Abstract
Morphogenesis of human cytomegalovirus (HCMV) is still only partially understood. We have characterized the role of HCMV tegument protein pUL71 in viral replication and morphogenesis. By using a rabbit antibody raised against the C terminus of pUL71, we could detect the protein in infected cells, as well as in virions showing a molecular mass of approximately 48 kDa. The expression of pUL71, detected as early as 48 h postinfection, was not blocked by the antiviral drug foscarnet, indicating an early expression. The role of pUL71 during virus replication was investigated by construction and analysis of a UL71 stop mutant (TBstop71). The mutant could be reconstituted on noncomplementing cells proving that pUL71 is nonessential for virus replication in human fibroblasts. However, the inhibition of pUL71 expression resulted in a severe growth defect, as reflected by an up to 16-fold reduced extracellular virus yield after a high-multiplicity infection and a small-plaque phenotype. Ultrastructural analysis of cells infected with TBstop71 virus revealed an increased number of nonenveloped nucleocapsids in the cytoplasm, many of them at different stages of envelopment, indicating that final envelopment of nucleocapsids in the cytoplasm was affected. In addition, enlarged multivesicular bodies (MVBs) were found in close proximity to the viral assembly compartment, suggesting that pUL71 affects MVBs during virus infection. The observation of numerous TBstop71 virus particles attached to MVB membranes and budding processes into MVBs indicated that these membranes can be used for final envelopment of HCMV.
INTRODUCTION
Human cytomegalovirus (HCMV) is an important opportunistic pathogen (16). HCMV infection causes considerable morbidity and mortality in individuals with an immature or compromised immune system, such as transplant recipients, cancer patients, AIDS patients, children in utero, and prematurely born infants.
HCMV is a member of the Betaherpesvirinae subfamily. Mature HCMV virions are comprised of four distinct structures identifiable in all herpesviruses: core, capsid, tegument, and envelope. The icosahedral capsid contains the core, which consists of the ∼240-kb linear double-stranded DNA genome. The envelope enclosing the entire virus particle is a lipid membrane derived from the host cell with incorporated virally encoded glycoproteins (33). The connection between the viral capsid and the envelope is mediated by the tegument. The tegument of HCMV is formed mostly by viral proteins but has been shown to also contain cellular proteins (46). From the 59 viral proteins which have been found in the tegument, only 39 proteins seem to be incorporated at significant levels into virus particles (1, 46). Tegument proteins have been described to possess specific functions, such as activation and regulation of viral gene expression, immune evasion, release of viral DNA into the nucleus, and regulation of cellular processes (reviewed in reference 21). Apart from their involvement in viral entry processes and aiding the establishment of infection, tegument proteins play important roles in viral morphogenesis, particularly as structural components and in envelopmental processes that result in infectious particles. The data mainly from alphaherpesviruses suggest that an intricate network of protein-protein interactions leads to the formation of the tegument and is equally important for secondary envelopment. In this context, many tegument proteins or functions seem to be conserved among herpesviruses (reviewed in reference 31). However, only few tegument proteins of HCMV have been studied in more detail, whereas the functions of the majority of tegument proteins remain largely unknown.
From sequence analysis it has been suggested that HCMV protein UL71 is one of the conserved herpesviral proteins (31). Homologous proteins can be found within all herpesvirus subfamilies. For example, the homolog of pUL71 in Epstein-Barr virus is BSRF1 (20) and in alphaherpesviruses UL51 (2, 26). Although there is not much known about the function of HCMV pUL71, it has been shown that UL51 in pseudorabies virus (PRV) and herpes simplex virus 1 (HSV-1) are not essential for virus replication. However, a UL51-null mutant of HSV-1 exhibited a growth defect reflected by reduced plaque sizes and a nearly 100-fold reduction in virus yield compared to wild-type virus (36). A similar growth defect has been reported for a UL51-deficient mutant in PRV (23). Large numbers of intracytoplasmic capsids lacking the envelope or at various stages of envelopment were found, suggesting that envelopmental processes were affected in the absence of PrV UL51 (23). Both PrV UL51 and HSV-1 UL51 are localized at the Golgi membranes in transfected cells (35). In infected cells, HSV-1 UL51 was found within cytoplasmic vesicles or in the viral envelope (35). From all of these studies, it has been suggested that UL51 may be important for the trafficking of Golgi-associated particles and/or for the targeting of capsids to their final envelopment site.
In both HSV-1 and PRV, UL51 has been identified as a viral protein incorporated into virus particles (10, 23). HCMV pUL71 has also been characterized as part of the tegument of virus particles (46). However, the specific function of pUL71 in the replication cycle of HCMV has not been investigated yet, and the published data about its requirement for viral replication in fibroblasts is contradictory. By deleting entire genes in a global HCMV genome mutagenesis approach, pUL71 was characterized to be essential for virus replication (13), while in another study using transposon mutagenesis, pUL71 was nonessential, but its inactivation resulted in a severe growth defect (48).
To clarify the role of pUL71 during HCMV replication, we constructed a mutant virus in which expression of pUL71 was abrogated by introduction of a stop codon 33 bp downstream from the UL71 start codon. In order to analyze the expression of pUL71 during infection, an antibody was produced in rabbits. Growth characteristics of the UL71 stop mutant, as well as of a corresponding revertant virus, were investigated. Finally, the function of pUL71 in virus morphogenesis was further characterized by electron microscopy and electron tomography of infected cells after high-pressure freezing and freeze substitution.
MATERIALS AND METHODS
Cells and viruses.
Human foreskin fibroblasts (HFFs) were used before passage 23 and maintained in minimal essential medium (Gibco-BRL) supplemented with 10% fetal calf serum (Gibco-BRL) and 1× nonessential amino acids (Biochrom AG). Permanent African green monkey kidney cells (Cos7) were maintained in minimal essential medium supplemented with 5% fetal calf serum. Human embryonic lung fibroblasts (MRC-5; European Collection of Cell Cultures) were maintained in Dulbecco modified Eagle medium (Gibco-BRL) supplemented with 5% fetal calf serum and used between passages 21 to 30. All media were supplemented with 2 mM l-glutamine (Biochrom AG), 100 U of penicillin, and 100 μg of streptomycin (both Gibco-BRL).
The parental virus, termed the wild-type virus, of all mutant viruses used in the present study was reconstituted from the HCMV bacterial artificial chromosome (BAC) clone TB40-BAC4 (41). Virus particles were purified from supernatants of infected cells by using a glycerol tartrate gradient as previously described (8).
Expression plasmids and transfection.
The open reading frame (ORF) of full-length UL71 was amplified with primers ex_71_for (TACGGGATCCATGCAGCTGGCCCAGCGC) and ex_71_rev (TACGGAATTCTTTTCCAAAACGTGCCAGGCTGT) and cloned into the eukaryotic expression vector pEF1/Myc-His C (Invitrogen) by using BamHI and EcoRI restriction sites, resulting in expression plasmid pEF-UL71. The primers ex_71_12-361_for (TACGGGATCCATGTGCCGTCGCAAAGCCG) and ex_71_rev were used for the generation of PCR product UL71aa12-361. The PCR product UL71aa-19-361 was amplified with the primers ex-seq_71_for (TACGGGATCCATGAGCAGAATCATACTCTGTTGCG) and ex_71_rev. These products were cloned similarly to plasmid pEF-UL71 resulting in plasmid pEF-UL71aa-19-361 containing a potential start codon located 57 nucleotides upstream of the annotated UL71 start codon (pUL71aa-19-361) and plasmid pEF-UL71aa12-361 harboring the UL71 sequence starting from the second ATG codon 34 nucleotides downstream of the predicted UL71 start codon (pUL71aa12-361).
For transfection, Cos-7 cells (3.3 × 104) were seeded into each well of a 24-well plate. The next day, the cells were transfected with 500 ng of the generated expression plasmids by using Lipofectamine LTX (Invitrogen) according to the manufacturer's protocol. After incubation for 6 h, the liposome-DNA mixture was removed, and cells were incubated with complete medium for 24 h.
