Abstract
Foamy virus (FV) capsid proteins have few lysines. Basic residues are almost exclusively represented by arginines indicating positive selective pressure. To analyze the possible functions of this peculiarity, we mutated an infectious molecular clone of the prototypic FV (PFV) to harbor lysines in the Gag protein at arginine-specifying positions and analyzed various aspects of the FV replication cycle. The majority of mutants replicated equally as well in permanent cell cultures as the original wild-type (wt) virus and were genetically stable in gag upon 10 cell-free passages. With respect to the features of late reverse transcription, nucleic acid content, and infectiousness of the virion DNA genome, the majority of mutants behaved like the wt. Several mutants of PFV were ubiquitinated in Gag but unable to generate virus-like particles (VLPs) or to undergo pseudotyping by a heterologous envelope. Using primary cells, however, a replicative disadvantage of the majority of mutants was disclosed. This disadvantage was enhanced upon interferon (IFN) treatment. We found no evidence that the lysine-bearing gag mutants showed more restriction than the wt virus by tetherin (CD317) or Trim5α. A single lysine in PFV Gag was found to be nonessential for transient replication in permanent cell culture if replaced by an arginine residue. Upon replication in primary cells, even without IFN treatment, this mutant was severely impaired, indicating the importance of specifying at least this lysine residue in PFV Gag. The paucity of lysines in FV Gag proteins may be a consequence of preventing proteasomal Gag degradation.
INTRODUCTION
Two subfamilies comprise the family of exogenous retroviruses: the orthoretroviruses, made up of six genera (α- to ε-retroviruses and lentiviruses), and the spumaretroviruses, comprised exclusively of the foamy virus (FV) genus (33). There are major differences in the replication strategies between both subfamilies which are responsible for this distinction (32, 48).
On a structural level, this is reflected by very unusual FV Gag proteins, which are not cleaved by the viral protease (PR) into matrix (MA), capsid (CA), and nucleocapsid (NC) subunits (32, 48). Instead, the FV PR cleaves only a small peptide (p3 in case of the prototypic FV [PFV]) from the C terminus of FV Gag proteins (17, 44). This C-terminal cleavage is essential for FV replication (13, 76). Complete proteolytic cleavage of the capsid occurs most likely in the process of uncoating after fusion with new cells and uses unconventional cleavage sites (19, 28).
The FV Env protein constitutes another peculiarity. In contrast to orthoretroviral Envs, which are—with surface (SU) and transmembrane (TM) subunits—bipartite in nature (22), the FV Env is a tripartite protein that bears, in addition to SU and TM, a membrane-spanning type II and particle-associated leader peptide (LP), which has a mass of 18 kDa in the case of PFV (30, 31). The LP of the PFV Env protein is oligo-ubiquitinated and appears to perform functions in viral replication which are confined to Gag in orthoretroviruses (62).
FV capsids do not bud spontaneously from the plasma membrane and do not naturally form virus-like particles (VLPs) (16). They require the presence of authentic Env for cellular export (45). Furthermore, pseudotyping of unmodified FV capsids with other (viral) glycoproteins appears to be impossible (45). On the other hand, pseudotyping of other viral capsids by FV Env and, in particular, by mutants in the ubiquitination sites of LP, is very efficient (72). In FVs, domains on the side of Env particularly located in LP are essential for a specific contact with Gag (31).
With Gag, the situation is less clear. Deletion studies suggested that approximately the first 100 amino acids of the 648 residues of PFV Gag are essential to mediate the specific Env interaction (6).
Some other domains in the PFV Gag protein have been characterized. A cytoplasmic targeting and retention signal (CTRS) is located in the N-terminal domain (amino acids [aa] 43 to 60) (12). A late (L) domain, specified by the motif PSAP (aa 284 to 287) interacts with the cellular export machinery (vacuolar protein sorting [VPS]) via TSG101 for complete release of virus particles from the plasma membrane (41, 61). However, ubiquitination of PFV Gag has not been found, while ubiquitination is a common feature of orthoretroviral capsids upon interaction with the VPS machinery (46, 77).
Four coiled-coil (CC) motifs (CC1 to CC4) are predicted to be present in PFV Gag. The CC1 motif is very N-terminally (aa 4 to 19) located, and mutations in this region indicated an interaction domain with Env (29). The CC2 (aa 133 to 146) is situated C-terminally adjacent to the CTRS and has been shown to be essential for correct Gag-Gag interactions (66) while CC3 (aa 161 to 174) enables PFV capsid cleavage products to interact with dynein motors of the microtubular network to travel to the microtubular organizing center (MTOC) (43). CC4 has not been characterized yet.
At the C terminus of PFV Gag, three boxes rich in glycines and arginines (GR boxes) have been identified (55). GR box I (aa 485 to 495) is believed to be nucleic acid-binding (74). The PFV GR box II (aa 547 to 557) has been reported to contain a basic nuclear localization signal (55) and is the main determinant of tethering incoming PFV to chromatin of the host cell by interacting with H2A and H2B core histones (67). No function has yet been assigned to GR box III (aa 601 to 611).
Further sequences in Gag, such as a conserved YEIL motif (aa 464 to 467), important for correct particle assembly and reverse transcription, have also been characterized (35). The known structural motifs of PFV Gag are summarized in Fig. S1 in the supplemental material.
FV Gag proteins present yet another highly unusual feature. As first noted by Zhadina et al. (77), they are extremely rare in lysine residues. Basic residues in FV Gags are almost exclusively represented by arginines. Therefore, we attempted to analyze functional aspects of these residues by mutating some arginine-specifying codons in PFV to lysine codons. In addition, we wanted to determine whether the single lysine at position 396 in PFV Gag is essential for replication by changing the respective sequence to an arginine-specifying codon.
MATERIALS AND METHODS
Cells.
HEK 293T (11), HT1080 (47), MRC-5 (23), TZM (10), and BHK/LTR(PFV)lacZ (56) indicator cells were maintained in Dulbecco's modified minimal essential medium supplemented with 10% fetal bovine serum and antibiotics. In addition, 1 mg/ml G418 (Invitrogen) was added to the BHK indicator cells.
Transfections and viruses.
If not otherwise stated, 2 × 106 HEK 293T cells in 6-cm dishes or 6 × 106 cells in 10-cm dishes were seeded the day before and transfected with a total of 6 μg or 16 μg, respectively, of plasmids (as specified in individual experiments). Transient transfections of cells, seeded in 6-cm dishes, were performed with 3 μg of proviral plasmid and 3 μg of pEGFP (where EGFP is enhanced green fluorescent protein) (Clontech) control plasmid to score the transfection efficiency. Adjustments of DNA amounts and negative controls were done with pcDNA3.1 (Invitrogen). For the “infectivity” analysis of virion DNA, 4 × 104 BHK/LTR(PFV)lacZ indicator cells, seeded in 12-well plates, were transfected with serial 10-fold dilutions of virion DNA. All transfections were performed using a polyethyleneimine protocol, as described previously (63). PFV titrations were done on BHK/LTR(PFV)lacZ cells, as previously described (56). To partially purify virus, 10 ml or 20 ml of viral supernatant, depending on the particular experiment, was centrifuged through 20% sucrose cushions in a Surespin 630 rotor (Sorvall) at 4°C and 27,000 rpm for 2 h. To take photomicrographs of transfected cells, HEK 293T were stained with hematoxylin-eosin (HE) stain, as described previously (18), 2 days after transfection and viewed with an Olympus BX50 microscope.
Infection of MRC-5 cells and IFN treatment.
Viruses were prepared by transfection of HEK 293T cells seeded in 6-cm dishes with 3 μg of proviral PFV plasmids. A 100-μl aliquot of the cell-free supernatant was used to infect 4 × 104 primary human fetal lung fibroblasts (MRC-5 from the European Collection of Cell Cultures) in 12-well plates at a multiplicity of infection (MOI) of 0.1. The cells were either left untreated or were incubated with medium containing 100 U/ml alpha interferon (α-IFN) or γ-IFN (Peprotech) 24 h prior to and during infection. After replication in MRC-5 cells for 3 days, fresh medium plus IFN, if appropriate, was added. The cell-free supernatant was titrated on BHK/LTR (PFV)lacZ cells after an additional 2-day-incubation.
