Abstract
The insect baculovirus chitinase (CHIA) and cathepsin protease (V-CATH) enzymes cause terminal host insect liquefaction, enhancing the dissemination of progeny virions away from the host cadavers. Regulated and delayed cellular release of these host tissue-degrading enzymes ensures that liquefaction starts only after optimal viral replication has occurred. Baculoviral CHIA remains intracellular due to its C-terminal KDEL endoplasmic reticulum (ER) retention motif. However, the mechanism for cellular retention of the inactive V-CATH progenitor (proV-CATH) has not yet been determined. Signal peptide cleavage occurs upon cotranslational ER import of the v-cath-expressed protein, and ER-resident CHIA is needed for the folding of proV-CATH. Although this implies that CHIA and proV-CATH bind each other in the ER, the putative CHIA–proV-CATH interaction has not been experimentally verified. We demonstrate that the amino-terminal 22 amino acids (aa) of Autographa californica multiple nucleopolyhedrovirus (AcMNPV) preproV-CATH are responsible for the entry of proV-CATH into the ER. Furthermore, the CHIA–green fluorescent protein (GFP) and proV-CATH-red fluorescent protein (RFP) fusion proteins colocalize in the ER. Using monomeric RFP (mRFP)-based bimolecular fluorescence complementation (BiFC), we determined that CHIA and proV-CATH interact directly with each other in the ER during virus replication. Moreover, reciprocal Ni/His pulldowns of His-tagged proteins confirmed the CHIA–proV-CATH interaction biochemically. The reciprocal copurification of CHIA and proV-CATH suggests a specific CHIA–proV-CATH interaction and corroborates our BiFC data. Deletion of the CHIA KDEL motif allowed for premature CHIA secretion from cells, and proV-CATH was similarly prematurely secreted from cells along with ΔKDEL-CHIA. These data suggest that CHIA and proV-CATH interact directly with each other and that this interaction aids the cellular retention of proV-CATH.
INTRODUCTION
Baculovirus infection is initiated after the ingestion of food contaminated with occlusion bodies (OBs) containing the infectious enveloped virions. OBs dissolve in the insect midgut and release the embedded virions (7). Baculoviruses in the Alphabaculovirus and Betabaculovirus genera cause systemic infections in their lepidopteran hosts, and progeny OBs are disseminated only after the death of the infected larvae. The release of alpha- and betabaculovirus OBs from the infected larval cadaver into the environment is enabled by the tightly regulated release and activation of the virus-encoded chitinase (CHIA) and cathepsin (V-CATH) enzymes, which act in concert to liquefy the host carcass. The subcellular retention, release, and activation of these two enzymes are coordinated to allow for dissolution only after optimal virus production (11, 21, 26). Once the host succumbs to the virus infection, viral OBs are liberated from the insect cadaver to facilitate horizontal transmission to other larvae. Most alpha- and betabaculoviruses contain homologues of chiA and v-cath, and several of these species maintain a conserved contiguous but antiparallel chiA–v-cath gene organization such that late mRNA transcription start sites for both open reading frames (ORFs) reside in a small (40- to 100-bp) intergenic region (27). In contrast, hymenopteran-specific gammabaculoviruses and dipteran-specific deltabaculoviruses do not contain homologues of either chiA or v-cath. Infection by gamma- and deltabaculoviruses is restricted to the host gastrointestinal tract and therefore does not cause terminal host liquefaction. Rather, the OBs of these viruses are disseminated by the sloughing off and release of OBs and infected midgut epithelial cells into the surroundings (1, 20).
The late-expressed chiA and v-cath genes of the Alphabaculovirus type species Autographa californica multiple nucleopolyhedrovirus (AcMNPV) lead to intracellular accumulation of both active CHIA and the inactive V-CATH progenitor (proV-CATH) (26, 28). Concomitant with the death of virus-infected cells, proV-CATH is proteolytically cleaved in a papain-like manner to active V-CATH (2, 14), and cell lysis occurs, simultaneously releasing both CHIA and V-CATH along with progeny OBs. Once several virus-infected cells lyse and release CHIA and V-CATH, the enzymatic dissolution of host cuticle and internal organs ensues, often reducing the host carcass to a progeny OB-laden, liquefied mass. The regulated and coordinated simultaneous release of active CHIA and V-CATH enzymes only at the terminal stages of infection allows for extensive virus replication in vital organs, which ultimately is the cause of death. The chiA and v-cath genes are auxiliary, since deletion of either or both does not affect virus replication or morbidity, even though both enzymes are critical for liquefaction (11). Furthermore, although CHIA and V-CATH are not considered virulence factors per se, increased or dysregulated expression of either gene can increase baculovirus virulence or affect the progression of the liquefaction process, respectively (15, 26, 29). Optimal induction of host cadaver liquefaction requires innate regulation of CHIA and V-CATH expression and activation. Therefore, the regulatory mechanisms governing the coordinated release of these enzymes from infected cells are of interest both fundamentally and possibly for the development of biorational pest control strategies.
Both CHIA and V-CATH are retained in the endoplasmic reticulum (ER) prior to cellular release (15, 28). AcMNPV v-cath is initially translated as a preproenzyme (13). The signal peptide of the nascent polypeptide is removed upon cotranslational import into the ER, where the polypeptide is posttranslationally modified at Asn65 with an N-linked glycan (15, 26). It has been postulated that the secretory signal peptide of preproV-CATH might be cleaved after Tyr11, unmasking a potential myristate lipid conjugation motif and a requisite N-terminal Gly12 myristate acceptor (13). The putative myristoylation of proV-CATH at Gly12 would offer a mechanism by which proenzyme secretion might be blocked, due to a hydrophobic interaction of myristic lipid with cellular lipid membranes or with viral CHIA or other ER proteins. However, it has been suggested recently that preproV-CATH is cleaved after Ala18, making the v-cath-encoded Val19 the N-terminal proV-CATH residue (6). This finding is inconsistent with putative signal cleavage after Y11 and subsequent myristoylation of Gly12.