Engineering BAC mutants.
Recombinant BAC clones were generated by using a markerless two-step RED-GAM recombination protocol (44), which was slightly modified for efficient mutagenesis of parental BAC TB40-BAC4 (accession no. EF999921.1 [41]) as described previously (8).
To generate the BAC for the UL71 stop mutant, TBstop71-BAC, a stop codon with a subsequent frameshift was introduced 33 bp downstream of the UL71 start codon as annotated in TB40-BAC4. The primers used for this mutagenesis were ep_71stop_for (GCACGATGCAGCTGGCCCAGCGCCTGTGCGAGCTGCTGTAGCCGTCGCAAAGCCGCGCCTGAGGATGACGACGATAAGTAGGG) and ep_71stop_rev (AGCAGCACGTAATCGGCCACAGGCGCGGCTTTGCGACGGCTACAGCAGCTCGCACAGGCGCCAACCAATTAACCAATTCTGATTAG). To generate the revertant BAC, termed TBrev71-BAC, RED-GAM mutagenesis was performed using the recombinant TBstop71-BAC. A silent mutation was introduced into TBrev71-BAC in order to enable discrimination of wild-type and revertant BAC. The primers used for the generation of TBrev71-BAC were ep_71stop_resc_for (GCACGATGCAGCTGGCCCAGCGCCTGTGCGAGCTGCTGATGTGTCGTCGCAAAGCCGCGCCTGAGGATGACGACGATAAGTAGGG) and ep_71stop_resc_rev (AGCAGCACGTAATCGGCCACAGGCGCGGCTTTGCGACGACACATCAGCAGCTCGCACAGGCGCCAACCAATTAACCAATTCTGATTAG). The primers used for the mutagenesis contain homology to the UL71 gene, both upstream and downstream of the sequence to be mutated, and homology to the pEPkan-S template plasmid (underlined) (44). Proper introduction of the point mutations in TBstop71-BAC and TBrev71-BAC were confirmed by sequencing. All BAC DNAs were further checked by restriction fragment length polymorphism.
Virus reconstitution.
For virus reconstitution, HCMV BAC DNA was isolated from Escherichia coli by using a Nucleobond AX Midi Kit (Macherey-Nagel). Recombinant viruses were reconstituted in MRC-5 cells by electroporation as described previously (8). HFFs were used for further virus propagation and generation of virus stocks. The recombinant viruses recovered from TB40-BAC4, TBstop71-BAC, and TBrev71-BAC were named wild type, TBstop71, and TBrev71, respectively.
Generation of a polyclonal antibody directed against pUL71.
The sequence of pUL71 was subjected to an in silico antibody epitope prediction analysis using free software Antigenicity Plot (http://www.bioinformatics.org/JaMBW/3/1/7/) which predicted high antigenicity for the C terminus of pUL71 encompassing amino acids 162 to 361. Therefore, the C-terminal region of pUL71 was fused N-terminally to the maltose-binding protein (MBP) and C-terminally to the His-Tag and was expressed using prokaryotic expression vector pTST101 (kindly provided by Guenther Muth, Tuebingen, Germany) in Escherichia coli strain BL21 (Novagen). The MBP-pUL71 fusion protein was purified from crude E. coli lysates by affinity chromatography against the MBP. Enzymatic digestion with factor Xa released the pUL71 fragment (15 kDa) from MBP (42 kDa), which was then separated from MBP by SDS-PAGE. After Coomassie blue staining the band corresponding to pUL71 was excised, resolved in phosphate-buffered saline (PBS), and confirmed by Western blotting with an antibody directed against the His-Tag (Penta-His antibody; Qiagen). The purified pUL71 fragment was used for the immunization of rabbits by standard protocols. The rabbits received two booster immunizations at intervals of 2 weeks. After a total of 8 weeks polyclonal sera were collected and used for the detection of pUL71.
Antibodies.
HCMV encoded and cellular proteins were detected with monoclonal antibodies (MAbs) in plaque assay, Western blot analysis or immunofluorescence experiments. The MAbs used in the present study included those directed against MCP (UL86, MAb 28-4), pp28 (UL99, MAb 41-28), pp65 (UL83, MAb 65-33), gB (UL55, MAb 27-287) (all kindly provided by W. Britt, University of Alabama, Birmingham), pp150 (UL32, XP-1) (19), IE1/2 + UL44 (clones CCH2 + DDG9; Dako), actin (Sigma), pp28 (clone 5C3; Santa Cruz), and human CD63 (clone H5C6; BD Pharmingen). Goat anti-mouse and goat anti-rabbit antibodies conjugated to Alexa Fluor 488 and 555, isotype-specific antibodies goat anti-mouse IgG2a conjugated to Alexa Fluor 488 and goat anti-mouse IgG1 conjugated to Alexa Fluor 555 (Invitrogen), Cy3 (Immunoresearch), or horseradish peroxidase (HRP; Millipore), and an HRP-conjugated rabbit anti-mouse antibody (Dako) were used.
Growth analysis and virus titration.
HFFs were infected with wild-type, TBstop71, and TBrev71 viruses at a multiplicity of infection (MOI) of 3 for single step growth kinetics or at an MOI of 0.01 for multistep growth kinetics. For high-MOI infections, cells were centrifuged at 1,900 rpm for 30 min and then incubated for 2 h at 37°C after inoculation. After incubation, the inoculum was removed, and cells were washed three times with PBS to remove remaining virus before adding cell-specific medium. Virus titers of the inocula used were controlled by titration on HFFs to verify the MOI. For low-MOI infections, HFFs were infected with the various viruses for 24 h at 37°C. Virus was removed by three washes with PBS before fresh medium was added. The medium was changed every 3 days. Equal infection was controlled by staining for immediate-early antigen of identically infected cells 48 h postinfection.
Virus-infected cells and supernatants were collected separately every 24 h for 7 days postinfection for high-MOI infections and every 3 days for 18 days for low-MOI infections. Supernatants were cleared of cells and cell debris by centrifugation at 4,000 rpm for 10 min before determination of virus yields on HFFs. Virus-infected cells were collected in fresh medium and stored at −80°C. Cell-associated infectious virus was released by mechanical lysis of cells (multiple uptake through a 1-mm syringe) and cell debris removed by low-speed centrifugation (4,000 rpm, 10 min).
Virus yields were determined on HFFs by titration of virus suspensions. HFFs infected with the different virus dilutions were fixed and permeabilized with ice-cold methanol at 2 days postinfection. Infected cells were stained with an antibody against HCMV proteins IE1/2 and UL44 and visualized by using an HRP-conjugated rabbit anti-mouse antibody (Dako) and a peroxidase assay.
Plaque assay.
Plaque assays were performed by infection of HFFs with the respective viruses, followed by incubation of the cells for 9 days at 37°C under a 0.6 to 0.7% methylcellulose overlay. The overlay was renewed once at day 5 postinfection. On day 9 postinfection, cells were washed two times with PBS to remove the methylcellulose and fixed with methanol prior staining for HCMV proteins IE1/2 and UL44 and visualization by using secondary goat anti-mouse antibody conjugated with Cy3 (Immunoresearch). Images of plaques of wild-type, TBstop71, and TBrev71 virus-infected cells were taken by using the Axio-Observer.Z1 fluorescence microscope (Zeiss) with the ×10 objective and Axiovision software (Zeiss). For each virus, plaque areas of at least 50 plaques were determined by using the software ImageJ (http://rsbweb.nih.gov/ij/index.html).
Electron microscopy.
For electron microscopy, HFF cells were grown on carbon-coated sapphire discs (3 mm in diameter; Engineering Office M. Wohlwend GmbH, Switzerland) and infected at an MOI of 0.5. At 5 days postinfection, the cells were frozen from the living state by high-pressure freezing with an HPF 01 apparatus (Engineering Office M. Wohlwend GmbH) and freeze substituted in acetone containing 0.1% (wt/vol) uranyl acetate, 0.2% (wt/vol) of osmium tetroxide, and 5% (vol/vol) of water, as described previously (4, 5). After being embedded in Epon, ultrathin sections were prepared on copper grids for transmission electron microscopy (TEM). Samples were imaged with the Zeiss EM10 transmission electron microscope at an acceleration voltage of 80 kV.
