. S., et al. 1995. Progression to AIDS in the absence of a gene for Vpr or Vpx. J. Virol. 69:2378–2383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Guo F., Cen S., Niu M., Saadatmand J., Kleiman L. 2006. Inhibition of formula-primed reverse transcription by human APOBEC3G during human immunodeficiency virus type 1 replication. J. Virol. 80:11710–11722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Harris R. S., et al. 2003. DNA deamination mediates innate immunity to retroviral infection. Cell 113:803–809 [DOI] [PubMed] [Google Scholar]
- 28. Hatziioannou T., et al. 2009. A macaque model of HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 106:4425–4429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hatziioannou T., et al. 2006. Generation of simian-tropic HIV-1 by restriction factor evasion. Science 314:95. [DOI] [PubMed] [Google Scholar]
- 30. Heeney J. L., Dalgleish A. G., Weiss R. A. 2006. Origins of HIV and the evolution of resistance to AIDS. Science 313:462–466 [DOI] [PubMed] [Google Scholar]
- 31. Himathongkham S., Luciw P. A. 1996. Restriction of HIV-1 (subtype B) replication at the entry step in rhesus macaque cells. Virology 219:485–488 [DOI] [PubMed] [Google Scholar]
- 32. Hoch J., et al. 1995. vpr deletion mutant of simian immunodeficiency virus induces AIDS in rhesus monkeys. J. Virol. 69:4807–4813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Holmes R. K., Koning F. A., Bishop K. N., Malim M. H. 2007. APOBEC3F can inhibit the accumulation of HIV-1 reverse transcription products in the absence of hypermutation: comparisons with APOBEC3G. J. Biol. Chem. 282:2587–2595 [DOI] [PubMed] [Google Scholar]
- 34. Hu S. L., et al. 1989. Protection of macaques against simian AIDS by immunization with a recombinant vaccinia virus expressing the envelope glycoproteins of simian type D retrovirus. Proc. Natl. Acad. Sci. U. S. A. 86:7213–7217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Igarashi T., et al. 2007. Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J. Virol. 81:11549–11552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Iwatani Y., et al. 2007. Deaminase-independent inhibition of HIV-1 reverse transcription by APOBEC3G. Nucleic Acids Res. 35:7096–7108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Izumi T. K., et al. 2010. HIV-1 viral infectivity factor interacts with TP53 to induce G2 cell cycle arrest and positively regulate viral replication. Proc. Natl. Acad. Sci. U. S. A. 107:20798–20803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Jia B., et al. 2009. Species-specific activity of SIV Nef and HIV-1 Vpu in overcoming restriction by tetherin/BST2. PLoS Pathog. 5:e1000429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Kamada K., et al. 2006. Generation of HIV-1 derivatives that productively infect macaque monkey lymphoid cells. Proc. Natl. Acad. Sci. U. S. A. 103:16959–16964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kao S., et al. 2003. Human immunodeficiency virus type 1 Vif is efficiently packaged into virions during productive but not chronic infection. J. Virol. 77:1131–1140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kent S. J., et al. 1995. Cytotoxic and proliferative T cell responses in HIV-1-infected Macaca nemestrina. J. Clin. Invest. 95:248–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kestler H. W., III, et al. 1991. Importance of the nef gene for maintenance of high virus loads and for development of AIDS. Cell 65:651–662 [DOI] [PubMed] [Google Scholar]
- 43. Khan M. A., et al. 2001. Human immunodeficiency virus type 1 Vif protein is packaged into the nucleoprotein complex through an interaction with viral genomic RNA. J. Virol. 75:7252–7265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kimata J. T., Kuller L., Anderson D. B., Dailey P., Overbaugh J. 1999. Emerging cytopathic and antigenic simian immunodeficiency virus variants influence AIDS progression. Nat. Med. 5:535–541 [DOI] [PubMed] [Google Scholar]