Cell death and cellular release of CHIA due to cell lysis temporally coincide with the enzymatic maturation and release of V-CATH (14). However, cell lysis is dependent on the normal expression and trafficking of proV-CATH and the catalytic maturation of V-CATH. If v-cath is not expressed or if proV-CATH is rendered insoluble due to chiA deletion, cell lysis is reduced, as measured in vitro by reduced polyhedron release or in vivo by reduced polyhedron content and turbidity of the hemolymph and a lack of liquefaction (11, 26, 29). In addition, glycosylation of AcMNPV, Bombyx mori NPV (BmNPV), and Choristoneura fumiferana multiple nucleopolyhedrovirus (CfMNPV) proV-CATH is required for its proper folding and/or maintenance in a soluble form (13, 15, 21). proV-CATH expressed without N-linked glycans, due to tunicamycin inhibition (15, 26) or the substitution of alanine for asparagine acceptor residues (19), formed insoluble aggregates, and consequently, mature, enzymatically active V-CATH was not produced. It was therefore postulated that these glycans are required for the binding of proV-CATH by the viral CHIA or by ER-resident folding chaperones (5, 15, 19). However, when AcMNPV proV-CATH was expressed normally (i.e., glycosylated) and was then deglycosylated in vitro with N-glycosidase F (PNGase F), it remained soluble and could be induced by sodium dodecyl sulfate (SDS) to produce the proteolytically processed, mature V-CATH form (15). This implies not only that CHIA and the proV-CATH glycan assist in proV-CATH trafficking but also that they are both integral to the regulation of the coordinated release of both the CHIA and V-CATH enzymes, since V-CATH maturation is linked to cell death and lysis (14).
The CHIA and proV-CATH proteins have to be temporally and spatially regulated in order to remain subcellular during infection, and proV-CATH remains in an inactive (zymogen) form. Only upon maximum OB accumulation and the death of the host does V-CATH mature enzymatically, at which point both CHIA and V-CATH are released from cells. Such regulation ensures the appropriate timing of liquefaction and the ensuing dissemination of OBs. The cellular ER retention of AcMNPV CHIA due to its C-terminal KDEL motif is well documented (10, 25, 28). However, although proV-CATH is directed to the ER, no ER retention mechanism for it has been identified (15, 21, 26). A molecular interaction between CHIA and proV-CATH within cells could foster the ER retention of both proteins. In this paper we provide biochemical and microscopy evidence for a molecular interaction between CHIA and proV-CATH. From our data, we propose that the coordinated intracellular localization of proV-CATH with CHIA is aided by their association in cells and governs the optimum timing of host liquefaction due to the intracellular retention, release, and coordinated enzyme activities, of these two enzymes.
MATERIALS AND METHODS
Cells and virus.
Monolayers of Sf21 or Hi5 cells were grown in Grace's insect medium supplemented with fetal bovine serum and penicillin-streptomycin as described previously (13). Cell monolayers were inoculated with AcMNPV at a multiplicity of infection (MOI) of 10 (unless otherwise specified) and were incubated for 1 h at room temperature, after which the inoculum was removed and replaced with growth medium. For time course experiments, the time when the inoculum was replaced with the growth medium was considered time zero (t = 0). Sf21 and Hi5 cells adapted (over 10 to 20 passages) to SFM900-III serum-free medium (Invitrogen) supplemented with penicillin-streptomycin were used for microscopy and protein secretion studies. This was because serum albumin, with a molecular mass of ∼60 kDa, obscures the immunodetection of the ∼58-kDa CHIA bands from samples of extracellular medium. Serum-free medium-adapted cells were also used for microscopy of the live virus-infected cells, because these cells adhered to the culture dishes better at late infection times (from 36 h postinfection [hpi]) than did cells grown in Grace's medium supplemented with fetal bovine serum. Virus titers (50% tissue culture infective doses [TCID50]) were determined by endpoint dilution (22).
Cloning methodology for generating viral constructs.
This section contains detailed cloning steps used to generate the various bacmid-based viral coexpression constructs. Corresponding schematics showing steps for generating the constructs described below are provided in Fig. S1 in the supplemental material as a guide. The primer-template combinations used for PCRs whose products were incorporated into the constructs are provided in Table S1 in the supplemental material. All preliminary cloning and subcloning required for constructing the various chiA and v-cath or related coding sequences was performed in the multiple-cloning site (MCS) of pBluescript (pBSK) or plasmids derived from pBSK. All pBSK-based cloning vectors described below are prefixed with a “p,” and the names of viruses derived from them, which are used throughout the text, lack that prefix.
We tagged CHIA with a FLAG epitope tag (CH) and proV-CATH with a hemagglutinin (HA) epitope tag (CA) to aid in immunoblotting. Unless otherwise stated, all chiA isoforms containing CH in their names are FLAG tagged and all v-cath isoforms containing CA in their names are HA tagged. The construction of the proV-CATH-HA fusion, which we call CA in this paper, has been described previously (12). The fusion of a FLAG tag in the penultimate position adjacent to the C-terminal chiA KDEL motif required the sequential cloning of two PCR amplicons (PCR A1 [KpnI/ApaI] and PCR A2 [ApaI/EcoRV]) into pBSK, generating pKpnI-FLAG-EcoRV, which has an ApaI site engineered in frame between the C-terminal FLAG-KDEL coding sequence and the upstream chiA sequence (see Fig. S1a in the supplemental material). Since v-cath naturally contains an ApaI site, the KpnI-FLAG-EcoRV cassette was subcloned by use of KpnI/EcoRV into pchiA.cathHA (13) to generate pCH/CA (see Fig. S1a).