For the quantification of the various virus morphogenesis stages, fully enveloped and budding virus particles, respectively, were counted in at least 25 images from at least three independent experiments for each virus. All micrographs used in this analysis were taken at a primary magnification of 10.000 in randomly selected regions of the viral assembly compartment. Statistical analysis was performed by using two-tailed Mann-Whitney-U test.
For electron tomography, 500-nm-thick sections were imaged in a Titan (FEI, Eindhoven) 300-kV field emission transmission electron microscope in scanning transmission mode using a high-angle annular dark field (HAADF) detector (Fischione Instruments, Horley, United Kingdom). A total of 71 images were recorded at tilt angles from −70° to +70°. The volume was reconstructed by weighted back projection using the IMOD software (24), with the help of 15-nm colloidal gold as fiducial markers for alignment.
Western blot and immunofluorescence microscopy.
Western blot analysis was performed exactly as described previously (8). For immunofluorescence microscopy, HFFs were seeded in μ-Slide eight-well plates (Ibidi, Martinsried, Germany) and infected with wild-type, TBstop71, and TBrev71 viruses at an MOI of 0.1 or 0.5. At 7 days postinfection, the cells were washed twice with PBS and fixed with 4% paraformaldehyde (PFA) in PBS for 10 min at 4°C. Cells were permeabilized with 0.1% Triton X-100 for 5 min, and nonspecific binding sites were blocked by incubation in blocking buffer (PBS supplemented with 1% bovine serum albumin [BSA] and 6% human serum) for at least 30 min. Primary antibodies were diluted in blocking buffer, followed by incubation for at least 45 min. The cells were then washed three times with washing solution (1% BSA and 0.1% Triton in PBS) and incubated for at least 45 min with the secondary antibodies conjugated with Alexa Fluor 488 or Alexa Fluor 555 (Invitrogen). The nuclei were stained with DAPI (4′,6′-diamidino-2-phenylindole; Roche). Confocal images were taken by using an Axio-Observer.Z1 fluorescence microscope equipped with the Apotome (Zeiss), Axiovision software, and a ×63 objective lens.
RESULTS
Identification and expression kinetics of the HCMV protein UL71.
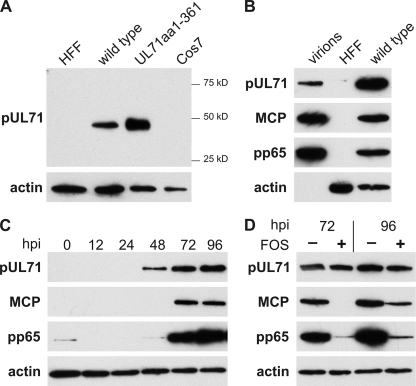
In order to investigate the expression profile of the HCMV protein UL71, a polyclonal antibody was raised in rabbits. The animals were immunized three times with a purified polypeptide encompassing the C-terminal amino acids 162 to 361 of pUL71. In Western blot analyses, the antibody specifically detected a protein of ∼48 kDa in lysates of HCMV-infected human fibroblasts and in lysates of Cos7 cells transiently transfected with the pUL71 expression plasmid pEF-UL71, confirming the specificity of our antibody (Fig. 1A). pUL71 has been previously identified as a protein of low abundance in virus particles by a two stage mass spectrometry approach (46). We therefore examined whether pUL71 could also be detected in gradient-purified virions. Again, a specific signal for pUL71 corresponding to the size of the pUL71 in lysates of virus-infected HFFs was detected in gradient-purified HCMV virions (Fig. 1B). These findings confirmed that pUL71 is a component of virus particles.
Fig. 1.
Characterization of HCMV pUL71 by Western blot analysis in infected and transfected cells, as well as in purified virus particles. (A) The presence of pUL71 was assayed by using a polyclonal pUL71 antibody directed against the C terminus of pUL71 in lysates of HFFs infected with HCMV wild-type virus at 5 days postinfection (wild type), mock-infected HFFs (HFF), and Cos7 cells transiently transfected with pEF-UL71 24 h posttransfection (UL71aa1-361) and control cells (Cos7). Values for the prestained molecular weight marker, Precision Plus (Bio-Rad), are displayed to the right. (B) Western blot analysis of lysates of gradient purified wild-type virus (virions), HFFs infected with wild-type virus 5 days postinfection (wild type), and mock-infected cells (HFF). Viral proteins pUL71, MCP, pp65, and cellular actin were detected with respective antibodies. (C) Expression kinetics of viral proteins in HFFs infected with wild-type virus. Viral proteins pUL71, MCP, and pp65 were detected at the indicated times postinfection. Detection of cellular actin was used to ensure comparable loading. (D) Expression of viral proteins pUL71, MCP, and pp65 in absence or presence of foscarnet (FOS, 400 μmol) in HFFs infected with wild-type virus at 72 and 96 h postinfection. Actin was used as loading control. Note that in the presence of foscarnet, the expression of MCP and pp65, but not of pUL71, is impaired.
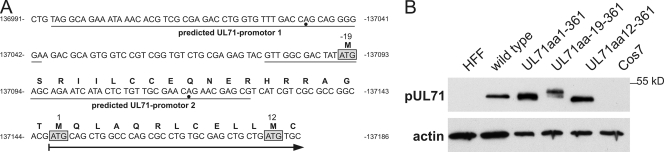
DNA sequence analysis of the UL71 region in the used HCMV strain TB40-BAC4, revealed two more possible start codons for the translation of pUL71: one upstream (resulting in a larger protein UL71aa19-361) and one downstream (resulting in a shorter protein UL71aa12-361) of the annotated start codon (resulting in protein UL71aa1-361). The promoter prediction software (Neural Network Promoter Prediction [http:www.fruitly.org/seq_tools]) predicted two promoters in the UL71 promoter region of TB40-BAC4 (Fig. 2A). To clarify which of the start codons is in fact used, expression plasmids were constructed containing UL71 sequences from all three start codons, respectively. Lysates of Cos7 cells transfected with these plasmids were analyzed by Western blotting and compared to lysates of HCMV-infected cells. As shown in Fig. 2B, expression of UL71aa1-361 resulted in a protein of similar size to that found in virus-infected cells. These data show that the already annotated start codon in the TB40-BAC4 sequence is used for the translation of UL71.
Fig. 2.
Characterization of the start codon of UL71. (A) Sequence of the promoter region of UL71 from nucleotides 136991 to 137186 of the TB40-BAC4 sequence (40). Predicted promoters within this sequence are underlined and putative transcriptional start is indicated by a dot. Possible start codons of pUL71 are boxed, and numbers above correspond to the amino acids relative to the annotated start codon indicated by the number 1. The arrow indicates the start and direction of pUL71 translation as annotated (accession number EF999921.1). (B) Western blot analysis of HFFs infected with HCMV wild-type virus at 5 days postinfection (wild type), mock-infected HFFs (HFF), Cos7 cells (Cos7), and Cos7 cells transiently transfected with the pUL71 expression plasmids in panel A as indicated.
In order to investigate the expression kinetics of pUL71, we performed Western blot analysis of lysates of wild-type virus-infected HFFs harvested at several time points postinfection and compared the expression of pUL71 to that of other HCMV proteins (Fig. 1C). The pUL71-specific band was first detected at 48 h postinfection. Starting from 72 h postinfection, increasing levels of pUL71 were detected, similar to early-late tegument protein pp65. However, pUL71 did not reach the high levels of the early-late tegument protein pp65 (Fig. 1C). This result suggested an early or early-late expression kinetics for pUL71. To verify this observation, viral DNA replication was blocked by treatment of infected cells with the antiviral drug foscarnet. As shown in Fig. 1D, pUL71 was detected in unchanged amounts in cell lysates of infected cells treated with foscarnet while the expression of the true-late major capsid protein (MCP) and of the early-late protein pp65 was clearly inhibited. These results show that expression of pUL71 is not dependent on viral DNA replication and that pUL71 is thus sharing expression characteristics of early proteins.