Recombinant DNA.
Standard molecular cloning procedures were employed to generate recombinant plasmids (2, 54). All new molecular clones were verified by nucleotide sequencing in the relevant parts.
Backbone for PFV mutants was the infectious plasmid pcHSRV2 (38), which is here denoted pcPFV2 to acknowledge the change in FV nomenclature (49). PCR mutagenesis (21) on a1.7-kb KasI/SwaI fragment was employed to introduce the amino acid changes shown in Fig. 1. Gene sequences are shown in Fig. S2 in the supplemental material. All oligonucleotides employed in this study are listed in Table S5 in the supplemental material.
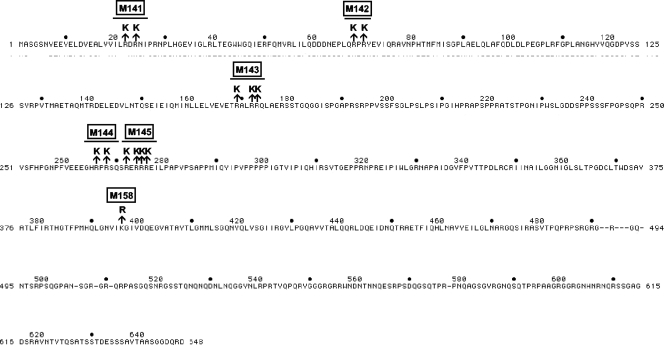
Fig. 1.
PFV Gag sequences and mutants analyzed in this study. The protein sequence of PFV Gag is shown, and the generated mutants are indicated.
Subgenomic plasmids of mutants M141 to M145 were made by inserting PFV gag gene sequences into the expression vector pCIgag-2 (20) via exchange of a 1.4-kb EcoRI/StuI fragment created by PCR with the oligonucleotide primer pair 4494/4495 from pcPFV2 M141-M145.
For the generation of DNA and cRNA probes, a fragment of approximately 0.5 kb was amplified by PCR from the pol gene of the PFV molecular clone pcPFV2 using the oligonucleotide primer pair 4499/4500 and inserted into the pCRII-Topo vector (Invitrogen) via HindIII/XhoI digestion. This produced the plasmid pDM39.
Using the primers hCD317HAs and hCD317r, the cDNA of human CD317 was amplified with Go-Taq polymerase (Promega) after reverse transcription with primer hCD317HAs from total RNA extracted with an RNeasy kit (Qiagen) from HeLa cells employing an RNase H mutant form of murine leukemia virus reverse transcriptase (MLV-RT RH−; Promega). The coding sequence of CD317 bearing a primer-encoded N-terminal hemagglutinin (HA) tag was inserted into a pcDNA3.1 directional Topo vector (Invitrogen) and characterized by restriction enzyme digestion and DNA sequencing, which revealed the identical sequence to the published one (accession number D281317). This resulted in the plasmid pchCD317-HA.
The plasmids pBJ5 HA-Ubi (where Ubi is ubiquitin) (64), the pLgatewayIRESEYFP gammaretroviral vectors carrying various chimeric (see below) Trim5α domains (73), pHIT60 (59), pCZIgag-2 (20), pCIgag-2 (20), pCpol-2 (20), pcziPol (20), pCenv-1 (20), pciEnv (20), pczVSV-G wt (where VSV-G wt is vesicular stomatitis virus G protein wild type) (20), pNL4-3 (1), pNL4-3 lacking Vpu (Vpu−) (5), and the PFV vectors pMD09 (20) and pTW01 (71) have been described previously.
Viral DNA sequencing.
Partially purified virus was resolved in 200 μl of phosphate-buffered saline (PBS), nucleic acids were extracted with a QIAamp MinElute Virus Spin kit (Qiagen), following the instructions of the manufacturer, and viral RNAs were destroyed by incubation with 0.2 mg/ml heated RNase A (Fermentas) at 37°C for 60 min. A seminested PCR was performed with 20 pm each of primer 4392/4513 for the first round and 4392/4514 in the second round, employing a final concentration of 200 nM each deoxynucleoside triphosphate (dNTP) and 1 U of Pwo polymerase (Peqlab) in a 20-μl reaction mixture using 35 standard cycles. A 10-μl aliquot of the first reaction product was amplified by nested PCR. Finally, oligonucleotide primers 4089, 4099, 4153, 4405, 4447, and 4515 were used to determine the PFV gag gene sequence by cycle sequencing with a Big Dye Terminator, version 1.1, Cycle Sequencing kit (Applied Biosystems). DNA sequences were automatically resolved on an ABI Prism 3100 Genetic Analyzer (Applied Biosystems).
AZT inhibition assay.
Taking advantage of the inhibition of the PFV reverse transcriptase by zidovudine ([AZT] GlaxoSmithKline), we performed AZT inhibition assays, as described previously (38, 52), to determine the time point of reverse transcription using 10 μM AZT and 6 μg of plasmid DNA (3 μg of viral and 3 μg of pEGFP plasmid DNAs) for transfection of 2 × 106 HEK 293T cells in 6-cm dishes. Viral titers were determined following transfer of the cell-free supernatant (0.45-μm filtrate) to BHK/LTR(PFV)lacZ-cells 2 days after transfection.
Quantitative PCR and reverse transcription-PCR (RT-PCR).
cRNA was transcribed in vitro using XbaI-linearized pDM39 with 10 U of SP6 RNA polymerase (Fermentas) using the buffer and nucleotides as recommended by the supplier at 37°C for 30 min. The template was destroyed by incubation with 0.5 U of RQ-DNase (Promega) at 37°C for 15 min. After spectrometric determination of the RNA concentration, serial 10-fold dilutions from 100 ng to 10−5 ng were employed to synthesize cDNA with 100 U/ml RNase H mutant MLV reverse transcriptase (Promega), using the conditions as recommended by the manufacturer, with the oligonucleotide primer 4500 at 54°C for 60 min. Similarly for DNA measurement, standard concentration dilutions were made from plasmid DNA from 1 ng to 10−5 ng. Virus was prepared from 10 ml of supernatant by ultracentrifugation through a sucrose cushion (partial purification). Virion nucleic acids were extracted using a QIAamp MinElute Virus Spin kit (Qiagen), taken up in 20 μl of nuclease-free water, and treated with 0.2 mg/ml RNase A or 20 U/ml RQ-DNase, both at 37°C for 60 min before determination of the DNA or RNA concentration, respectively. The DNase was inactivated by addition of EDTA up to a final concentration of 2 mM and heating at 65°C for 10 min. Reverse transcription was performed on 1/25th of the eluted viral RNA, using the identical conditions and oligonucleotide primers described above for the generation of cDNA probe. From all samples an approximate 200-bp fragment was amplified on an iCycler iQ (Bio-Rad) employing 10 pmol of each primer 4497/4507 and SYBR Green (Qiagen) as a fluorophore for 40 cycles at 95°C (10 s) for denaturation and 60°C (30 s) for annealing and elongation after an initial denaturation step at 95°C for 5 min with HotStarTaq Plus DNA polymerase (Qiagen). After specificity control by melting point analysis, the sample nucleic acid content was calculated with the iCycler iQ Optical System software, version 3.1 (Bio-Rad).
Infectivity analysis of virion DNA.
Partially purified PFV from 20 ml of supernatant of transiently transfected HEK 293T cells, resolved in 200 μl of PBS, was treated with 4 U of DNase I (Fermentas) at 37°C overnight to eliminate remaining plasmid contamination. After inactivation of the enzyme, DNA was extracted, as above, and resolved in 24 μl of water. BHK indicator cells were transfected as described with serially 10-fold diluted virion DNA (corresponding to 0.8 ml of supernatant up to 0.8 × 10−3 ml of supernatant). Positive controls were produced by transfection of cells with pcPFV2 plasmid DNA, and negative controls were produced by transfection of cells with DNase I-treated (4 U at 37°C overnight) infectious plasmid and with empty pcDNA3.1 vector. Following transfer of the supernatant 3 days later to fresh indicator cells, these were stained after 3 days to determine virus replication.
Vector transfer and protein detection.