Three viral constructs were generated for the CHIA/proV-CATH colocalization experiment. A viral construct that expresses green fluorescent protein (GFP)-tagged CH and native v-cath (CH-GFP/v-cath) and another virus that expresses red fluorescent protein (RFP)-tagged CA and native chiA (chiA/CA-RFP) were produced to independently localize CH-GFP or CA-RFP. A third virus, which coexpresses GFP-tagged CH and RFP-tagged CA, was generated (CH-GFP/CA-RFP) to simultaneously colocalize CH-GFP and CA-RFP expressed by the same virus. The monomeric mRFP1(Q66T) (23) coding sequence was fused to the C terminus of v-cath to replace DsREDHA in pchiA.cathDsREDHA (13), thus generating pchiA/CA-RFP (see Fig. S1b in the supplemental material). This was done by SpeI cloning of the PCR B1 amplicon (mRFPHA) in place of DsREDHA in pchiA.cathDsREDHA. To fuse GFP between CHIA and the downstream FLAG-KDEL sequence, a gfp amplicon (PCR B2) was cloned by use of ApaI into pKpnI-FLAG-EcoRV, generating pKpnI-GFP-EcoRV (see Fig. S1b). The KpnI-GFP-EcoRV construct was subcloned by use of KpnI/EcoRV either into pCCnative (13) to generate pCH-GFP/v-cath or into pchiA/CA-RFP to generate pCH-GFP/CA-RFP (see Fig. S1b). To generate the monomeric RFP (mRFP)-based and GFP-based endoplasmic reticulum (ER) markers, PCR amplicons of the ER-targeted pmGFP5-ER (PCR C1) or pmRFP-ER (PCR C2) construct (23) were cloned by use of XbaI/SstI into pchiA.HAcathHA (13) to generate pchiA/ER-GFP and pchiA/ER-RFP, respectively (see Fig. S1c in the supplemental material). These fluorescent ER marker genes were cloned in place of v-cath and therefore were transcribed from the native v-cath promoter adjacent to chiA.
Three virus constructs, two experimental and one control, were generated to map the v-cath-encoded amino terminus responsible for the ER translocation of a GFP reporter. The PCR amplicons of the GFP control (PCR C3), N12-GFP (PCR C4), and N22-GFP (PCR C5) were cloned by use of XbaI/SstI into pchiA.HAcathHA to generate pchiA/GFP, pchiA/N12-GFP, and pchiA/N22-GFP, respectively (see Fig. S1c in the supplemental material). These GFP constructs, like the pER-GFP and pER-RFP constructs, were cloned in place of v-cath and were transcribed from the v-cath promoter adjacent to chiA. The N22-GFP amplicon was amplified in three sequential PCRs in order to fuse the entire primer-incorporated (see Table S1 in the supplemental material, F 1, 2, and 3), v-cath-encoded N-terminal 22 amino acids to gfp (see Fig. S1c).
Four virus constructs were generated for the bimolecular fluorescence complementation (BiFC) assay (16). The engineering of a SpeI recognition site in frame between the chiA-encoded N-terminal signal sequence cleavage site (Ala17) and downstream chiA required sequential cloning of two PCR amplicons (PCR D1 [EcoRV/SpeI] and D2 [SpeI/SstI]) into pCH/CA to generate the intermediate plasmid pSPECH/v-cath (see Fig. S1d in the supplemental material). A PCR amplicon (PCR D3) of the C-terminal portion of mRFP (mRFPC) portion (17) was cloned into the engineered chiA SpeI site of pSPECH/v-cath to generate pCH-mRFPC/v-cath (see Fig. S1d). The MYC-tagged mRFPN coding sequence was subcloned by use of SpeI/XbaI from pBatTL-smRFPN (17) into the SpeI site of pchiA.cathDsREDHA (in place of DsREDHA) in frame with the C terminus of v-cath to generate pchiA/CA-mRFPN (see Fig. S1d). The v-cath-myc-mRFPN fusion sequence was subcloned by use of EcoRI/SstI from pchiA/CA-mRFPN into pCH-mRFPC/v-cath to generate pCH-mRFPC/CA-mRFPN (see Fig. S1d). The ER-targeted MYC-mRFPN-KDEL PCR amplicon (PCR D4) used for a BiFC negative control was cloned by use of KpnI/SpeI into pSPECH/v-cath to generate pMYC-mRFPN-KDEL/v-cath (see Fig. S1d).
Two viral constructs were generated to assess the CHIA–proV-CATH interaction biochemically by reciprocal Ni/His pulldowns. A third virus, CH/CA, was used as a negative control. A PCR amplicon (PCR E1) generated to fuse a 6× His sequence after the FLAG sequence of the pKpnI-FLAG-EcoRV construct was cloned by use of KpnI/EcoRV into pCH/CA to generate pCH-HIS/CA (see Fig. S1e in the supplemental material). To fuse the HA-His sequence in frame with v-cath, a PCR amplicon (PCR E3) was cloned by use of EcoRI/SpeI into pCH/CA to generate pCH/CA-HIS (see Fig. S1e). Another virus was generated to compare the coretention/cosecretion of CHIA and proV-CATH due to CHIA KDEL deletion to that of the control virus CH/CA. To delete the chiA-encoded KDEL motif, a PCR amplicon (PCR E2) with a deletion of the C-terminal KDEL sequence after the FLAG sequence (of the pKpnI-FLAG-EcoRV construct) was generated and cloned by KpnI/EcoRV into pCH/CA to generate p(ΔKDEL)CH/CA (see Fig. S1e).
Manipulation of viral genomes and generation of engineered viruses.
The pBSK-based constructs were all subcloned by use of KpnI/SstI into the polh locus at the MCS of the previously described modified pFastBAC plasmid (13) with its polh promoter and downstream N-terminal 6× His tag fusion deleted. These pFastBAC vectors were used to integrate the various gene constructs into the bacmid genome by using standard bacmid technology to generate the corresponding recombinant AcBACΔCC-derived AcMNPVs, as summarized in Table 1. The AcBACΔCC bacmid had its native chiA-v-cath locus deleted (18), so that the various chiA and v-cath gene constructs were located only in the bacmid polh locus engineered into AcBACΔCC and were expressed under the control of their native intergenic promoters by these viruses. The integrity of all pFastBAC clones was verified by DNA sequencing, and that of the corresponding AcBACΔCC-derived viral constructs was confirmed by PCR.