Construction and propagation of a UL71 stop mutant.
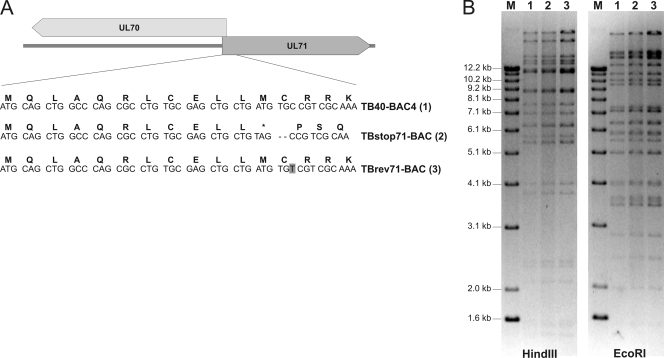
There have been contradictory reports showing that the product of the UL71 ORF is either essential for (13) or nonessential but very important for viral growth in fibroblast (48). To further elucidate this point, we sought to generate a UL71-deficient recombinant virus. Our analysis of the genomic region in which the UL71 ORF is located revealed that the promoter of the UL70 ORF lies within UL71. Therefore, it was not possible to delete the entire UL71 sequence without the risk of affecting the expression of ORF UL70. To minimize the risk of affecting expression of ORF UL70, a UL71 stop mutant (TBstop71-BAC) was generated based on BAC TB40-BAC4, which is the infectious clone of the highly endotheliotropic HCMV strain TB40E (41). As depicted in Fig. 3A, the codon for methionine (M12) 33 bp downstream from the start codon was mutated to a stop codon, followed by the introduction of a frameshift mutation using RED-mediated recombination. As a control, a revertant BAC mutant (TBrev71-BAC) was generated based on TBstop71-BAC, in which the mutation was restored to the wild-type sequence. In order to be able to distinguish between TBrev71-BAC and wild-type BAC TB40-BAC4, a silent mutation was introduced in TBrev71-BAC (Fig. 3A). The successful introduction of the stop codon mutation in TBstop71-BAC, as well as its restoration in TBrev71-BAC was confirmed by sequence analysis of the mutated regions. Furthermore, restriction fragment length polymorphism analysis revealed identical restriction patterns for all three BACs, confirming that no spurious recombination of the DNA sequences had occurred (Fig. 3B).
Fig. 3.
Generation of UL71 stop mutant TBstop71. (A) Position of the introduced stop codon and the frameshift mutation at the 5′ prime of UL71. In TBrev71-BAC, the stop and frameshift mutation was restored to the original UL71 sequence with a silent mutation (indicated by the gray box). (B) Restriction fragment length polymorphism analysis of BAC-DNAs of TB40-BAC4 (wild type), TBstop71-BAC, and TBrev71-BAC with restriction enzymes EcoRI and HindIII. The three BAC-DNAs were used to reconstitute the respective viruses. Lanes M, molecular marker (1-kb DNA ladder; Invitrogen).
Wild-type virus, mutant virus TBstop71, and the revertant virus TBrev71 were reconstituted from the respective BAC-DNAs in MRC-5 cells for further analysis. TBstop71 was replication competent in MRC-5 cells, proving that pUL71 is not essential for virus replication.
Protein expression of TBstop71.
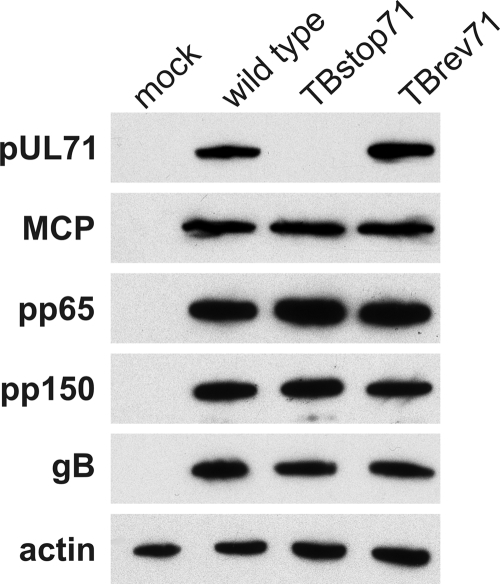
We next verified that the introduction of the stop codon in UL71 indeed prevents its expression. Lysates of HFFs infected with wild-type, TBstop71, and TBrev71 viruses were subjected to Western blot analysis using our polyclonal antiserum against pUL71 (Fig. 4). pUL71 was not detected in lysates of TBstop71 virus-infected cells but was clearly present in lysates of wild-type and TBrev71 virus-infected cells. This confirmed that expression of pUL71 in the TBstop71 virus mutant was abrogated by the introduction of the stop codon and further ensured the specificity of our antiserum against pUL71. The lysates were additionally probed for a number of other HCMV proteins, such as MCP, pp150, pp65, and glycoprotein B (gB). Expression levels of these viral proteins were unaltered in lysates of TBstop71 virus-infected cells compared to lysates of wild-type and revertant virus (Fig. 4). These data suggested that the expression of these other viral proteins was not affected by the lack of pUL71 in cells infected with the TBstop71 virus.
Fig. 4.
Western blot analysis of HFFs infected with wild-type, TBstop71, and TBrev71 viruses at 5 days postinfection and mock-infected control cells. Cell lysates were separated on a 10% sodium dodecyl sulfate-polyacrylamide gel and analyzed by Western blot analysis. Actin and viral proteins were detected with the respective antibodies.
Virus growth is impaired in the absence of pUL71.
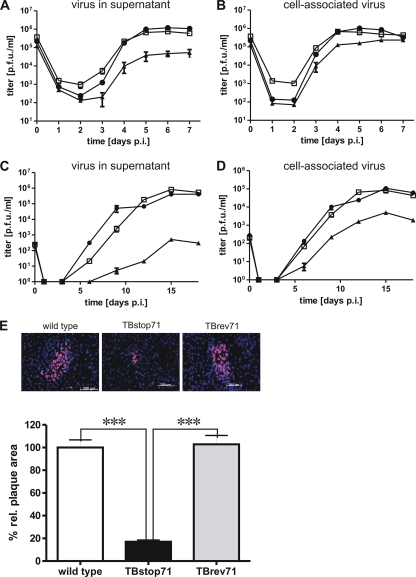
As noted earlier, propagation of the TBstop71 virus was possible in noncomplementing cells, which showed that pUL71 is not essential for virus replication. To investigate the growth properties of TBstop71 virus in more detail, we analyzed the ability of wild-type, TBstop71, and TBrev71 viruses to spread from cell to cell in HFFs by measuring plaque sizes at 9 days postinfection. While plaques formed by TBrev71 virus were comparable in size and morphology to those formed by wild-type virus, infection with TBstop71 virus produced only small plaques with plaque areas ca. 20% the size of those formed by wild-type and revertant viruses (Fig. 5).
Fig. 5.