HEK 293T cells, seeded in 10-cm dishes, were transfected employing the four-plasmid PFV vector system, as detailed previously (20), with a total of 17 μg of plasmid DNAs consisting of 10 μg of pTW01 vector, 5 μg of pCIgag-2 or derivatives, 1 μg of pCpol-2, and 1 μg of pCenv-1 or pczVSV-G wt. When constructs directing the expression of genes encoding viral glycoproteins were omitted, the total DNA amount was adjusted with pcDNA3.1. The clarified supernatant was transferred to HT1080 cells which were analyzed by fluorescence-activated cell sorting (FACS) 48 h later. For determination of ubiquitination of viral proteins, the cells were transfected with 8 μg of proviral constructs and 8 μg of pBJ5. Cellular lysates were prepared in detergent-containing buffer (20) and centrifuged through a QIAshredder (Qiagen). Partially purified virus was also lysed in detergent-containing buffer. Finally, proteins were resolved in sodium dodecyl sulfate-containing gels containing 8 or 10% polyacrylamide (SDS-PAGE), blotted to nitrocellulose, and reacted with polyclonal rabbit antisera against PFV Gag (20), the HA tag (Sigma), and glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Sigma) as described previously (20). After reaction with secondary α-rabbit antibody (Dako), the blots were developed using the ECL reagent (Amersham Biosciences). For the detection of VSV-G protein, we used the mouse monoclonal antibody I.1, as described previously (26, 27), and anti-mouse secondary antibody (Dako).
Tetherin susceptibility testing.
The cocktail for transfection of HEK 293T cells contained 2.4 μg of either pNL4-3 or pNL4-3 Vpu− together with 0.5 μg of pUC or pchCD317-HA or the four-plasmid PFV vector system detailed above (gag, 1.2 μg; pol, 0.5 μg; env, 0.5 μg; GFP-encoding pMD9 vector, 2.5 μg; 1.5 μg of pUC or pchCD317-HA) employing wt or mutated gag packaging plasmids. The cell-free supernatant was transferred to HT1080 cells which were scored for GFP expression by FACS analysis, or to TZM (JC53-BL) cells that were scored for HIV replication, as described previously (10). Adequate plasmid expression in transfected cells was determined by immunoblotting using HA-, PFV Gag-, or HIV Gag-specific antisera (data not shown).
Trim5α restriction assay.
The two-color restriction assay for Trim5α was essentially performed as described previously (4, 73). Briefly, HT1080 cells were rendered positive for enhanced yellow fluorescent protein (EYFP) and human Trim5α (huTrim5α) or chimeric huTrim5α constructs carrying the B30.2 domain from Old World gorilla and orangutan or New World capuchin and Goeldi's marmoset by gammaretroviral transduction. These recipient cells were then challenged with a defined amount of EGFP-expressing FV vector virus produced in HEK 293T cells employing wt or the mutated gag packaging plasmids, resulting in transduction efficiencies of 5 to 30% EGFP-expressing cells in wt HT1080 cells. Controls included the wt macaque simian foamy virus (SFVmac) gag packaging plasmid (73) that is restricted by the two applied New World monkey factors (73). The calculation of restriction was performed as described after determination of the transduction rates by two-color FACS analysis (4, 73).
Bioinformatics and statistical analysis.
The GenBank and EMBL databank accession numbers used in this study are AY046058.1 for HIV type 1, NC_001722.1 for HIV type 2, NC_001550.1 for Mason-Pfizer monkey virus (MPMV), AF228552.1 for mouse mammary tumor virus (MMTV), AF033811 for MLV, NC_001436.1 for primate T-lymphotropic virus type I (PTLV-I), X85262 for human hepatitis B virus (HBV), Y07724 for PFV, NC_001364.1 for SFV from chimpanzee (SFVcpz), HM245790 for SFV from gorilla (SFVgor), AJ544579 for SFV from orangutan (SFVora), NC_010819.1 for SFV from macaque (SFVmac), NC_010820.1 for SFV from African green monkey (SFVagm), EU010385 for SFV from spider monkey (SFVspm), GU356395 for SFV from marmorset (SFVmar), and NC-002201.1 for equine FV (EFV), AY134750.1 for bovine FV (BFV), AJ564746 for feline FV (FFV), and the consensus sequence of endogenous FV from sloth (SloEFV), following the publication of Katzourakis et al. (25). We did not use the published squirrel monkey SFV (SFVsqu) sequence because this sequence probably contains a frameshift in the gag gene. We used the EMBnet Coils server (www.ch.embnet.org/software/COILS_form.html) and the EMBL server (www.russell.embl-heidelberg.de/cgi-bin/coils-svr.pl) to locate coiled-coil sequences in the PFV Gag protein. In addition, the Ensembl Identifier (www.ensembl.org) database was used to identify other viral proteins devoid of lysines.
RESULTS
Amino acid composition of retroviral Gag proteins.
Characteristic of the eukaryotic genome (3), orthoretroviral Gag proteins are composed of an average number of basic amino acids, made up of both lysines and arginines. Even the Gag homologous core protein of HBV is composed of both lysine and arginine residues (Table 1) although arginines dominate due to the arginine-rich C terminus of the HBV core (58).
Table 1.
Basic residues in retroviral Gag proteins and in the HBV core protein
| Virus and/or family or genome | Protein length (aa) | No of lysines (%) | No. of arginines (%) |
|---|---|---|---|
| Orthoretrovirinae | |||
| HIV-1 | 492 | 39 (7.9) | 28 (5.7) |
| HIV-2 | 521 | 39 (7.5) | 36 (6.9) |
| MPMV | 657 | 58 (8.8) | 22 (3.3) |
| MMTV | 591 | 61 (10.3) | 28 (4.7) |
| MLV | 538 | 31 (5.8) | 50 (9.3) |
| PTLV-I | 429 | 20 (4.7) | 21 (4.9) |
| Average | (7.5) | (5.8) | |
| HBV | 185 | 03 (1.6) | 23 (12.4) |
| Eucaryotic genome | (6.0) | (5.2) | |
| Spumaretrovirinae | |||
| PFV | 648 | 1 (<0.2) | 64 (9.9) |
| SFVcpz | 653 | 2 (0.3) | 60 (9.2) |
| SFVgor | 644 | 1 (<0.2) | 61 (9.5) |
| SFVora | 625 | 1 (<0.2) | 60 (9.6) |
| SFVmac | 647 | 0 (0.0) | 56 (8.7) |
| SFVagm | 643 | 0 (0.0) | 57 (8.9) |
| SFVspm | 624 | 4 (0.6) | 49 (7.9) |
| SFVmar | 613 | 6 | 48 (7.8) |
| EFV | 559 | 1 (<0.2) | 50 (8.9) |
| BFV | 544 | 0 (0.0) | 48 (8.8) |
| FFV | 514 | 8 (1.6) | 46 (8.9) |
| Average | (0.4) | (8.9) | |
| SloEFV | 503 | 4 (0.8) | 28 (5.6) |
In sharp contrast to orthoretroviruses, the Gag proteins of exogenous FVs bear only a very limited number of lysines (Table 1). The FFV Gag protein may be an exception because it contains eight lysines, but even in this case the vast majority of basic residues in Gag are represented by 46 arginines. Furthermore, five of the eight FFV lysines are located in the C-terminal peptide that is cleaved from the main body of Gag during processing. According to the consensus sequence of the only known endogenous FV (25), the respective Gag protein harbors four lysine residues. However, since only fragments of open reading frames (ORFs) are known, the SloEFV sequence has to be considered with caution. The dominance of arginines over lysines in FV Gag proteins is all the more remarkable since it is not seen in FV Pol and Env proteins (see Table S2 in the supplemental material).
Overall, it appears that despite the considerable length of FV Gag proteins, basic residues are represented almost exclusively by arginines which, as in the case of PFV, may account for almost 10% of all residues.
FV Gag arginine residues were not maintained by chance.
Four lines of theoretical evidence argue against the arbitrariness of arginines in FV Gag proteins, as follows.
(i) Orthoretroviral Gag proteins are usually more conserved than the respective Env proteins. The sequence swarm observed in HIV-1 Env is an instructive example for this. Contrary to this, FVs show a higher conservation in Env than in Gag (48). However, despite this high variability in Gag, no or almost no lysines appear.