Table 1.
Summary of virus constructs generated,a chiA and v-cath isoforms (or other ORFs), their novel features, and the corresponding experiments and figures
| Virus name | CHIA isoform | V-CATH isoform | Other ORF(s) | Other feature(s) | Expt(s) |
|---|---|---|---|---|---|
| CH/CA | CH | CA | CH has a FLAG tag; CA has an HA tag | CH/CA profiles (Fig. 1), Ni/His (Fig. 5), cosecretion (Fig. 6) | |
| CH-GFP/v-cath | CH-GFP | v-cathb | CH-GFP has a FLAG tag | Colocalization (Fig. 2) | |
| chiA/CA-RFP | chiAb | CA-RFP | CA-RFP has an HA tag | Colocalization (Fig. 2) | |
| CH-GFP/CA-RFP | CH-GFP | CA-RFP | Colocalization (Fig. 2) | ||
| chiA/ER-GFP | chiA | ER-GFP-HDEL | ER-GFP is under the control of the v-cath promoter | Colocalization (Fig. 2), GFP translocation (Fig. 3), BiFC (4) | |
| chiA/ER-RFP | chiA | ER-RFP-HDEL | ER-RFP is under the control of the v-cath promoter | Colocalization (Fig. 2), GFP translocation (Fig. 3) | |
| chiA/GFP | chiA | EGFP | EGFP is under the control of the v-cath promoter | GFP translocation (Fig. 3) | |
| chiA/N12-GFP | chiA | CA N12-GFP fusionc | N12-GFP | N12-GFP is under the control of the v-cath promoter | GFP translocation (Fig. 3) |
| chiA/N22-GFP | chiA | CA N22-GFP fusionc | N22-GFP | N22-GFP is under the control of the v-cath promoter | GFP translocation (Fig. 3) |
| CHmRFPC/v-cath | CH-mRFPC (+FLAG)d | v-cath | CH-mRFPC has a FLAG tag | BiFC (Fig. 4) | |
| chiA/CA-mRFPN | chiA | CA-mRFPN (+MYC)d | mRFPN has a MYC tag | BiFC (Fig. 4) | |
| MYC-mRFPN-KDEL/v-cath | CH-mRFPN-KDELe | v-cath | CH-mRFPN has a MYC tag and a KDEL motif under the control of the chiA promoter | BiFC (Fig. 4) | |
| CH-mRFPC/CA-mRFPN | CH-mRFPC (+FLAG) | CA-mRFPN (+MYC) | CH-mRFPC has a FLAG tag; CA-mRFPN has a MYC tag | BiFC (Fig. 4) | |
| CH-HIS/CA | CH-HIS (+FLAG) | CA | CH-HIS has a FLAG tag; CA has an HA tag | Ni/His (Fig. 5) | |
| CH/CA-HIS | CH | CA-HIS (+HA) | CH has a FLAG tag; CA-HIS has an HA tag | Ni/His (Fig. 5) | |
| (ΔKDEL)CH/CA | (ΔKDEL)CH | CA | (ΔKDEL)CH has a FLAG tag; CA has an HA tag | Cosecretion (Fig. 6) |
All viruses were constructed in the AcBACΔCC bacmid (Fig. 1a) as described in Materials and Methods.
v-cath and chiA refer to native, unmodified v-cath and chiA, respectively.
CA N12-GFP and CA N22-GFP refer to GFP containing the N-terminal 12 or 22 amino acids, respectively, derived from preproV-CATH.
mRFPC and mRFPN refer to the carboxy- and amino-terminal portions of mRFP, respectively.
CH-mRFPN-KDEL refers to a MYC-tagged mRFPN construct that has been fused to the N-terminal chiA signal sequence in order to target the protein to the ER and that is modified at the C terminus with a KDEL motif so that the protein accumulates in the ER, as does CH-mRFPC.
Final construct schematics are provided in the figures, and a summary of the chiA and v-cath isoforms (or other genes) encoded by each bacmid-derived virus is provided in Table 1. Unless otherwise stated, all chiA isoforms containing CH in their names are FLAG tagged and all v-cath isoforms containing CA in their names are HA tagged. All bacmids were developed and correspondingly named as chiA–v-cath isoform coexpression constructs. Since we wanted to preserve the native-like transcription of both genes, their native intergenic promoters were retained in the bacmid polh locus (see Fig. 1a). Construct names containing chiA or v-cath indicate that the virus encodes native, unmodified chiA or v-cath.
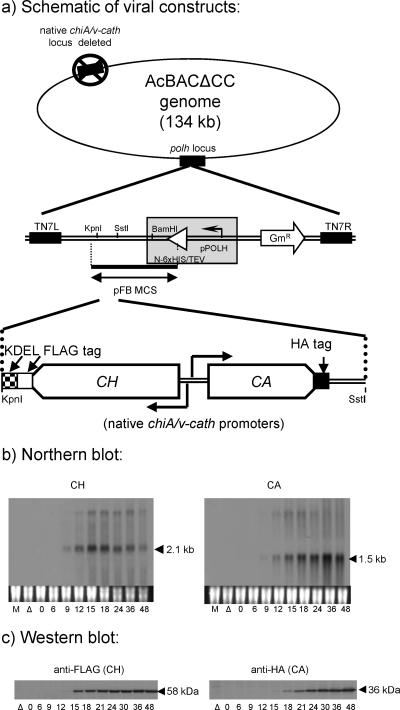
Fig. 1.