- 45. Kimata J. T., Mozaffarian A., Overbaugh J. 1998. A lymph node-derived cytopathic simian immunodeficiency virus Mne variant replicates in nonstimulated peripheral blood mononuclear cells. J. Virol. 72:245–256 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kimata J. T., Wilson J. M., Patel P. G. 2004. The increased replicative capacity of a late-stage simian immunodeficiency virus mne variant is evident in macrophage- or dendritic cell-T-cell cocultures. Virology 327:307–317 [DOI] [PubMed] [Google Scholar]
- 47. Kimball L. E., Bosch M. L. 1998. In vitro HIV-1 infection in Macaca nemestrina PBMC is blocked at a step beyond reverse transcription. J. Med. Primatol. 27:99–103 [DOI] [PubMed] [Google Scholar]
- 48. Kirchhoff F., et al. 1999. The human immunodeficiency virus type 1 nef gene can to a large extent replace simian immunodeficiency virus nef in vivo. J. Virol. 73:8371–8383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Knoepfel S. A., et al. 2008. Comparison of G-to-A mutation frequencies induced by APOBEC3 proteins in H9 cells and peripheral blood mononuclear cells in the context of impaired processivities of drug-resistant human immunodeficiency virus type 1 reverse transcriptase variants. J. Virol. 82:6536–6545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Koning F. A., et al. 2009. Defining APOBEC3 expression patterns in human tissues and hematopoietic cell subsets. J. Virol. 83:9474–9485 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Lee S. J., Garza L., Yao J., Notkins A. L., Zhou P. 2004. A nonneutralizing anti-HIV-1 antibody turns into a neutralizing antibody when expressed on the surface of HIV-1-susceptible cells: a new way to fight HIV. J. Immunol. 173:4618–4626 [DOI] [PubMed] [Google Scholar]
- 52. Liu B., Yu X., Luo K., Yu Y., Yu X. F. 2004. Influence of primate lentiviral Vif and proteasome inhibitors on human immunodeficiency virus type 1 virion packaging of APOBEC3G. J. Virol. 78:2072–2081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Liu H., et al. 1995. The Vif protein of human and simian immunodeficiency viruses is packaged into virions and associates with viral core structures. J. Virol. 69:7630–7638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Malim M. H., Emerman M. 2008. HIV-1 accessory proteins: ensuring viral survival in a hostile environment. Cell Host Microbe 3:388–398 [DOI] [PubMed] [Google Scholar]
- 55. Mandell C. P., et al. 1999. SIV/HIV Nef recombinant virus (SHIVnef) produces simian AIDS in rhesus macaques. Virology 265:235–251 [DOI] [PubMed] [Google Scholar]
- 56. Mangeat B., et al. 2003. Broad antiretroviral defense by human APOBEC3G through lethal editing of nascent reverse transcripts. Nature 424:99–103 [DOI] [PubMed] [Google Scholar]
- 57. Mariani R., et al. 2003. Species-specific exclusion of APOBEC3G from HIV-1 virions by Vif. Cell 114:21–31 [DOI] [PubMed] [Google Scholar]
- 58. Mbisa J. L., et al. 2007. Human immunodeficiency virus type 1 cDNAs produced in the presence of APOBEC3G exhibit defects in plus-strand DNA transfer and integration. J. Virol. 81:7099–7110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. McCormick-Davis C., et al. 2000. A molecular clone of simian-human immunodeficiency virus (DeltavpuSHIV(KU-1bMC33)) with a truncated, non-membrane-bound vpu results in rapid CD4+ T cell loss and neuro-AIDS in pig-tailed macaques. Virology 272:112–126 [DOI] [PubMed] [Google Scholar]
- 60. McDonald D., et al. 2002. Visualization of the intracellular behavior of HIV in living cells. J. Cell Biol. 159:441–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Miyagi E., et al. 2007. Enzymatically active APOBEC3G is required for efficient inhibition of human immunodeficiency virus type 1. J. Virol. 81:13346–13353 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Newman E. N., et al. 2005. Antiviral function of APOBEC3G can be dissociated from cytidine deaminase activity. Curr. Biol. 15:166–170 [DOI] [PubMed] [Google Scholar]