(ii) Arginines are specified by the codons CGG, CGA, CGC, CGU, and AGG and AGA while only AAG and AAA encode lysines. Thus, the mutation of the second nucleotide of the last two codons would result in a corresponding amino acid change from lysine to arginine. In FVs almost half of the arginine-specifying codons are represented by AGAs, which could be derived from AAA lysine codons. Since the overall frequency of lysines in the PFV proteome (made up of the Gag, Pol, Env, Tas, and Bet proteins), P(K), is 0.058, the probability (P) of a protein of 648-aa length to contain just one lysine by chance, as in the Gag of PFV, adds up to a very unlikely value of P: P = P(K) × [1 − P(K)]647 ≈ 1.8 × 10−18.
(iii) The gag genes of retroviruses have an AT content of around 52%. In FV gag genes, the AT content, at an average of 54%, is even slightly higher (data not shown). Despite the high AT content, the mutations from supposedly previously lysine-specifying to now arginine-specifying codons in the gag genes of FVs, implying A to G transitions, were maintained.
(iv) The codons in question are located in the amino terminal half of Gag (Fig. 1 and Fig. S1 in the supplemental material). Therefore, reasons other than functions that have been confined to the arginine-rich GR boxes at the C terminus of FV Gag must be responsible for the dominance of arginines over lysines at these positions.
Mutagenesis of PFV Gag.
The Gag sequence of PFV was changed, as shown in Fig. 1 (for genomic sequence, see Fig. S2 in the supplemental material). Care was taken not to disrupt known PFV Gag motifs or the partially characterized Gag cleavage sites being used after infection (17, 28, 44). However, mutant M143 (R169K R172K R173K) is located within CC3, shown to be required for PFV intracellular traffic (43). Since of all arginine-specifying codons only AGG and AGA could be derived from lysine-specifying codons by a single nucleotide exchange, we preferentially mutated these to AAG and AAA and left the other codons of wt sequence. This strategy limited the number of codons of interest to 41 out of 64. Since in PFV several of the respective codons are in close proximity, we constructed mutants altering two or three codons (Fig. 1). In addition, some combinatory mutants of PFV were also made.
Mutants were generated initially in the context of human cytomegalovirus immediate-early gene promoter-driven proviral constructs, which then were analyzed in assays monitoring replication competence and nucleic acid content. However, to enable the study of a potential VLP formation, vector transfer, and pseudotyping, we also inserted the altered sequences into a subgenomic gag gene expression vector.
Replication of mutated proviruses in permanent cells.
HEK 293T cells were transiently transfected with the mutant PFV constructs along with the control plasmid pEGFP in order to measure transfection efficiency (data not shown). The supernatants were harvested after 2 days, and infectivity was assayed on the BHK indicator cell line (56). The results from at least three independent experiments are summarized in Table 2. Surprisingly, we noticed that all PFV mutants were fully replication competent in this transient assay and that several mutants including M152 and M154 that contain six and seven lysine residues instead of arginines, respectively, replicated approximately as well as wt virus.
Table 2.
PFV titers after cell-free passages 1 and 10
| Virus | Passage 1 data |
Passage 10 data |
||
|---|---|---|---|---|
| No. of expts | Titer (per ml) | No. of expts | Titer (per ml) | |
| pcPFV2 (wt) | 18 | (9.3 ± 2.0) × 106 | 3 | (1.2 ± 0.2) × 107a |
| M141 (R22K R24K) | 10 | (1.1 ± 0.3) × 105 | 3 | (8.0 ± 1.4) × 105 |
| M142 (R68K R70K) | 3 | (9.6 ± 4.00 × 106 | 3 | (1.9 ± 0.3) × 107 |
| M143 (R169K R172K R173K) | 10 | (1.1 ± 0.3) × 107 | 3 | (1.7 ± 0.5) × 107 |
| M144 (R266K R268K) | 9 | (5.3 ± 1.5) × 106 | 3 | (2.0 ± 0.7) × 107 |
| M145 (R272K R274K R275K R276K) | 9 | (9.7 ± 5.2) × 106 | 3 | (1.6 ± 0.2) × 107 |
| M146 (M141 + M142) | 3 | (9.6 ± 4.8) × 104 | NDb | |
| M147 (M141 + M143) | 3 | (5.4 ± 1.8) × 104 | ND | |
| M148 (M141 + M144) | 3 | (1.5 ± 0.6) × 105 | ND | |
| M149 (M141 + M145) | 3 | (1.5 ± 0.4) × 105 | ND | |
| M150 (M142 + M143) | 3 | (8.6 ± 1.5) × 106 | ND | |
| M151 (M142 + M144) | 3 | (1.0 ± 0.3) × 107 | ND | |
| M152 (M142 + M145) | 3 | (7.9 ± 2.6) × 106 | ND | |
| M153 (M143 + M144) | 3 | (6.5 ± 1.7) × 106 | ND | |
| M154 (M143 + M145) | 3 | (6.5 ± 1.1) × 106 | ND | |
| M155 (M144 + M145) | 3 | (5.7 ± 1.7) × 106 | ND | |
| M156 (M146 + M144) | 3 | (7.6 ± 3.3) × 104 | ND | |
| M157 (M156 + M143) | 3 | (7.2 ± 4.7) × 104 | ND | |
| M158 (K396R) | 3 | (7.1 ± 1.0) × 106 | ND | |
This value was derived from a lytically-infected culture.
ND, not done.
M141 and derivates containing M141 were the poorest mutants in this assay and gave rise to titers reduced up to 2 orders of magnitude compared to wt titers. In addition, M141 induced a massive cytopathic effect (CPE) upon transfection of HEK 293T cells, which normally produce insignificant CPE upon transfection with wt FV constructs (see Fig. S3 in the supplemental material). The reason for this is currently not clear.
Analysis of PFV revertants and pseudo-revertants in gag.
To determine the possible occurrence of revertants and pseudo-revertants at different sites in gag, we transfected HEK 293T cells with mutated constructs and transferred the cell-free virus to indicator cells. CPE developed after 3 to 4 days, and new cell cultures were infected by simultaneous transfer of cell-free supernatants. The gag sequences were amplified after the first and 10th cell-free passage from extracellular viral DNA by seminested PCR and subjected to nucleic acid sequencing. This analysis revealed that all mutants were genetically stable at the modified positions and in the remainder of gag.
Nucleic acid content of lysine-containing viruses.
One major difference between orthoretroviruses and FVs concerns the virion nucleic acid content, at least upon cell culture replication (32, 48). We therefore speculated that also the avoidance of lysines in Gag might influence this. By using an AZT inhibition assay (38, 52), the time point of reverse transcription during replication of wt PFV and mutants was determined. This revealed no significant difference between wt virus and mutants (see Fig. S4 in the supplemental material). In addition, using quantitative real-time PCR and RT-PCR, we measured the virion nucleic acid content of wt PFV and mutant particles (see Table S2). Results using pol gene-specific primers indicated that some viral mutants contained more DNA in relation to RNA than the wt virus, which has been shown to harbor from equal amounts up to approximately six times as much RNA as DNA, depending on the genomic region (long terminal repeat [LTR] versus gag) that has been looked at (52, 75). It appeared that with all mutants a slight tendency to harbor more DNA than the amount harbored by wt virus was observed. However, no clear picture emerged in that the better-replicating mutants contained more DNA than the slower-replicating viruses. Furthermore, the poorly replicating PFV M141 mutant harbored less nucleic acid (DNA as well as RNA) than wt or the better-replicating viruses.
Relevance of the DNA genome.
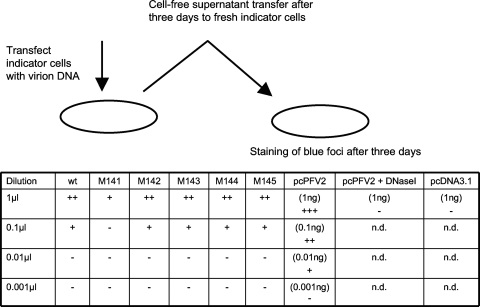
A further indication that in FVs DNA is the relevant genome comes from experiments where cells were transfected with DNA extracted from virions (52, 75). To analyze this aspect in a semiquantitative way for the PFV gag mutants, we transfected BHK indicator cells with serially 10-fold diluted DNAs extracted from partially purified virions and transferred identical amounts of the cell-free supernatants to fresh indicator cells for staining 3 days later. As illustrated in Fig. 2 with the exception of M141, that appeared to contain less infectious DNA, all other mutants behaved like wt virus.