Temporal patterns of chiA and v-cath expression by the CH/CA bacmid-derived virus. (a) Schematic of the virus construct showing the chiA-v-cath gene locus in the modified (12) polh locus (lacking a polh promoter or 6× His/tobacco etch virus [TEV] fusion sequence) of a bacmid (AcBACΔCC) with its native chiA–v-cath locus deleted (18). (b) Total RNA (5 μg/lane) was probed with DIG-labeled, strand-specific ssDNA probes complementary to chiA or v-cath RNA, as labeled. Lanes M, mock-infected cells; lanes Δ, the chiA-v-cath deletion AcBACΔCC bacmid with no chiA or v-cath. Cells were infected at an MOI of 10. The mock-infected and chiA-v-cath deletion virus RNA and protein samples were collected at 24 hpi. (c) Soluble intracellular proteins were detected with an anti-FLAG (for CH) or anti-HA (for CA) antibody.
Northern blot analysis.
Sf21 cells grown in Grace's medium were infected at an MOI of 10. Total RNA was isolated over a time course (0 to 48 hpi), and 5 μg of RNA per lane was electrophoresed in denaturing (2.2 M formaldehyde) morpholinepropanesulfonic acid (MOPS)-agarose gels (1.2%) and hybridized to digoxigenin (DIG)-labeled strand-specific single-stranded DNA (ssDNA) chiA or v-cath probes. The 470-nucleotide chiA-specific probe comprised residues complementary to the chiA transcript from nucleotide −3 to 467 in the chiA ORF. The 536-nucleotide v-cath-specific probe comprises residues complementary to the v-cath transcript from nucleotide −21 to 525 in the v-cath ORF. Hybridized probes were detected with a chemiluminescent substrate (CSPD) as described previously (12). The chiA–v-cath deletion virus RNA sample was collected at 24 hpi.
Western blot analyses.
For temporal intracellular protein analysis, 1 million Sf21 cells infected at an MOI of 10 in a 35-mm-diameter dish were resuspended in growth medium at each time point from 0 to 48 hpi, and cells were collected by centrifugation (5 min, 500 × g). The chiA–v-cath deletion virus protein sample was collected at 24 hpi. Cells were lysed by iterative pipetting and were then incubated on ice for 5 min in cell lysis buffer (100 mM NaCl, 20 mM Tris, 0.5% NP-40 [pH 7.5]) supplemented with cysteine protease inhibitor (E64) to 50 μM. Soluble protein was obtained from supernatants after centrifugation (4,000 × g, 5 min) of crude lysates at 4°C and was stored at −70°C. Equal lysate sample volumes were separated by 12% polyacrylamide gel electrophoresis (PAGE) and were electroblotted to polyvinylidene difluoride (PVDF) membranes. Equal protein loading was verified based on similar intensities of silver-stained gels. Antibody solutions and membrane wash solutions were composed of 50 mM Tris, 150 mM NaCl, and 0.1% Tween 20 (pH 7.5). Proteins on membranes were probed with either anti-HA, anti-FLAG, or anti-MYC monoclonal murine antibodies (Sigma). A horseradish peroxidase-conjugated anti-mouse secondary antibody (Sigma) and a chemiluminescent substrate (Pierce) were used to detect specific proteins on X-ray film (Bioflex).
Colocalization of CHIA and proV-CATH in virus-infected cells.
For intracellular localization of CHIA and proV-CATH individually in live, virus-infected cells, we generated two viruses, one that expressed CHIA fused with GFP (CH-GFP/v-cath) and one that expressed proV-CATH fused to mRFP (chiA/CA-RFP). A third virus that coexpressed CH-GFP and CA-RFP (CH-GFP/CA-RFP) was generated to colocalize CHIA and proV-CATH expressed by the same virus (see Fig. 2).
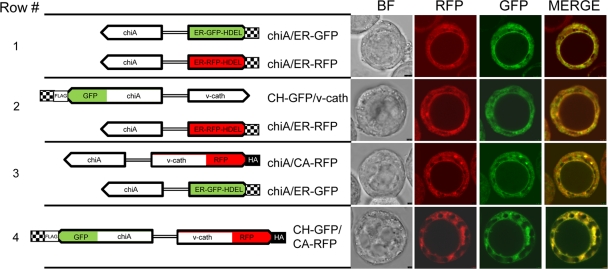
Fig. 2.
Colocalization of CHIA and proV-CATH in the ER of virus-infected Hi5 cells. To the left, the chiA/ER-GFP and chiA/ER-RFP viral constructs and the positions of gfp fusion to chiA in CH-GFP and mrfp fusion to v-cath in CA-RFP are shown. Overlapping fluorescence patterns are produced by the chiA/ER-GFP and chiA/ER-RFP ER marker viruses (row 1). When assessed individually, chiA/ER-RFP and CH-GFP/v-cath exhibit overlapping fluorescence patterns (row 2), as do chiA/ER-GFP and chiA/CA-RFP (row 3). When the fusions are assessed simultaneously, the CH-GFP/CA-RFP virus produces CH-GFP and CA-RFP fluorescence patterns that overlap (row 4). Virus-infected cells (MOI, 10) were photographed at 36 hpi using confocal laser scanning microscopy. Image panels show bright-field images (BF), the red (RFP) and green (GFP) fluorescent channels, and an overlay of the RFP and GFP channels (Merge). The images shown are representative of those used for deriving Pearson coefficients.