- 63. Newman R. M., et al. 2008. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 4:e1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Okumura A., et al. 2008. HIV-1 accessory proteins VPR and Vif modulate antiviral response by targeting IRF-3 for degradation. Virology 373:85–97 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Overbaugh J., Anderson R. J., Ndinya-Achola J. O., Kreiss J. K. 1996. Distinct but related human immunodeficiency virus type 1 variant populations in genital secretions and blood. AIDS Res. Hum. Retrovir. 12:107–115 [DOI] [PubMed] [Google Scholar]
- 66. Patel P. G., Yu Kimata M. T., Biggins J. E., Wilson J. M., Kimata J. T. 2002. Highly pathogenic simian immunodeficiency virus mne variants that emerge during the course of infection evolve enhanced infectivity and the ability to downregulate CD4 but not class I major histocompatibility complex antigens. J. Virol. 76:6425–6434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Pido-Lopez J., et al. 2007. Stimulation of cell surface CCR5 and CD40 molecules by their ligands or by HSP70 up-regulates APOBEC3G expression in CD4+ T cells and dendritic cells. J. Immunol. 178:1671–1679 [DOI] [PubMed] [Google Scholar]
- 68. Polacino P., et al. 2007. Immunogenicity and protective efficacy of Gag/Pol/Env vaccines derived from temporal isolates of SIVmne against cognate virus challenge. J. Med. Primatol. 36:254–265 [DOI] [PubMed] [Google Scholar]
- 69. Refsland E. W., et al. 2010. Quantitative profiling of the full APOBEC3 mRNA repertoire in lymphocytes and tissues: implications for HIV-1 restriction. Nucleic Acids Res. 38:4274–4284 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rose K. M., Marin M., Kozak S. L., Kabat D. 2004. Transcriptional regulation of APOBEC3G, a cytidine deaminase that hypermutates human immunodeficiency virus. J. Biol. Chem. 279:41744–41749 [DOI] [PubMed] [Google Scholar]
- 71. Rose P. P., Korber B. T. 2000. Detecting hypermutations in viral sequences with an emphasis on G→A hypermutation. Bioinformatics 16:400–401 [DOI] [PubMed] [Google Scholar]
- 72. Rudensey L. M., Kimata J. T., Long E. M., Chackerian B., Overbaugh J. 1998. Changes in the extracellular envelope glycoprotein of variants that evolve during the course of simian immunodeficiency virus SIVMne infection affect neutralizing antibody recognition, syncytium formation, and macrophage tropism but not replication, cytopathicity, or CCR-5 coreceptor recognition. J. Virol. 72:209–217 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Saito A., et al. 2011. Improved capacity of a monkey-tropic HIV-1 derivative to replicate in cynomolgus monkeys with minimal modifications. Microbes Infect. 13:58–64 [DOI] [PubMed] [Google Scholar]
- 74. Sakai K., Dimas J., Lenardo M. J. 2006. The Vif and Vpr accessory proteins independently cause HIV-1-induced T cell cytopathicity and cell cycle arrest. Proc. Natl. Acad. Sci. U. S. A. 103:3369–3374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Sarkis P. T., Ying S., Xu R., Yu X. F. 2006. STAT1-independent cell type-specific regulation of antiviral APOBEC3G by IFN-alpha. J. Immunol. 177:4530–4540 [DOI] [PubMed] [Google Scholar]
- 76. Sayah D. M., Sokolskaja E., Berthoux L., Luban J. 2004. Cyclophilin A retrotransposition into TRIM5 explains owl monkey resistance to HIV-1. Nature 430:569–573 [DOI] [PubMed] [Google Scholar]
- 77. Schumacher A. J., Hache G., Macduff D. A., Brown W. L., Harris R. S. 2008. The DNA deaminase activity of human APOBEC3G is required for Ty1, MusD, and human immunodeficiency virus type 1 restriction. J. Virol. 82:2652–2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Sheehy A. M., Gaddis N. C., Choi J. D., Malim M. H. 2002. Isolation of a human gene that inhibits HIV-1 infection and is suppressed by the viral Vif protein. Nature 418:646–650 [DOI] [PubMed] [Google Scholar]