Fig. 2.
Semiquantitative analysis of the infectiousness of virion DNA from PFV. DNAs were extracted from virions, and indicator cells were transfected with 1 μl of virion DNA corresponding to 0.8 ml of supernatant or 10-fold dilutions thereof, as indicated. Controls included the transfection of cells with the infectious plasmid pcPFV2, with pcPFV2 treated with DNase I, and with pcDNA3.1. After the transfection of cells, 1 ml of the cell-free supernatant was transferred to fresh indicator cells, which were subsequently stained for blue foci, scored as follows: +++, approximately 10%; ++, approximately 5%; +, up to 0.1%; −, no blue cell foci; n.d., not determined. A representative example of two individual experiments yielding virtually similar results is shown.
In summary, there was a match between the results obtained from titer determination, measurement of the virion nucleic acid content, and the transfection of permanent cells with virion DNA: in all assays M141 was poor, while the other mutants reached approximately wt values.
Ubiquitination of mutated Gag.
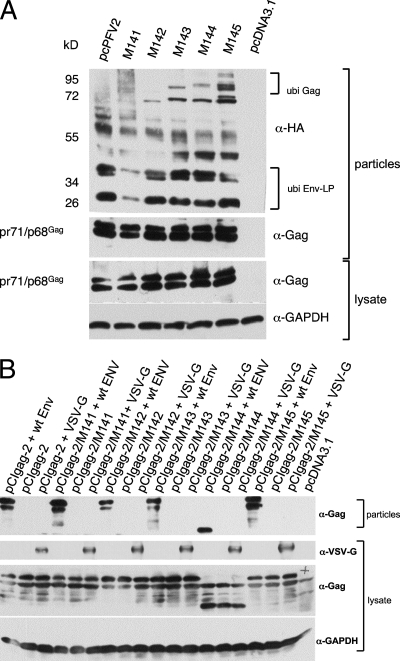
Lysine residues can be posttranslationally altered, for instance, by adding ubiquitin (70). As far as structural viral proteins are concerned, mono- or oligo-ubiquitination is regularly observed when proteins are interacting with the VPS machinery although the specific role of this Gag modification in viral particle export is still ill defined (36, 46, 77). By cotransfecting cells with proviral plasmids together with an expression construct for HA-tagged ubiquitin (64), we analyzed our mutants for the presence of ubiquitinated Gag. As shown in Fig. 3A, Gag as well as Env-LP ubiquitination was demonstrated for the majority of mutant particles analyzed while wt PFV displayed only Env-LP ubiquitination (62). The data from the immunoblotting experiment were suggestive of an oligo-ubiquitination of the altered PFV Gag proteins (except for M141, that was not found to be unequivocally ubiquitinated in Gag and was possibly reduced in Env-LP ubiquitination) at least when generated in HEK 293T cells.
Fig. 3.
Ubiquitination and analysis of pseudotyping and VLP formation by PFV Gag. (A) The ubiquitination of wt and mutated PFV was analyzed by transfecting HEK 293T cells with the respective proviral constructs together with the HA-tagged ubiquitin-encoding plasmid pBJ5 HA-Ubi. Cellular lysates and extracellular viral particles were prepared and probed with antisera against GAPDH and PFV Gag (cellular lysate) or the HA-tag and Gag (viral particles). Oligo-ubiquitination is readily visible for the Env-LP and for the lysine mutants (as indicated). (B) A potential pseudotyping or VLP formation of altered PFV capsids was analyzed by transfecting the cells with the vector pTW01 and subgenomic constructs specifying wt or mutant PFV gag, wt pol, wt env, the heterologous glycoprotein VSV-G, or no glycoprotein (VLP formation), as indicated. The immunoblot was developed using antisera against Gag, VSV-G, and GAPDH. All mutants still required authentic Env for particle export. In contrast, when inserted into the proviral plasmid backbone, M144 led to premature cleavage or expression of a truncated Gag when inserted into the subgenomic expression plasmid. α, anti.
Assay for VLP formation and pseudotyping of viral mutants.
The finding of ubiquitinated Gag prompted us to analyze the dependence on authentic Env for particle export of the mutated PFVs. To do this, we cotransfected HEK 293T with the four-plasmid PFV vector system described previously (20) using the wt and the five mutated gag gene expression constructs M141 to M145 in the pCIgag-2 background. Aside from the invariant pTW01 vector and the pol gene expression construct pCpol-2, two different env gene expression plasmids were individually employed for cotransfection of cells: wt Env corresponds to the basic PFV env plasmid pCenv-1, and the envelope protein of vesicular stomatitis virus (VSV-G) was used for the analysis of pseudotyping PFV Gag. The transfection of cells in the absence of any env construct was used for the analysis of VLP formation. As indicated in Fig. 3B, all mutants still required the presence of the authentic Env for particle export. Similar results were obtained when the supernatant was analyzed for infectivity by vector transfer (data not shown). The expression of VSV-G was monitored by immunoblotting using a monoclonal antibody against this heterologous envelope protein (Fig. 3B), and its functionality was determined by successful transfer of an HIV-1 vector construct (data not shown). Despite ubiquitination, the mutant capsid proteins were neither able to be pseudotyped by the heterologous VSV-G envelope nor to form VLPs.
Interestingly, a discrepancy between the results obtained with the M144 mutant inserted into a replication-competent molecular clone and into the subgenomic gag expression vector was observed. Introduction of M144 in the subgenomic gag expression plasmid led to either a premature cleavage or the generation of truncated Gag (Fig. 3B) and a largely reduced infectivity upon the respective vector transfer (data not shown). Since the DNA sequence of the expression construct appeared to be correct and since the inability of the respective plasmid to generate full-length Gag was observed with three independent molecular clones, it was unlikely to be occurring by chance. A potential explanation for this unusual feature would be alternative splicing in the context of this particular expression construct. However, we did not analyze this matter further.
Analysis in primary cells and of IFN sensitivity.
To determine the replication competence of the PFV mutants in a different cell type and under altered conditions, we used viruses prepared by transfection of HEK 293T cells with full-length constructs, infected MRC-5 cells at a MOI of 0.1, and determined their cell-free titers after replication for 5 days. The primary human fibroblasts were either left untreated or were incubated with 100 U/ml α-IFN or γ-IFN (incubation period from −24 h until transfer of supernatant), respectively.
Early reports on the IFN-dependence of FV replication (50, 51) were the rationale behind this experiment. Specifically, it was reported that γ-IFN delayed the cultivation of FVs and, furthermore, that the likelihood of isolating FV from naturally infected host cells was strongly enhanced by the addition of antibodies to γ-IFN to the culture medium (15). These findings point to the vulnerability of FV replication to the IFN system although FVs are not regarded as superb IFN inducers (51).
As shown in Table 3, we noted a difference in replication competence of all mutants compared to wt virus. The titer reduction of M141 was not surprising since it was already observed in permanent cells. However, the other mutants, with the exception of M142, replicated slightly less well upon cultivation in primary cells. In particular, M154, having seven arginine residues changed to lysine, appeared to be affected. Furthermore, upon IFN treatment this defect was severely enhanced. In particular, γ-IFN treatment almost abolished the replication of mutants; however, a significant reduction was also observed with unmutated virus.
Table 3.
Infection of MRC-5 cells in the presence or absence of interferon
| Virus (no. of additional lysines in Gag) | Titer in MRC-5 cells (per ml)a |
||
|---|---|---|---|
| Naive | With α-IFN (100 U/ml) | With γ-IFN (100 U/ml) | |
| pcPFV2 | 6.3 × 104 | 2.8 × 103 | 12 |
| M141 (2) | 3.0 × 103 | 1.1 × 102 | <1 |
| M142 (2) | 6.6 × 104 | 4.2 × 102 | 1 |
| M143 (3) | 1.1 × 104 | 4.3 × 102 | 3 |
| M144 (2) | 3.1 × 104 | 5.8 × 101 | 1.5 |
| M145 (4) | 2.9 × 104 | 3.1 × 101 | 1 |
| M154 (7) | 9.8 × 103 | 8 | <1 |
| M158 | 7.2 × 101 | 6 | <1 |
Mean values of two independent experiments.