Hi5 cells were used due to their larger size and higher expression of chiA and v-cath, which allowed CA-RFP to be visualized and localized more easily. Serum-free medium was used, because more cells remained attached at late times of infection than with serum-containing Grace's medium. One million Hi5 cells in serum-free medium were seeded into 35-mm-diameter dishes and were allowed to attach for 1 h; then they were infected (MOI, 10) with the different viruses. The virus inoculum was removed, and the monolayers were rinsed (twice) with growth medium and were incubated at 27°C until the appropriate time for microscopic analysis. Growth medium from infected monolayers was aspirated, and cells were rinsed (twice) with phosphate-buffered saline (PBS) (pH 6.2) at room temperature. Three milliliters of room-temperature PBS (pH 6.2) was added to rinsed culture dishes, in which a 63× lens of a Leica DM 6000B microscope was immersed. The GFP fluorophores were excited with an argon laser (50 mW), and the RFP fluorophores were excited with a green HeNe laser (1.2 mW). Photographs of bright-field (BF) images and the RFP and GFP fluorophores were captured sequentially because the argon laser excited both the GFP and RFP fluorophores. Images were photographed with a Leica digital camera (DFC350FX). Fluorescent images were merged using Leica software and were optimized for brightness and contrast using ImageJ software (http://rsbweb.nih.gov/ij/index.html). Colocalization analysis was performed by comparing the Pearson coefficient ImageJ intensity correlation analysis plugin with those calculated by using the Student t test function of Microsoft Excel. Colocalization analysis based on Pearson coefficients determines the relative intensities of GFP- and RFP-induced fluorescence in the same groups of pixels delimited by the region of interest (ROI) selected. For the colocalization analyses, the entire cell image was used as a global ROI. When it was necessary to abrogate the incorporation of fluorescence from neighboring cells in the colocalization analysis, an elliptical ROI surrounding only the entire cell of interest was used. Confocal images were acquired utilizing the Leica 3× line average mode.
GFP ER translocation assay.
To functionally define the N-terminal signal sequence of v-cath that is responsible for ER localization, peptides comprising either 12 or the 22 N-terminal v-cath-encoded amino acids were fused to gfp to determine if either peptide promotes ER localization of normally diffuse GFP (Fig. 3). Control viruses expressing either GFP or ER-targeted GFP (ER-GFP) were also generated for comparison of GFP fluorescence patterns. None of these four viruses carry v-cath, because the GFP isoform-encoded genes were cloned in place of v-cath (adjacent to chiA) and were expressed from the native v-cath promoter. The ER-GFP cassette (9) has an N-terminal secretory signal, so that the protein enters the ER, and a C-terminal HDEL ER retention motif, allowing it to accumulate in the ER of infected cells (13). The GFP expressed by the GFP virus is from an unmodified egfp gene. An MOI of 10 was used for infection with each virus.
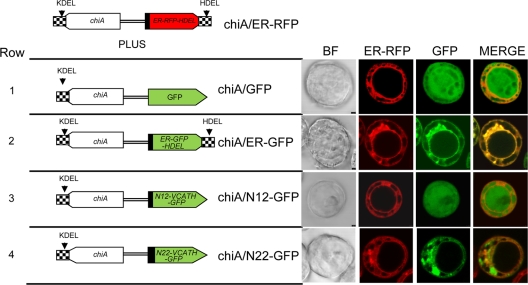
Fig. 3.
Fluorescence patterns of V-CATH-GFP fusion proteins from coinfection of Hi5 cells with chiA/ER-RFP (top) and the constructs shown on the left. Bright-field (BF) and fluorescence patterns of virus-expressed V-CATH-GFP fusion proteins (chiA/N12-GFP or chiA/N22-GFP [rows 3 and 4]) were compared to those of the diffuse GFP control virus (chiA/GFP [row 1]) or the ER-targeted GFP control virus (chiA/ER-GFP [row 2]) in infected Hi5 cells. In all rows, the ER was labeled with red fluorescence by coinfection with the chiA/ER-RFP virus. For N12-GFP and N22-GFP, the first 12 or 22 amino acids, respectively, encoded by the v-cath ORF are fused to the amino terminus of GFP. Hi5 cells were infected at an MOI of 10. Virus-infected cells were photographed at 40 hpi using confocal laser scanning microscopy.
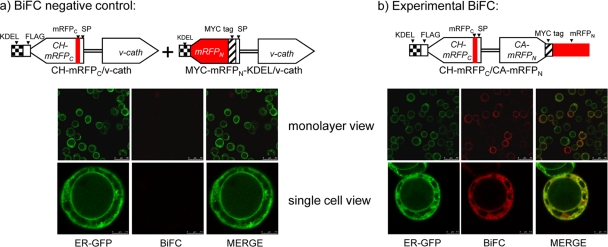
mRFP-based BiFC assay.
To probe the CHIA–proV-CATH interaction in live virus-infected cells, we developed an mRFP-based (17) BiFC (16) system. One million Hi5 cells in serum-free medium were seeded in 35-mm-diameter dishes and were allowed to attach for 1 h before infection. Cells either were infected at an MOI of ∼100 with the BiFC virus (CH-mRFPC/CA-mRFPN) that expressed both portions of the split mRFP (fused to CH and v-cath as described above) or were coinfected with two other viruses (CH-mRFPC/v-cath and MYC-mRFPN-KDEL/v-cath), each at an MOI of ∼100, to provide negative controls for the BiFC assay. The cellular ER was identified by GFP-induced fluorescence produced by infection with the chiA/ER-GFP virus. At 48 hpi, microscopy was performed as described above except that for the acquisition of low-level red fluorescence in the BiFC assay, the Leica 3× frame accumulation mode was utilized (for both the BiFC negative controls and the experimental BiFC).
Ni-agarose affinity copurification of CHIA and proV-CATH.