Growth characterization of TBstop71 virus. (A and B) Growth kinetics of wild-type (□), TBstop71 (▴), and TBrev71 (•) viruses. HFFs were infected with an MOI of 3. Culture supernatants (A) and cell-associated virus particles (B) were sampled at the indicated times postinfection, and virus yields were determined by titration on HFFs. Day 0 values represent the inocula. Each data point and error bar represents the mean ± the standard error of at least three independent experiments. (C and D) Multistep growth kinetic of wild-type (□), TBstop71 (▴), and TBrev71 (•) viruses. HFFs were infected with an MOI of 0.01. Culture supernatants (C) and cell-associated virus particles (D) were sampled at the indicated times postinfection, and virus yields were determined by titration on HFFs. Day 0 values represent the inocula. Each data point and error bar represents the mean ± the standard error of at least three independent experiments. (E) Plaque assay of wild-type, TBstop71, and TBrev71 virus-infected HFFs. Cells were infected with 100 PFU. After 24 h, the cells were overlaid with methylcellulose and fixed with methanol at 9 days postinfection. Infected cells were detected by an anti-IE1/2 and UL44 monoclonal antibody. The proteins were visualized by a Cy3-conjugated secondary goat anti-mouse antibody. Nuclei were marked with DAPI. The plaque areas of wild-type, TBstop71, and TBrev71-virus infected cells were analyzed with the Axio-Observer.Z1 fluorescence microscope using a ×10 objective lens. For the statistical analysis, the relative plaque areas of wild-type, TBstop71, and TBrev71 virus-infected cells were measured by the program ImageJ. The mean percentage of the areas and standard error were determined by counting at least 50 plaques in each experiment. (The experiments were repeated at least two times; ***, P < 0.0001.)
Next, we compared the replication kinetics of wild-type, TBstop71, and TBrev71 viruses in HFFs using an MOI of 3. The amount of cell-associated infectious particles, as well as virus yield, in the supernatants was determined at different times postinfection. Virus yield in the supernatant of TBstop71 virus-infected cells on day 7 postinfection was ∼16-fold reduced compared to that of wild-type or revertant virus (Fig. 5A). Yields of cell-associated virus of TBstop71 determined from cell lysates were only slightly reduced compared to those of wild-type and TBrev71 viruses (Fig. 5B). However, a pronounced growth defect of TBstop71 was observed in multistep replication kinetics using an MOI of 0.01. Both cell-associated virus yields and virus accumulation in the supernatant were delayed and severely impaired compared to wild-type and TBrev71 viruses (Fig. 5C and D).
In summary, similar properties of wild-type and TBrev71 viruses with respect to replication kinetics or plaque formation indicated that the mutation in TBstop71 was successfully repaired and that the observed growth defects of TBstop71 virus were caused by the absence of pUL71.
Absence of pUL71 affects the morphology of multivesicular bodies.
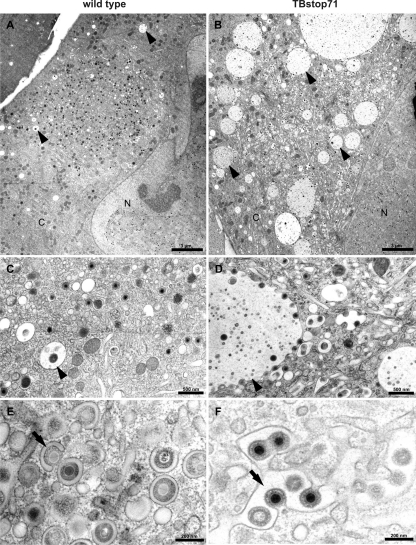
By electron microscopy we investigated whether the observed growth defect of TBstop71 virus is based on a defect in virus morphogenesis. Fibroblasts were grown on sapphire disks, infected with wild-type or TBstop71 viruses using an MOI of 0.5, fixed by high-pressure freezing on day 5 postinfection, and further processed for TEM.
In the cytoplasm of wild-type virus-infected cells we regularly found small vesicles in close proximity to the assembly compartment containing several intraluminal vesicles (Fig. 6A). According to the size of the intraluminal vesicles (∼50 nm in diameter), their ultrastructure, and their numbers inside of the large vesicles, the latter were phenotypically classified as multivesicular bodies (MVBs) (22, 32). This is in accordance with previous reports describing endosomes with characteristics of MVBs in HCMV-infected cells (15). In wild-type virus-infected cells, MVBs exhibited diameters of usually considerably less than 1 μm and only rarely contained virions or dense bodies (Fig. 6C). In contrast to that, considerably enlarged MVBs with diameters of up to several μm in close proximity to the viral assembly compartment were found in cells infected with TBstop71 virus (Fig. 6B). Interestingly, numerous TBstop71 virus particles were attached to membranes of these enlarged MVBs, and many of these MVBs contained virions (Fig. 6D). The accumulation of viral particles at the MVB membranes, as observed in TBstop71 virus-infected cells, was not found in wild-type virus-infected cells. These findings indicated that pUL71 influences the morphology of MVBs during HCMV infection.
Fig. 6.
Ultrastructural analysis of wild-type (A, C, and E) and TBstop71 (B, D, and F) virus-infected MRC-5 cells at 5 days postinfection. (A) A typical assembly compartment of an HCMV wild-type infected cell in close proximity to the nucleus (N). MVBs (arrowheads) with diameters of not more than 1 μm can be observed in the cytoplasm (C). (B) In a TBstop71 virus-infected cell, many enlarged MVBs (arrowhead) with diameters of several μm can be seen located around the assembly compartment. (C) No virus particles are attached at the MVB membrane, although in some cases viral particles or dense bodies were found inside of MVBs. (D) Many viral particles are attached to MVB membranes. Some of them are in a budding process into or are already inside the lumen of the MVB. (E) Few viral particles in a budding process into small crescent-shaped vesicles (arrow). Note that the majority of virus particles are fully enveloped within small vesicles. (F) Larger vesicles (arrow) are used as multiple budding sites by TBstop71 virus particles.
pUL71 is important for efficient secondary envelopment of HCMV.
We further concentrated our electron microscopic investigations on the viral morphogenesis at 5 days postinfection. Although nuclear stages of virus morphogenesis were indistinguishable between the TBstop71 and wild-type viruses (data not shown), we found differences regarding the secondary envelopment of these viruses in the cytoplasm. Final envelopment occurs in a virally induced compartment in the perinuclear region, termed assembly compartment. Whereas most of the particles found in the assembly compartment of wild-type virus-infected cells were fully enveloped (Fig. 6E), a large number of nucleocapsids in the process of secondary envelopment were observed in cells infected with TBstop71 virus (Fig. 6F). To allow a quantitative comparison, enveloped, budding, and naked cytoplasmic nucleocapsids in the assembly compartment were counted using TEM images of TBstop71 and wild-type virus-infected cells (Table 1) . The quantification confirmed our first observation that, in contrast to wild-type virus, the majority of TBstop71 nucleocapsids is not yet fully enveloped, indicating that pUL71 is important for an efficient secondary envelopment of HCMV particles.
Table 1.
Quantification of HCMV envelopment in the cytoplasmic assembly compartment at 5 days postinfection
| Virus | No. of cells analyzed | Mean % particles ± SDa |
||
|---|---|---|---|---|
| Enveloped particles | Budding particles | Naked particles | ||
| Wild type | 27 | 86.64 ± 4.82 | 12.95 ± 14.68 | 0.41 ± 1.66 |
| TBstop71 | 26 | 26.90 ± 6.99 | 70.14 ± 9.40 | 2.97 ± 3.83 |
The percent values of enveloped virus particles, nonenveloped virus particles attached to membranes (budding particles), and nonenveloped virus particles not connected to membranes (naked particles) are given.
In addition to the already described morphological differences in TBstop71 virus-infected cells, it is noteworthy that in an increased number of cells the viral assembly compartment was composed of enlarged vesicles or vesicles of a more tubular shape into which budding of virus particles took place (Fig. 6D and F). These alterations could only rarely be seen in wild-type virus-infected cells in which budding of nucleocapsids occurred mainly into small vesicles (Fig. 6E). In addition, these enlarged vesicles or tubules (Fig. 6F), as well as membranes of vesicles ultrastructurally defined as MVBs (Fig. 6D), were used as budding sites by numerous nucleocapsids at the same time in TBstop71 virus-infected cells.
Electron tomography of TBstop71 virus-infected cells.