These results left the possible explanation for us that the rareness of lysines in FV Gag proteins is an evolutionary strategy to avoid proteasomal degradation resulting from an antiviral innate immune response. To gain some insight, we investigated the role of antiretroviral restriction factors on the replication of our mutants.
Role of known cellular restriction factors.
FV replication has been reported to be sensitive to a variety of known cellular antiretroviral restriction factors, namely, to proteins belonging to the APOBEC3 (9, 34, 42, 53), TRIM5α (73), and tetherin (CD317) families (24). The incorporation of APOBEC3 proteins is a feature of the viral capsid (8). However, the target of APOBEC3 proteins is the viral nucleic acid, and therefore these proteins are very unlikely to shape the FV Gag protein (8). Furthermore, the FV Bet protein already counteracts cellular APOBEC3 proteins (34, 42, 53). We did not, therefore, further investigate this and concentrated on the potential role of tetherin and Trim5α in being responsible for the particular composition of FV Gag proteins.
The target of tetherin is the enveloped virion (14). Since tetherin is induced by IFN (39), we determined its role on the FV gag gene mutants as follows: HEK 293T cells were transfected with the four-plasmid FV vector system described above employing the pMD9 vector, env, and pol expression constructs and either wt or the mutated gag packaging plasmids together with pchCD317HA, an expression plasmid for human tetherin, or an irrelevant control plasmid. As a further control we performed a similar experiment using the pNL4-3 and pNL4-3 Vpu− HIV-1 proviral constructs. The cell-free supernatants were analyzed for infectivity 2 days later on TZM indicator cells (in case of HIV-1 molecular clones) or on HT1080 human fibroblasts by FACS (in case of PFV constructs). As reported previously (24), we observed a drop in infectivity upon coexpression of human tetherin for both, Vpu− HIV-1 and the PFV vector system. However, we did not detect any significant difference between wt and mutated PFV gag constructs except the reduction in vector transfer seen with the M144 subgenomic gag expression plasmid, already described above (see Fig. S5 in the supplemental material).
The target of Trim5α proteins is the incoming viral capsid (14, 40, 68). The Trim5 family members are also induced by IFN (68). Furthermore, the N-terminal half of Gag, the region harboring the residues in question here, also contains the FV region conferring susceptibility to Trim5α proteins (73). Concordant with results obtained with other retroviruses, the restriction was conferred by the B30.2 domain of TRIM5α (60, 73). FVs are vulnerable to only a very minor fraction of heterologous Trim5α proteins and, in particular, the human variant restricted none of the FV isolates tested (73). To analyze a potential involvement of Trim5α in the evolution of basic residues in FV Gags, we employed a similar approach, as described recently (73). Human HT1080 fibroblastoid cells were modified to express chimeric Trim5α genes with B30.2 domains from various species together with EYFP by gammaretroviral vector transfer. The challenge vector, EGFP expressing pMD9, was produced by transfecting HEK 293T cells with the four-plasmid FV vector system, already described using wt and mutated gag packaging constructs. Equal amounts of challenge virus were used to transduce the recipient HT1080 cells. However, we noted no difference with respect to the Trim5α restriction pattern between the wt and gag-mutated virus vectors (see Table S3 in the supplemental material).
Role of K396Gag in PFV replication.
To get a first hint of the role in replication of the single lysine at position 396 of PFV Gag (K396Gag), we changed the corresponding nucleic acid sequence to specify an arginine codon (Fig. 1; see also Fig. S1 in the supplemental material). Analysis of the resulting mutant M158 for replication in a transient assay using HEK 293T cells for virus production and BHK indicator cells as recipients revealed that it behaved almost like wt virus (Table 2). However, when primary MRC-5 fibroblasts were allowed to replicate M158, we noted a very strong reduction in titer development (Table 3). This reduction was found to be most prominent in naive MRC-5 cells (approximately 3 logs), and we regard the further drop in infectivity observed upon cultivation of cells in the presence of IFN to be nonsignificant relative to the drop also observed with unmutated virus (Table 3).
DISCUSSION
Before the HIV-1 pandemic, FVs were probably the most successful of all retroviruses. With the exception humans and rodents, FVs naturally infect a very wide spectrum of mammalian hosts (37). Recent evidence indicates that FVs are found in some wild Artiodactyla species (M. Materniak and J. Kuzmak, Poland, personal communication), and it can be assumed that FVs are even more widespread.
In evolutionary terms FVs can be regarded as successful since they have maintained their genome for long periods of time and only marginally evolved faster than their natural hosts (57, 65, 69). It is likely that the benign nature of infection and the peculiar FV replication strategy (32, 48) contribute considerably to this success. It is further likely that the avoidance of lysine residues in the FV Gag proteins adds to this replication strategy.
Database searches showed that the avoidance of lysines, as in FV Gag proteins, is a rare feature in virology. Among over 2,400 sequenced viruses, we found only six non-foamy viral proteins larger than 500 aa that were devoid of lysines and harbored arginines (see Table S5 in the supplemental material). Among these were three herpesviral and three poxviral proteins, all proteins of large DNA-containing viruses. The function of these proteins is largely unknown, and it remains to be investigated whether the lack of lysine is an indication of comparable selective pressure acting on these proteins.
When we modified PFV molecular clones to harbor lysines instead of arginines in their Gag proteins, we expected to observe a gradual drop in viral replication competence with every newly introduced lysine residue. Surprisingly, except for one mutant (M141 and derivatives) that was shown to incorporate less nucleic acid, the other mutants replicated like wt virus and showed no tendency to revert within the period of the experiment, if assayed in a permanent cell line. Furthermore, investigation of other aspects, such as late reverse transcription or DNA content, revealed no significant differences between these mutants and the wt.
With respect to M141, the immunoblot showing equal amounts of cellular expression and cell-free virus compared to wt levels, together with the strong CPE we observed upon transfecting HEK 293T cells with this mutant, argues for a premature activation of the Env fusion activity and underscores the idea that budding of FVs requires a specific Gag-Env interaction (45). M141 capsid assembly may be completed before sufficient amounts of viral nucleic acids have been incorporated. The lower nucleic acid content after introduction of this mutant into HEK 293T cells is consistent with this view.
The introduction of lysine residues in PFV Gag resulted in posttranslational modifications of what appeared to be oligo-ubiquitination (except for M141). This modification did not result in the independence of PFV capsids from its authentic Env to be exported. However, it may have influenced the outcome of the replication experiment in primary cells.
While in untreated MRC-5 cells the difference in replication competence between the gag mutants (except M141) and wt virus ranged from nonexisting (M142), to marginal (M143, M144, and M145), to a maximum of 10-fold (M154), IFN treatment resulted in a more severe drop in infectivity. This was hardly visible upon γ-IFN treatment, probably due to the sharp drop of infectivity observed even with wt virus that did not allow the detection of a significant further titer reduction of the mutants. However, α-IFN treatment resulted in a clearly discernible effect of all gag mutants. Furthermore, there was at least for some viral mutants a clearly detectable dependence on the number of lysine residues introduced; e.g., M154 (bearing seven lysines) was more impaired than the parental viruses M143 (three lysines) and M145 (four lysines). It remains to be seen whether upon IFN treatment of permanent cell lines, a similar reduction in PFV titers can be detected. In addition, the IFN treatment probably only enhances the visibility of the replication disadvantage of the lysine mutants because we mutated only 7 out of 41 residues in question. Thus, changing more lysine residues may reveal the detection of further defects even in the absence of IFN treatment and may depend on the actual residues changed because in this respect not all basic amino acids may be of identical importance.