- 79. Shibata R., Sakai H., Kawamura M., Tokunaga K., Adachi A. 1995. Early replication block of human immunodeficiency virus type 1 in monkey cells. J. Gen. Virol. 76(Pt. 11):2723–2730 [DOI] [PubMed] [Google Scholar]
- 80. Simon J. H., Miller D. L., Fouchier R. A., Malim M. H. 1998. Virion incorporation of human immunodeficiency virus type-1 Vif is determined by intracellular expression level and may not be necessary for function. Virology 248:182–187 [DOI] [PubMed] [Google Scholar]
- 81. Stephens E. B., et al. 2002. Deletion of the vpu sequences prior to the env in a simian-human immunodeficiency virus results in enhanced Env precursor synthesis but is less pathogenic for pig-tailed macaques. Virology 293:252–261 [DOI] [PubMed] [Google Scholar]
- 82. Stopak K. S., Chiu Y. L., Kropp J., Grant R. M., Greene W. C. 2007. Distinct patterns of cytokine regulation of APOBEC3G expression and activity in primary lymphocytes, macrophages, and dendritic cells. J. Biol. Chem. 282:3539–3546 [DOI] [PubMed] [Google Scholar]
- 83. Stremlau M., et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 84. Sui Y., et al. 2010. Innate and adaptive immune correlates of vaccine and adjuvant-induced control of mucosal transmission of SIV in macaques. Proc. Natl. Acad. Sci. U. S. A. 107:9843–9848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Thielen B. K., et al. 2007. T cells contain an RNase-insensitive inhibitor of APOBEC3G deaminase activity. PLoS Pathog. 3:1320–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Toohey K., Wehrly K., Nishio J., Perryman S., Chesebro B. 1995. Human immunodeficiency virus envelope V1 and V2 regions influence replication efficiency in macrophages by affecting virus spread. Virology 213:70–79 [DOI] [PubMed] [Google Scholar]
- 87. Vetter M. L., Johnson M. E., Antons A. K., Unutmaz D., D'Aquila R. T. 2009. Differences in APOBEC3G expression in CD4+ T helper lymphocyte subtypes modulate HIV-1 infectivity. PLoS Pathog. 5:e1000292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Virgen C. A., Hatziioannou T. 2007. Antiretroviral activity and Vif sensitivity of rhesus macaque APOBEC3 proteins. J. Virol. 81:13932–13937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Virgen C. A., Kratovac Z., Bieniasz P. D., Hatziioannou T. 2008. Independent genesis of chimeric TRIM5-cyclophilin proteins in two primate species. Proc. Natl. Acad. Sci. U. S. A. 105:3563–3568 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wei X., et al. 2002. Emergence of resistant human immunodeficiency virus type 1 in patients receiving fusion inhibitor (T-20) monotherapy. Antimicrob. Agents Chemother. 46:1896–1905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Wilson S. J., et al. 2008. Independent evolution of an antiviral TRIMCyp in rhesus macaques. Proc. Natl. Acad. Sci. U. S. A. 105:3557–3562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Wyand M. S., Manson K. H., Garcia-Moll M., Montefiori D., Desrosiers R. C. 1996. Vaccine protection by a triple deletion mutant of simian immunodeficiency virus. J. Virol. 70:3724–3733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ying S., Zhang X., Sarkis P. T., Xu R., Yu X. 2007. Cell-specific regulation of APOBEC3F by interferons. Acta Biochim. Biophys. Sin (Shanghai) 39:297–304 [DOI] [PubMed] [Google Scholar]
- 94. Zhang F., et al. 2009. Nef proteins from simian immunodeficiency viruses are tetherin antagonists. Cell Host Microbe 6:54–67 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Zhang H., et al. 2003. The cytidine deaminase CEM15 induces hypermutation in newly synthesized HIV-1 DNA. Nature 424:94–98 [DOI] [PMC free article] [PubMed] [Google Scholar]