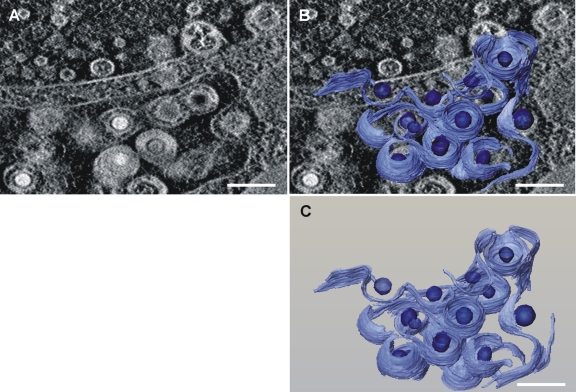
To obtain a better view of the envelopmental defect of TBstop71 virus, electron tomography was performed on 500-nm thin sections of virus-infected cells at 5 days postinfection. As also shown in Fig. 7, the majority of the cytoplasmic nucleocapsids of TBstop71 virus are in the process of secondary envelopment at membranes of large vesicles or vesicles morphologically defined as MVBs (see movie S1 [http://www.uniklinik-ulm.de/struktur/institute/virologie/home/forschung/publikationen/supp1.html]). As observed in ultrathin sections, numerous budding events into these vesicles were observed in cells infected with TBstop71 virus. Together with the results of the growth kinetics, these data indicated that in the absence of pUL71 secondary envelopment is less efficient or proceeds slower than in wild-type virus-infected cells.
Fig. 7.
Three-dimensional reconstruction of membranes and viral capsids from tomography of TBstop71 virus-infected HFFs at 5 days postinfection. (A) Single slice from the three-dimensional tomogram with a large vesicle in which multiple budding processes of TBstop71 virus particles are observed. (B) Membranes used as budding sites as well as viral capsids were reconstructed and superimposed on the original micrograph. Membranes are depicted in light blue; viral capsids are indicated in dark blue. (C) Reconstruction of the budding processes only. Bars, 200 nm. Note that areas with high mass density, such as membranes, occur bright in the dark-field STEM tomograms. See Movie S1 at http://www.uniklinik-ulm.de/struther/institute/virologie/home/forschung/publikationen/suppl.html.
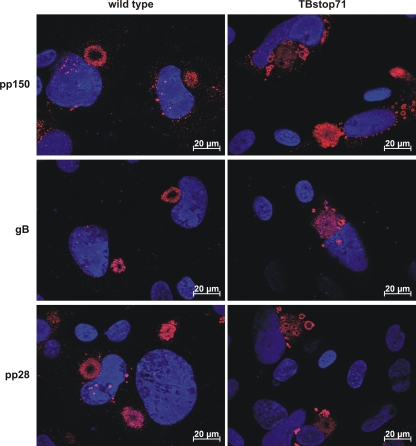
pp150, pp28, and gB are also localized to the enlarged vesicles in TBstop71 virus-infected HFFs.
Based on our observation of the association and budding processes of numerous virus particles at membranes of large MVBs in TBstop71 virus-infected cells, we hypothesized that other viral proteins important for viral budding must be also localized to these membranes. We therefore investigated the intracellular localization of pp150, pp28, and gB at 7 days postinfection (Fig. 8). In wild-type or TBrev71 virus-infected cells, we found an accumulation of pp150, pp28, and gB at the viral assembly compartment. In TBstop71 virus-infected cells, pp150 and pp28, as well as gB, additionally accumulated at large intracellular vesicles in close proximity to the assembly compartment. These large vesicles could very well correspond to the enlarged MVBs observed in ultrastructural analysis.
Fig. 8.
Subcellular localization of gB, pp28, and pp150 in wild-type and TBstop71 virus-infected HFFs (MOI = 0.1 to 0.5). Cells were fixed with 4% PFA in PBS at 7 days postinfection. Proteins were detected by the respective primary antibodies visualized by goat anti-mouse antibodies conjugated to Alexa Fluor 555. Nuclei were marked with DAPI.
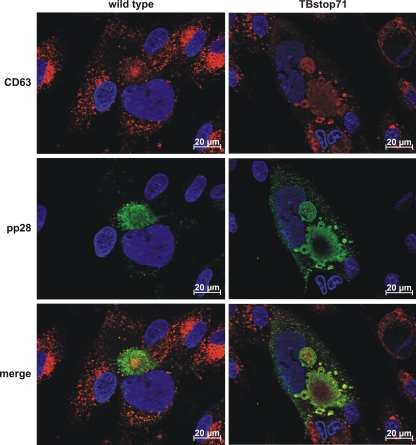
To further demonstrate that these large vesicles are indeed MVBs, we used cellular CD63 as a marker protein. CD63 is a tetraspanin abundantly present in late endosomes and lysosomes and is enriched on intraluminal vesicles (reviewed in reference 37). In TBstop71 virus-infected cells, CD63 signals accumulated in the lumens of large vesicles which were also stained positive for pp28, indicating that these enlarged structures are MVBs (Fig. 9).
Fig. 9.
Subcellular localization of MVB marker CD63 and viral pp28 in wild-type and TBstop71 virus-infected HFFs (MOI = 0.1 to 0.5). Cells were fixed with 4% PFA in PBS at 7 days postinfection. CD63 and pp28 were detected by the respective MAbs and visualized by isotype-specific secondary antibodies conjugated to Alexa Fluor 488 (for pp28) and 555 (for CD63). Cell nuclei were visualized by using DAPI.
DISCUSSION
The function of many HCMV proteins during viral morphogenesis is still unknown, but it is assumed that tegument proteins form a structured network by protein-protein interactions that is important for the production of infectious viral progeny. We performed the present study to characterize the tegument protein pUL71 and to gain information on its role during HCMV infection. From previous HCMV genome analyses the kinetics of pUL71 expression were not clear (7). Our data show that pUL71 has to be classified as a viral protein expressed with an early kinetic, since pUL71 expression was not inhibited by the antiviral drug foscarnet, whereas expression of the early-late protein pp65 or true-late protein MCP was clearly blocked. By using an antiserum produced against a bacterially expressed C-terminal pUL71 polypeptide, we show that pUL71 is expressed as a protein of ∼48 kDa in infected cells and is also detectable as a component of purified virions. The latter finding supports previous data from a mass spectrometry-based study (46) indicating that pUL71 is incorporated into HCMV particles during infection.
Naturally, many components of the complex herpesvirus particles are important for viral morphogenesis, but genome-wide mutagenesis studies were not able to clarify the specific function and contribution of pUL71 to HCMV replication (13, 48). By generation of the UL71 stop mutant virus, TBstop71, which does not express pUL71, we were able to show that—although not essential for viral replication in fibroblasts—the absence of pUL71 in the TBstop71 virus resulted in a severe growth defect compared to wild-type and revertant viruses. This defect is reflected by a dramatically impaired cell-to-cell spread and viral growth in both low- and high-MOI infections with TBstop71 virus. Similar to our result is the phenotype of a mutant based on HCMV strain AD169 carrying a transposon insertion in UL71 which is characterized by a small-plaque phenotype and impaired viral growth (48). A small-plaque phenotype and an impaired virus release has also been described for deletion mutants of the pUL71 homolog UL51 in the alphaherpesviruses HSV-1 (36) and PRV (23).
Interestingly, in low-MOI infections, cell-associated virus yields of TBstop71 virus were severely reduced, whereas cell-associated virus yields in high-MOI infections revealed only a slight reduction compared to wild-type and revertant viruses. This was unexpected in light of the pronounced envelopment defect and the strong reduction of virus yields in the supernatant of TBstop71 virus-infected cells. The reason for this is not entirely clear. One explanation might be that the transport of virus containing vesicles is impaired when pUL71 is not expressed. Another explanation could be that infectious virus particles get trapped in large vesicles in the cytoplasm of infected cells. There is ultrastructural evidence for the latter showing virus particles within MVBs and (in some cases) an accumulation of virions in large vesicles in cells infected with TBstop71 virus.