The observed effect could be attributed neither to tetherin nor to Trim5α, two factors known to restrict FV replication (24, 73). Thus, it appears possible that the lack of lysines is an elegant evolutionary strategy of FV to avoid proteasomal degradation of the Gag protein that is made in great excess over the other structural virus proteins in infected cells (our unpublished observation). In following this argument, it will be instructive to analyze ubiquitination of the gag mutants in MRC-5 cells. However, other possible posttranslational lysine modifications (7, 70), such as sumoylation, acetylation, or methylation, also remain to be investigated.
We initially thought that the codon specifying the single lysine at position 396 of PFV Gag did not convert to an arginine codon, simply because of the lack of evolutionary pressure. However, the replication behavior in untreated primary cells indicated that this is not the case. For unknown reasons, there is evolutionary pressure on PFV to keep this lysine residue in place. It would be interesting to know how other FVs bearing lysine residues in their Gag proteins behave in this respect and what the functions of these residual lysines might be.
Supplementary Material
ACKNOWLEDGMENTS
We are indebted to Paul Bieniasz (New York, NY) for very helpful discussions on the FV replication strategy, to Jörg Schultz (Würzburg, Germany) for help with the bioinformatical analysis, and to Ben Berkhout (Amsterdam, Netherlands), Valerie Bosch (Heidelberg, Germany), Heinrich Göttlinger (Worcester, MA), Jeremy Luban (Geneva, Switzerland), Didier Trono (Lausanne, Switzerland), and Klaus Überla (Bochum, Germany) for discussions or gifts of reagents. We thank B. Pacheco and J. Sodroski (Boston, MA) for making available the SFVsqu and SFVmar sequences prior their appearance in GenBank, Benedikt Weissbrich (Würzburg, Germany) for MRC-5 cells, and the sequencing unit of the institute for their help with DNA sequencing, Stefan Gattenlöhner (Würzburg, Germany) for invaluable help with cell staining, Myra O. McClure (London, United Kingdom) for critical reading of the manuscript, and Sylvia Geubig for its preparation.
This work was supported by DFG (SFB 479 and Re 627/8) and BMBF (FONEFA).
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Adachi A., et al. 1987. Productive, persistent infection of human colorectal cell lines with human immunodeficiency virus. J. Virol. 61:209–213 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Ausubel F. M., et al. 1987. Current protocols in molecular biology. John Wiley, New York, NY [Google Scholar]
- 3. Bender A., van Dooren G. G., Ralph S. A., McFadden G. I., Schneider G. 2003. Properties and prediction of mitochondrial transit peptides from Plasmodium falciparum. Mol. Biochem. Parasitol. 132:59–66 [DOI] [PubMed] [Google Scholar]
- 4. Bock M., Bishop K. N., Towers G., Stoye J. P. 2000. Use of a transient assay for studying the genetic determinants of Fv1 restriction. J. Virol. 74:7422–7430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bosch V., Pawlita M. 1990. Mutational analysis of the human immunodeficiency virus type 1 env gene product proteolytic cleavage site. J. Virol. 64:2337–2344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cartellieri M., et al. 2005. N-terminal Gag domain required for foamy virus particle assembly and export. J. Virol. 79:12464–12476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chen Y., et al. 2007. Lysine propionylation and butyrylation are novel post-translational modifications in histones. Mol. Cell Proteomics 6:812–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cullen B. R. 2006. Role and mechanism of action of the APOBEC3 family of antiretroviral resistance factors. J. Virol. 80:1067–1076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Delebecque F., et al. 2006. Restriction of foamy viruses by APOBEC cytidine deaminases. J. Virol. 80:605–614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Derdeyn C. A., et al. 2000. Sensitivity of human immunodeficiency virus type 1 to the fusion inhibitor T-20 is modulated by coreceptor specificity defined by the V3 loop of gp120. J. Virol. 74:8358–8367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. DuBridge R. B., et al. 1987. Analysis of mutation in human cells by using an Epstein-Barr virus shuttle system. Mol. Cell. Biol. 7:379–387 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Eastman S. W., Linial M. L. 2001. Identification of a conserved residue of foamy virus Gag required for intracellular capsid assembly. J. Virol. 75:6857–6864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Enssle J., et al. 1997. Carboxy-terminal cleavage of the human foamy virus Gag precursor molecule is an essential step in the viral life cycle. J. Virol. 71:7312–7317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Evans D. T., Serra-Moreno R., Singh R. K., Guatelli J. C. 2010. BST-2/tetherin: a new component of the innate immune response to enveloped viruses. Trends Microbiol. 18:388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Falcone V., Schweizer M., Toniolo A., Neumann-Haefelin D., Meyerhans A. 1999. Gamma interferon is a major suppressive factor produced by activated human peripheral blood lymphocytes that is able to inhibit foamy virus-induced cytopathic effects. J. Virol. 73:1724–1728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Fischer N., et al. 1998. Foamy virus particle formation. J. Virol. 72:1610–1615 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Flügel R. M., Pfrepper K. I. 2003. Proteolytic processing of foamy virus Gag and Pol proteins. Curr. Top. Microbiol. Immunol. 277:63–88 [DOI] [PubMed] [Google Scholar]
- 18. Gattenloehner S., et al. 2007. Novel RUNX1 isoforms determine the fate of acute myeloid leukemia cells by controlling CD56 expression. Blood 110:2027–2033 [DOI] [PubMed] [Google Scholar]
- 19. Giron M. L., Colas S., Wybier J., Rozain F., Emanoil-Ravier R. 1997. Expression and maturation of human foamy virus Gag precursor polypeptides. J. Virol. 71:1635–1639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Heinkelein M., et al. 2002. Improved primate foamy virus vectors and packaging constructs. J. Virol. 76:3774–3783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Higuchi R. 1990. Recombinant PRC, p. 177–183 In Innis M. A., Gelfand D. H., White T. J. (ed.), PCR protocols: a guide to methods and applications. Academic Press, San Diego, CA [Google Scholar]
- 22. Hunter E. 1997. Viral entry and receptors, p. 71–121 In Coffin J. M., Hughes S. H., Varmus H. E. (ed.), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY: [PubMed] [Google Scholar]
- 23. Jacobs J. P., Jones C. M., Baille J. P. 1970. Characteristics of a human diploid cell designated MRC-5. Nature 227:168–170 [DOI] [PubMed] [Google Scholar]
- 24. Jouvenet N., et al. 2009. Broad-spectrum inhibition of retroviral and filoviral particle release by tetherin. J. Virol. 83:1837–1844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Katzourakis A., Gifford R. J., Tristem M., Gilbert M. T., Pybus O. G. 2009. Macroevolution of complex retroviruses. Science 325:1512. [DOI] [PubMed] [Google Scholar]
- 26. Lefrancois L., Lyles D. S. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. I. Analysis of neutralizing epitopes with monoclonal antibodies. Virology 121:157–167 [PubMed] [Google Scholar]
- 27. Lefrancois L., Lyles D. S. 1982. The interaction of antibody with the major surface glycoprotein of vesicular stomatitis virus. II. Monoclonal antibodies of nonneutralizing and cross-reactive epitopes of Indiana and New Jersey serotypes. Virology 121:168–174 [DOI] [PubMed] [Google Scholar]
- 28. Lehmann-Che J., et al. 2005. Protease-dependent uncoating of a complex retrovirus. J. Virol. 79:9244–9253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Life R. B., Lee E. G., Eastman S. W., Linial M. L. 2008. Mutations in the amino terminus of foamy virus Gag disrupt morphology and infectivity but do not target assembly. J. Virol. 82:6109–6119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Lindemann D., Goepfert P. A. 2003. The foamy virus envelope glycoproteins. Curr. Top. Microbiol. Immunol. 277:111–129 [DOI] [PubMed] [Google Scholar]
- 31. Lindemann D., et al. 2001. A particle-associated glycoprotein signal peptide essential for virus maturation and infectivity. J. Virol. 75:5762–5771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Linial M. L. 2007. Foamy viruses. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 33. Linial M. L., et al. 2005. Retroviridae, p. 421–440 In Fauquet C. M., Mayo M. A., Maniloff J., Desselberger U., Ball L. A. (ed.), Virus taxonomy: classification and nomenclature of viruses. Eighth report of the International Committee on Taxonomy of Viruses. Elsevier/Academic Press, London, United Kingdom [Google Scholar]