Twenty million Sf21 cells (in 150-mm-diameter dishes) were infected (MOI, ∼100) with one of three viruses: CH-HIS/CA, CH/CA-HIS, or the negative control CH/CA, which expresses CH and CA, both lacking His tags. At 40 hpi, monolayers were scraped into culture medium and were collected by centrifugation (500 × g, 5 min). Cell pellets were washed by suspension in 20 ml PBS (pH 6.2) and were collected by centrifugation (500 × g, 5 min). Cell pellets were lysed by iterative pipetting in 1 ml of cell lysis buffer (100 mM NaCl, 20 mM Tris, 0.5% NP-40 [pH 7.5]) supplemented with 10 mM imidazole, cysteine protease inhibitor (E64) to 50 μM, and 250 U of benzonase (Novagen) to reduce the viscosity of the lysate, followed by incubation on ice for 10 min. Lysates were then centrifuged (twice at 12,000 × g and 4°C) to pellet insoluble material, and the cleared lysate was recovered. Ni-agarose (Qiagen) was equilibrated with cell lysis buffer (three times) and was used for small-batch His affinity purifications from each cell lysate. Fifty microliters of equilibrated Ni-agarose was added to 1 ml of the cleared lysates, mixed (by inversion on a rotating device) for 1 h at ambient temperature (20 to 25°C), and then washed (four times, for 5 min each time, at 4°C) with cold PBS (pH 8) containing 20 mM imidazole, as suggested by the Ni-agarose manufacturer's protocol (Qiagen), to reduce nonspecific protein binding. Washed Ni-agarose containing bound protein complexes was resuspended in 80 μl of SDS-PAGE loading buffer (containing 100 μM cysteine protease inhibitor E64) and was heated to 95°C for 5 min. Ni-agarose was then pelleted by brief centrifugation (30 s), and the supernatant containing the protein sample was decanted to a new tube and was analyzed by Western blotting as described above. Equal volumes of lysate samples were loaded onto gels (Fig. 5b, lanes 1, 2, and 3). Three microliters of the Ni-agarose-purified protein samples was loaded to detect the His-tagged proteins, and 9 μl of the same sample was loaded to detect the copurified proteins. For example, when CH-HIS was pulled down from the CH-HIS/CA lysate, 3 μl of the Ni-agarose-purified proteins was loaded to detect CH-HIS (Fig. 5b, lane 4, 58-kDa CHIA band) and 9 μl of the same purified protein sample was loaded to detect CA (Fig. 5b, lane 4, 36-kDa proV-CATH band). Nine microliters of the Ni-agarose-purified proteins from the CH/CA negative-control lysate was loaded for the detection of both CH and CA (Fig. 5b, lane 5).
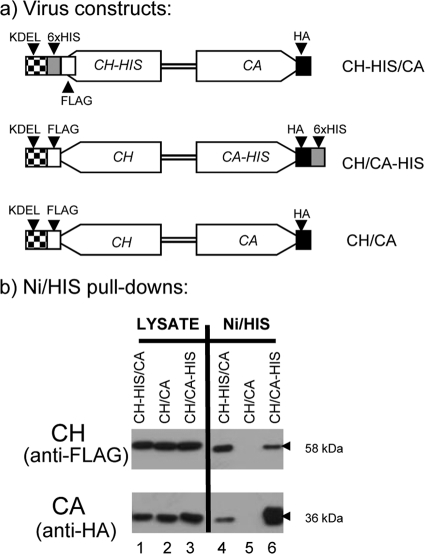
Fig. 5.
Reciprocal Ni affinity copurification of 6× His-tagged CHIA and proV-CATH. (a) Viral constructs. The CH-HIS/CA virus coexpresses His-tagged CH and non-His-tagged CA. The CH/CA-HIS virus coexpresses His-tagged CA and non-His-tagged CH. The CH/CA negative-control virus coexpresses native, non-His-tagged CH and CA counterparts. (b) Western blot analysis of purified proteins using anti-HA (to detect CA) and anti-FLAG (to detect CH). Lanes 1 to 6, lysates of cells infected with the virus indicated above each lane. Proteins from the input lysate (Lysate) and those remaining on washed Ni-agarose beads (Ni/His) were analyzed. Lysates from 2 × 107 Sf21 cells infected at an MOI of ∼ 100 at 40 hpi were used for the purifications.
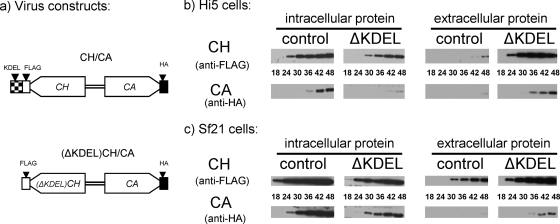
Temporal analysis of CHIA and proV-CATH coretention/cosecretion.
To assess the coretention and cosecretion of CHIA and proV-CATH during virus replication, we compared the intracellular and extracellular distributions of CH and CA proteins produced by infection at an MOI of 10 with the CH/CA and (ΔKDEL)CH/CA viruses. For the analysis of proteins secreted into serum-free medium, at each time point for each virus-cell system, 500 μl of medium from a single 60-mm-diameter dish containing 3 million infected cells was removed and centrifuged (2,000 × g, 5 min), and the upper 100 μl was collected, supplemented with E64 (to 50 μM), and stored at −70°C until analysis by Western blotting as described above. The soluble intracellular proteins from the infected monolayers of the corresponding 60-mm-diameter dishes were then prepared as described above.
Equivalent volumes of these extracellular and intracellular samples were monitored for retention (intracellular) and secretion (extracellular) of CA and the two CH isoforms (with or without KDEL) during virus infection (18 to 48 hpi) by Western blotting as described above. Two microliters of the intracellular proteins, from both the infected Hi5 and Sf21 cultures, was loaded. Three microliters of the Hi5 extracellular samples was loaded, and 5 μl of the Sf21 extracellular samples was loaded. Equal protein loading was verified for the intracellular samples by the silver-staining intensities of gels. The same experiment using both Hi5 and Sf21 cultures was performed in parallel. Four individual gels/blots were processed simultaneously so that we could effectively compare the patterns by which CA and the isoforms of CH accumulated intra- and extracellularly. That is, the intracellular CH/CA- and (ΔKDEL)CH/CA-infected Hi5 samples were separated on a single gel and were blotted to a single membrane. The same was done for the infected Hi5 extracellular medium samples and the corresponding infected Sf21 samples. Membranes containing the intracellular samples were bisected horizontally (at the 43-kDa position of the protein ladder), and the top (for CH) and bottom (for CA) portions were incubated with anti-FLAG and anti-HA antibodies, respectively, in order to detect the 58-kDa CH isoforms and the 36-kDa CA. Membranes containing the extracellular samples were not bisected but rather were incubated with solutions containing both the anti-FLAG and anti-HA antibodies, each at the same concentration as when used individually. For exposure to X-ray film, bisected membranes were reassembled, and all four antibody-probed blots were exposed simultaneously to a single piece of film. This allowed for a more direct semiquantitative comparison of band intensities between the samples. The same X-ray film was used for all the blots in Fig. 6 except for the extracellular Sf21 CH samples, for which a more (∼50%) exposed film was substituted.