The quantitative electron microscopy results from ultrathin sections and the use of electron tomography clearly revealed a defect in secondary envelopment in the absence of pUL71. Wild-type virus particles appeared significantly more often at a fully enveloped state compared to the TBstop71 virus. Most of the TBstop71 particles were observed at various stages of envelopment, while envelopment processes could less frequently be seen in wild-type virus-infected cells. Obviously, virion formation is impaired or delayed in TBstop71 virus-infected cells, which in turn may be the underlying cause for the impaired virus yields of TBstop71 virus. Klupp et al. reported similar findings for a UL51 deletion mutant of PrV, indicating that UL51 is involved in virion morphogenesis (23). In the case of UL51 of HSV-1, an involvement at an early step of viral morphogenesis downstream of the primary envelopment has been reported (36). Altogether, an association with envelopmental processes seems to be common for all pUL71 homologs investigated thus far, although the underlying molecular mechanisms remain to be elucidated.
HCMV morphogenesis is completed in a virally induced compartment often termed as “assembly compartment” or “assembly complex” (18, 38). The assembly compartment represents a complex juxtanuclear structure where many virally encoded tegument, envelope, and nonstructural proteins have been found to accumulate during infection (11, 17, 18, 25, 27, 38, 39, 43). In agreement with the function as the site of final envelopment, the assembly compartment has been reported to contain many vesicles derived from the trans-Golgi network, secretory, and endosomal pathways (9, 12, 15, 38, 45). It has been reported that HCMV infection induces significant changes in the cellular membrane system such as a reorientation of components of the endosomal pathway that leads to the formation of the assembly compartment (12). This reorientation also includes other cellular proteins such as the endoplasmic reticulum chaperone BiP for which a contribution to the formation, integrity, and function of the assembly compartment has been proposed (3). Although the exact origin of the membranes used by HCMV for its final envelopment is still under debate, recent data support the hypothesis that the virus may induce a novel membrane compartment containing both TGN and endosomes (6). Ultrastructural analysis of HCMV-infected cells revealed that most of the viral particles acquire their final envelope by wrapping into small vesicles that turn into crescent-shaped cisternae as the budding process proceeds (40; the present study). This eventually results in small vesicles containing a single enveloped virus particle (15; the present study). In contrast to this, we noticed an unusual enlargement of tubular vesicles within the viral assembly compartment when pUL71 is not present and multiple budding events into those tubules. These data suggest that in the absence of pUL71 the formation of tubules and/or vesicles used for viral budding may also be affected.
The most striking morphological abnormality in cells infected with TBstop71 was the appearance of enlarged vesicles containing multiple intraluminal vesicles in close proximity to the viral assembly compartment (Fig. 6B). Such an appearance is well described in the relevant literature and strongly suggests that these are late endosomes also termed as MVBs (22, 32). Our observation that numerous virus particles were found (i) in close association with membranes of large MVBs, (ii) in envelopmental processes into MVBs, or (iii) to be enveloped within MVBs underlines previous data that membranes of MVBs or endosomal membranes can be used as a final budding site (12, 14). The cellular processes of membrane budding that are normally part of endosome sorting and MVB formation are controlled by the cellular endosomal sorting complex required for transport (ESCRT). Recent data showed that the ESCRT ATPase Vps4 is recruited to the HCMV assembly compartment, thereby suggesting that ESCRT machinery is involved in final maturation steps (42). In the light of our TBstop71 phenotype and the importance of the ESCRT machinery for MVB formation, it is reasonable to speculate that pUL71 has the ability to interfere with MVBs or to manipulate ESCRT functions, thus causing abnormal MVBs. Alternatively, it is conceivable that the delayed envelopment of TBstop71 resulting in the accumulation of virus particles in the AC may cause the development of enlarged vesicles as a downstream effect of the envelopment defect. Further investigations are needed to clarify this.
It is assumed that final envelopment requires the presence of all necessary viral and cellular proteins (reviewed in references 28, 29, 30, and 31). This at least seems to be the case for the HCMV tegument proteins pp28 and pp150, as well as for gB, which were accumulated at large vesicular structures in close proximity to the assembly compartment in cells infected with TBstop71 virus. These structures are positive for CD63, an MVB marker protein (37), verifying together with the ultrastructural analysis that these vesicular structures are late endosomes of the MVB type. However, the role of MVBs for HCMV maturation or egress is still unclear. There are at least two possible fates of virus particles inside MVBs: (i) degradation by fusion of the MVBs with the lysosome or (ii) transport within MVBs to the cell membrane along the exosomal pathway and egress from the cell by fusion of the MVB membrane with the plasma membrane. Evidence for both possibilities has been published. A slightly increased virus production can be observed upon treatment of HCMV-infected cells with protease inhibitors to inhibit endolysosomal proteolysis, suggesting that lysosomal degradation of at least a small portion of virus particles takes place during infection (14). It has recently been described that mature virions of the betaherpesvirus human herpesvirus 6 can be found in MVBs and are released from the cell by fusion of the MVB membrane with the plasma membrane, demonstrating that MVBs are used as exit route by betaherpesviruses (34). However, during our intensive ultrastructural investigations, no evidence of MVBs fusing with the plasma membrane could be found in HCMV-infected cells. Thus, we hypothesize that MVBs are not hijacked by HCMV to support viral egress from fibroblasts. It is conceivable that the importance of MVBs for viral morphogenesis or egress can be different in the various cell types that are productively infected by HCMV.
In conclusion, our results indicate an important role of pUL71 for virus spread and release and in particular in envelopmental processes that lead to the formation of infectious particles. The absence of pUL71 results in a delayed completion of the envelopment process that goes along with enlarged vesicles within the viral assembly compartment. During the revision process of the present study, the phenotype of our pUL71stop mutant was confirmed by another group using a different HCMV strain (47). Furthermore, our data show that MVB membranes can be used as viral budding site during HCMV infection. In addition, we could show that the absence of the viral protein UL71 leads to an enlargement of MVBs, indicating for an interaction of HCMV with the cellular machinery responsible for MVB formation.
ACKNOWLEDGMENTS
The technical assistance of Jutta Hegler, Monika Duerre, and Anke Lueske is gratefully acknowledged.
This study was supported by the Deutsche Forschungsgesellschaft through SPP1175 (grant ME 1740/2-1) and by the Landesgraduiertenförderung Baden-Württemberg.