- 34. Löchelt M., et al. 2005. The antiretroviral activity of APOBEC3 is inhibited by the foamy virus accessory Bet protein. Proc. Natl. Acad. Sci. U. S. A. 102:7982–7987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mannigel I., Stange A., Zentgraf H., Lindemann D. 2007. Correct capsid assembly mediated by a conserved YXXLGL motif in prototype foamy virus Gag is essential for infectivity and reverse transcription of the viral genome. J. Virol. 81:3317–3326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Martin-Serrano J. 2007. The role of ubiquitin in retroviral egress. Traffic 8:1297–1303 [DOI] [PubMed] [Google Scholar]
- 37. Meiering C. D., Linial M. L. 2001. Historical perspective of foamy virus epidemiology and infection. Clin. Microbiol. Rev. 14:165–176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Moebes A., et al. 1997. Human foamy virus reverse transcription that occurs late in the viral replication cycle. J. Virol. 71:7305–7311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Neil S. J., Zang T., Bieniasz P. D. 2008. Tetherin inhibits retrovirus release and is antagonized by HIV-1 Vpu. Nature 451:425–430 [DOI] [PubMed] [Google Scholar]
- 40. Nisole S., Stoye J. P., Saib A. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3:799–808 [DOI] [PubMed] [Google Scholar]
- 41. Patton G. S., Morris S. A., Chung W., Bieniasz P. D., McClure M. O. 2005. Identification of domains in Gag important for prototypic foamy virus egress. J. Virol. 79:6392–6399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Perkovic M., et al. 2009. Species-specific inhibition of APOBEC3C by the prototype foamy virus protein bet. J. Biol. Chem. 284:5819–5826 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Petit C., et al. 2003. Targeting of incoming retroviral Gag to the centrosome involves a direct interaction with the dynein light chain 8. J. Cell Sci. 116:3433–3442 [DOI] [PubMed] [Google Scholar]
- 44. Pfrepper K. I., et al. 1999. Molecular characterization of proteolytic processing of the Gag proteins of human spumavirus. J. Virol. 73:7907–7911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Pietschmann T., et al. 1999. Foamy virus capsids require the cognate envelope protein for particle export. J. Virol. 73:2613–2621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Pincetic A., Leis J. 2009. The mechanism of budding of retroviruses from cell membranes. Adv. Virol. 2009:6239691–6239699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Rasheed S., Nelson-Rees W. A., Toth E. M., Arnstein P., Gardner M. B. 1974. Characterization of a newly derived human sarcoma cell line (HT-1080). Cancer 33:1027–1033 [DOI] [PubMed] [Google Scholar]
- 48. Rethwilm A. 2007. Foamy virus vectors: an awaited alternative to gammaretro- and lentiviral vectors. Curr. Gene Ther. 7:261–271 [DOI] [PubMed] [Google Scholar]
- 49. Rethwilm A. 2003. The replication strategy of foamy viruses. Curr. Top. Microbiol. Immunol. 277:1–26 [DOI] [PubMed] [Google Scholar]
- 50. Rhodes-Feuillette A., et al. 1990. Effects of human recombinant alpha and gamma and of highly purified natural beta interferons on simian Spumavirinae prototype (simian foamy virus 1) multiplication in human cells. Res. Virol. 141:31–43 [DOI] [PubMed] [Google Scholar]
- 51. Rhodes-Feuillette A., Saal F., Lasneret J., Santillana-Hayat M., Peries J. 1987. Studies on in vitro interferon induction capacity and interferon sensitivity of simian foamy viruses. Arch. Virol. 97:77–84 [DOI] [PubMed] [Google Scholar]
- 52. Roy J., et al. 2003. Feline foamy virus genome and replication strategy. J. Virol. 77:11324–11331 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Russell R. A., et al. 2005. Foamy virus Bet proteins function as novel inhibitors of the APOBEC3 family of innate antiretroviral defense factors. J. Virol. 79:8724–8731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Sambrook J., Russell D. W. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY [Google Scholar]
- 55. Schliephake A. W., Rethwilm A. 1994. Nuclear localization of foamy virus Gag precursor protein. J. Virol. 68:4946–4954 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Schmidt M., Rethwilm A. 1995. Replicating foamy virus-based vectors directing high level expression of foreign genes. Virology 210:167–178 [DOI] [PubMed] [Google Scholar]
- 57. Schweizer M., Neumann-Haefelin D. 1995. Phylogenetic analysis of primate foamy viruses by comparison of Pol sequences. Virology 207:577–582 [DOI] [PubMed] [Google Scholar]
- 58. Seeger C., Zoulim F., Mason W. S. 2007. Hepadnaviruses, p. 2977–3029 In Knipe D. M., et al. (ed.), Fields virology, 5th ed. Lippincott Williams, Philadelphia, PA [Google Scholar]
- 59. Soneoka Y., et al. 1995. A transient three-plasmid expression system for the production of high titer retroviral vectors. Nucleic Acids Res. 23:628–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Song B., et al. 2005. Retrovirus restriction by TRIM5alpha variants from Old World and New World primates. J. Virol. 79:3930–3937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Stange A., et al. 2005. Characterization of prototype foamy virus Gag late assembly domain motifs and their role in particle egress and infectivity. J. Virol. 79:5466–5476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Stanke N., Stange A., Luftenegger D., Zentgraf H., Lindemann D. 2005. Ubiquitination of the prototype foamy virus envelope glycoprotein leader peptide regulates subviral particle release. J. Virol. 79:15074–15083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Stirnnagel K., et al. 2010. Analysis of prototype foamy virus particle-host cell interaction with autofluorescent retroviral particles. Retrovirology 7:45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Strack B., Calistri A., Gottlinger H. G. 2002. Late assembly domain function can exhibit context dependence and involves ubiquitin residues implicated in endocytosis. J. Virol. 76:5472–5479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Switzer W. M., et al. 2005. Ancient co-speciation of simian foamy viruses and primates. Nature 434:376–380 [DOI] [PubMed] [Google Scholar]
- 66. Tobaly-Tapiero J., et al. 2001. Human foamy virus capsid formation requires an interaction domain in the N terminus of Gag. J. Virol. 75:4367–4375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Tobaly-Tapiero J., et al. 2008. Chromatin tethering of incoming foamy virus by the structural Gag protein. Traffic 9:1717–1727 [DOI] [PubMed] [Google Scholar]
- 68. Towers G. J. 2007. The control of viral infection by tripartite motif proteins and cyclophilin A. Retrovirology. 4:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Verschoor E. J., et al. 2004. The phylogeography of orangutan foamy viruses supports the theory of ancient repopulation of Sumatra. J. Virol. 78:12712–12716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Walsh C. T., Garneau-Tsodikova S., Gatto G. J., Jr 2005. Protein posttranslational modifications: the chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 44:7342–7372 [DOI] [PubMed] [Google Scholar]
- 71. Wiktorowicz T., Peters K., Armbruster N., Steinert A. F., Rethwilm A. 2009. Generation of an improved foamy virus vector by dissection of cis-acting sequences. J. Gen. Virol. 90:481–487 [DOI] [PubMed] [Google Scholar]
- 72. Wurm M., et al. 2010. The influence of semen-derived enhancer of virus infection on the efficiency of retroviral gene transfer. J. Gene Med. 12:137–146 [DOI] [PubMed] [Google Scholar]
- 73. Yap M. W., et al. 2008. Restriction of foamy viruses by primate Trim5α. J. Virol. 82:5429–5439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Yu S. F., et al. 1996. The carboxyl terminus of the human foamy virus Gag protein contains separable nucleic acid binding and nuclear transport domains. J. Virol. 70:8255–8262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Yu S. F., Sullivan M. D., Linial M. L. 1999. Evidence that the human foamy virus genome is DNA. J. Virol. 73:1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Zemba M., et al. 1998. The carboxy-terminal p3Gag domain of the human foamy virus Gag precursor is required for efficient virus infectivity. Virology 247:7–13 [DOI] [PubMed] [Google Scholar]
- 77. Zhadina M., McClure M. O., Johnson M. C., Bieniasz P. D. 2007. Ubiquitin-dependent virus particle budding without viral protein ubiquitination. Proc. Natl. Acad. Sci. U. S. A. 104:20031–20036 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.