Footnotes
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Baldick C. J., Shenk T. 1996. Proteins associated with purified human cytomegalovirus particles. J. Virol. 70:6097–6105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baumeister J., Klupp B. G., Mettenleiter T. C. 1995. Pseudorabies virus and equine herpesvirus 1 share a nonessential gene which is absent in other herpesviruses and located adjacent to a highly conserved gene cluster. J. Virol. 69:5560–5567 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Buchkovich N. J., Maguire T. G., Paton A. W., Paton J. C., Alwine J. C. 2009. The endoplasmic reticulum chaperone BiP/GRP78 is important in the structure and function of the human cytomegalovirus assembly compartment. J. Virol. 83:11421–11428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Buser C., et al. 2007. Quantitative investigation of murine cytomegalovirus nucleocapsid interaction. J. Microsc. 228:78–87 [DOI] [PubMed] [Google Scholar]
- 5. Buser C., Walther P., Mertens T., Michel D. 2007. Cytomegalovirus primary envelopment occurs at large infoldings of the inner nuclear membrane. J. Virol. 81:3042–3048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Cepeda V., Esteban M., Fraile-Ramos A. 2010. Human cytomegalovirus final envelopment on membranes containing both trans-Golgi network and endosomal markers. Cell Microbiol. 12:386–404 [DOI] [PubMed] [Google Scholar]
- 7. Chambers J., et al. 1999. DNA microarrays of the complex human cytomegalovirus genome: profiling kinetic class with drug sensitivity of viral gene expression. J. Virol. 73:5757–5766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Chevillotte M., et al. 2009. Major tegument protein pp65 of human cytomegalovirus is required for the incorporation of pUL69 and pUL97 into the virus particle and for viral growth in macrophages. J. Virol. 83:2480–2490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Crump C. M., Hung C. H., Thomas L., Wan L., Thomas G. 2003. Role of PACS-1 in trafficking of human cytomegalovirus glycoprotein B and virus production. J. Virol. 77:11105–11113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Daikoku T., Ikenoya K., Yamada H., Goshima F., Nishiyama Y. 1998. Identification and characterization of the herpes simplex virus type 1 UL51 gene product. J. Gen. Virol. 79:3027–3031 [DOI] [PubMed] [Google Scholar]
- 11. Das S., Pellett P. E. 2007. Members of the HCMV US12 family of predicted heptaspanning membrane proteins have unique intracellular distributions, including association with the cytoplasmic virion assembly complex. Virology 361:263–273 [DOI] [PubMed] [Google Scholar]
- 12. Das S., Vasanji A., Pellett P. E. 2007. Three-dimensional structure of the human cytomegalovirus cytoplasmic virion assembly complex includes a reoriented secretory apparatus. J. Virol. 81:11861–11869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Dunn W., et al. 2003. Functional profiling of a human cytomegalovirus genome. Proc. Natl. Acad. Sci. U. S. A. 100:14223–14228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fraile-Ramos A., et al. 2007. The ESCRT machinery is not required for human cytomegalovirus envelopment. Cell Microbiol. 9:2955–2967 [DOI] [PubMed] [Google Scholar]
- 15. Fraile-Ramos A., et al. 2002. Localization of HCMV UL33 and US27 in endocytic compartments and viral membranes. Traffic 3:218–232 [DOI] [PubMed] [Google Scholar]
- 16. Gerna G., Baldanti F., Revello M. G. 2004. Pathogenesis of human cytomegalovirus infection and cellular targets. Hum. Immunol. 65:381–386 [DOI] [PubMed] [Google Scholar]
- 17. Hensel G., Meyer H., Gartner S., Brand G., Kern H. F. 1995. Nuclear localization of the human cytomegalovirus tegument protein pp150 (ppUL32). J. Gen. Virol. 76:1591–1601 [DOI] [PubMed] [Google Scholar]
- 18. Homman-Loudiyi M., Hultenby K., Britt W., Soderberg-Naucler C. 2003. Envelopment of human cytomegalovirus occurs by budding into Golgi-derived vacuole compartments positive for gB, Rab 3, trans-Golgi network 46, and mannosidase II. J. Virol. 77:3191–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Jahn G., et al. 1990. Generation and application of a monoclonal antibody raised against a recombinant cytomegalovirus-specific polypeptide. Klin. Wochenschr. 68:1003–1007 [DOI] [PubMed] [Google Scholar]
- 20. Johannsen E., et al. 2004. Proteins of purified Epstein-Barr virus. Proc. Natl. Acad. Sci. U. S. A. 101:16286–16291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kalejta R. F. 2008. Tegument proteins of human cytomegalovirus. Microbiol. Mol. Biol. Rev. 72:249–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kleijmeer M. J., et al. 2001. Antigen loading of MHC class I molecules in the endocytic tract. Traffic 2:124–137 [DOI] [PubMed] [Google Scholar]
- 23. Klupp B. G., et al. 2005. Functional analysis of the pseudorabies virus UL51 protein. J. Virol. 79:3831–3840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kremer J. R., Mastronarde D. N., McIntosh J. R. 1996. Computer visualization of three-dimensional image data using IMOD. J. Struct. Biol. 116:71–76 [DOI] [PubMed] [Google Scholar]
- 25. Landini M. P., Severi B., Badiali L., Gonczol E., Mirolo G. 1987. Structural components of human cytomegalovirus: in situ localization of the major glycoprotein. Intervirology 27:154–160 [DOI] [PubMed] [Google Scholar]
- 26. Lenk M., Visser N., Mettenleiter T. C. 1997. The pseudorabies virus UL51 gene product is a 30-kilodalton virion component. J. Virol. 71:5635–5638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mach M., Kropff B., Kryzaniak M., Britt W. 2005. Complex formation by glycoproteins M and N of human cytomegalovirus: structural and functional aspects. J. Virol. 79:2160–2170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Mettenleiter T. C. 2006. Intriguing interplay between viral proteins during herpesvirus assembly or: the herpesvirus assembly puzzle. Vet. Microbiol. 113:163–169 [DOI] [PubMed] [Google Scholar]
- 29. Mettenleiter T. C. 2002. Herpesvirus assembly and egress. J. Virol. 76:1537–1547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mettenleiter T. C., Klupp B. G., Granzow H. 2009. Herpesvirus assembly: an update. Virus Res. 143:222–234 [DOI] [PubMed] [Google Scholar]
- 31. Mettenleiter T. C. 2004. Budding events in herpesvirus morphogenesis. Virus Res. 106:167–180 [DOI] [PubMed] [Google Scholar]
- 32. Mobius W., et al. 2003. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic 4:222–231 [DOI] [PubMed] [Google Scholar]
- 33. Mocarski E. S., Shenk T., Pass R. F. 2007. Cytomegaloviruses, p. 2701–2772 In Knipe D. M., et al. (ed.), Fields virology, vol. 2 Lippincott/The Williams & Wilkins Co., Philadelphia, PA [Google Scholar]
- 34. Mori Y., et al. 2008. Human herpesvirus-6 induces MVB formation, and virus egress occurs by an exosomal release pathway. Traffic 9:1728–1742 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nozawa N., et al. 2003. Subcellular localization of herpes simplex virus type 1 UL51 protein and role of palmitoylation in Golgi apparatus targeting. J. Virol. 77:3204–3216 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nozawa N., et al. 2005. Herpes simplex virus type 1 UL51 protein is involved in maturation and egress of virus particles. J. Virol. 79:6947–6956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Pols M. S., Klumperman J. 2009. Trafficking and function of the tetraspanin CD63. Exp. Cell Res. 315:1584–1592 [DOI] [PubMed] [Google Scholar]
- 38. Sanchez V., Greis K. D., Sztul E., Britt W. J. 2000. Accumulation of virion tegument and envelope proteins in a stable cytoplasmic compartment during human cytomegalovirus replication: characterization of a potential site of virus assembly. J. Virol. 74:975–986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Seo J. Y., Britt W. J. 2006. Sequence requirements for localization of human cytomegalovirus tegument protein pp28 to the virus assembly compartment and for assembly of infectious virus. J. Virol. 80:5611–5626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Severi B., Landini M. P., Govoni E. 1988. Human cytomegalovirus morphogenesis: an ultrastructural study of the late cytoplasmic phases. Arch. Virol. 98:51–64 [DOI] [PubMed] [Google Scholar]
- 41. Sinzger C., et al. 2008. Cloning and sequencing of a highly productive, endotheliotropic virus strain derived from human cytomegalovirus TB40/E. J. Gen. Virol. 89:359–368 [DOI] [PubMed] [Google Scholar]
- 42. Tandon R., AuCoin D. P., Mocarski E. S. 2009. Human cytomegalovirus exploits ESCRT machinery in the process of virion maturation. J. Virol. 83:10797–10807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Theiler R. N., Compton T. 2002. Distinct glycoprotein O complexes arise in a post-Golgi compartment of cytomegalovirus-infected cells. J. Virol. 76:2890–2898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Tischer B. K., von Einem J., Kaufer B., Osterrieder N. 2006. Two-step red-mediated recombination for versatile high-efficiency markerless DNA manipulation in Escherichia coli. Biotechniques 40:191–197 [DOI] [PubMed] [Google Scholar]
- 45. Tooze J., Hollinshead M., Reis B., Radsak K., Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur. J. Cell Biol. 60:163–178 [PubMed] [Google Scholar]
- 46. Varnum S. M., et al. 2004. Identification of proteins in human cytomegalovirus (HCMV) particles: the HCMV proteome. J. Virol. 78:10960–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Womack A., Shenk T. 2010. Human cytomegalovirus tegument protein pUL71 is required for efficient virion egress. MBio 1:e00282–e00210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yu D., Silva M. C., Shenk T. 2003. Functional map of human cytomegalovirus AD169 defined by global mutational analysis. Proc. Natl. Acad. Sci. U. S. A. 100:12396–12401 [DOI] [PMC free article] [PubMed] [Google Scholar]