Fig. 6.
Effects of CHIA KDEL deletion on subcellular cotrafficking of CHIA and proV-CATH. (a) Viral constructs. The control CH/CA-coexpressing virus is the same as that described in Table 1. The (ΔKDEL)CH/CA virus coexpresses CH lacking its native C-terminal KDEL motif and CA. (b and c) Western blot analyses with anti-FLAG (CH) and anti-HA (CA) antibodies, as indicated, of intracellular and extracellular proteins from Hi5 (b) or Sf21 (c) cultures infected with the CH/CA (control) or (ΔKDEL)CH/CA (ΔKDEL) virus expressing either control CH and CA or (ΔKDEL)CH and CA, respectively.
RESULTS
Bacmid chiA and v-cath temporal expression profiles.
The temporal profiles for the expression of bacmid-derived CH and CA RNAs and epitope-tagged proteins from their native promoters (Fig. 1a) were examined by Northern and Western blotting, respectively. The temporal patterns of both CH and CA RNAs (Fig. 1b) resembled those of native AcMNPV chiA and v-cath (12) in that both the expected 2.1-kb CH and 1.5-kb CA RNAs were detectable starting from 9 hpi and transcript levels of each gene increased temporally to 24 or 36 hpi. This indicated that temporal transcriptional regulation of these two antiparallel genes was not affected by epitope tagging or by cloning of the chiA–v-cath locus into the polh region of the bacmid otherwise lacking its native chiA and v-cath genes.
The size of the CA mRNA transcript (∼1.5 kb) was the same as that of AcMNPV v-cath, because its native termination site (at position 108127 in the AcMNPV genome) (12) was incorporated into the polh locus of the bacmid. However, the native chiA mRNA termination site (at position 104537 in AcMNPV) was not incorporated into the bacmid polh locus. Consequently, the observed 2.1-kb CH mRNA is the size expected considering the termination and polyadenylation of the RNA transcript at the simian virus 40 (SV40) polyadenylation signal sequences cloned downstream of the CH construct.
Anti-FLAG and anti-HA antibodies were used to monitor temporal CH and CA production, respectively. The data in Fig. 1c show that both proteins were translated with kinetics similar to those with native AcMNPV (12). The ∼58-kDa CH protein was first detected from 12 to 15 hpi, whereas the ∼36-kDa CA protein was detected about 3 h later, at 15 to 18 hpi. The delay in the translation of CA relative to that of CH, as described for the native proteins (12), was maintained for the epitope-tagged proteins expressed from the polh locus in the bacmid system.
Colocalization of CHIA and proV-CATH in virus-infected cells.
In order to localize CHIA and proV-CATH individually in live, virus-infected cells, we generated two viruses, one that expresses CHIA fused with GFP (CH-GFP/v-cath) and one that expresses proV-CATH fused to mRFP (chiA/CA-RFP) (Fig. 2). A third virus, coexpressing CH-GFP and CA-RFP (CH-GFP/CA-RFP), was generated in order to follow the colocalization of CHIA and proV-CATH expressed by the same virus (Fig. 2).
Cells coinfected with the chiA/ER-GFP and chiA/ER-RFP viruses produced red and green fluorescence patterns that colocalized in the ER (Fig. 2, row 1), indicating that either virus could be used to identify the ER in coinfection experiments. The CH-GFP/v-cath infection produced a GFP fluorescence pattern that overlapped precisely with that of the ER-RFP marker (Fig. 2, row 2). The chiA/CA-RFP virus produced an RFP fluorescence pattern that overlapped with that of the ER-GFP marker (Fig. 2, row 3). This independent localization of both CH-GFP and CA-RFP to the ER corroborated the observation that native CHIA and proV-CATH colocalize to the ER and that fusion with the fluorescent proteins did not disrupt the ER targeting of either. When CH-GFP and CA-RFP were coexpressed by the CH-GFP/CA-RFP virus (Fig. 2, row 4), the resultant GFP and RFP fluorescence patterns colocalized, producing a pattern that resembled that produced by either the CH-GFP or CA-RFP fusion expressed individually, or by the ER-GFP or ER-RFP marker. This suggested that CH-GFP and CA-RFP colocalize in the ER and that neither a native chiA nor a native v-cath gene is required for the ER localization of CA-RFP or CH-GFP, respectively.
We also assessed the ER colocalization of CH-GFP and CA-RFP based on the Pearson correlation coefficient to assess if the pixel intensities from global (n = 5) GFP and RFP ER fluorescence were correlated spatially in the ER. We determined that the Pearson correlation coefficient (Rr) for ER-localized fluorescence of CH-GFP and ER-RFP was 0.565 (±0.802) and that of CA-RFP and ER-GFP was 0.531 (±0.055). However, a significantly (P < 0.01) higher Rr of 0.735 (±0.028) was obtained for the ER colocalization of CH-GFP and CA-RFP coexpressed by the CH-GFP/CA-RFP virus. The higher Rr value for CH-GFP/CA-RFP colocalization is more suggestive of a molecular interaction between CH-GFP and CA-RFP than the lower Rr values obtained for simple CH-GFP or CA-RFP localization in the ER with the ER markers. In addition, when we assessed several (n = 10) discrete regions of interest within a single fluorescent CH-GFP/CA-RFP colocalization image, Rr values similar to those obtained for the global cellular ER fluorescence analysis described above were obtained (data not shown).
ER localization of GFP fused to 12 or 22 N-terminal pre-proV-CATH amino acids.
Since proV-CATH is targeted to the ER (13), we tested if either the N-terminal 12- or 22-amino-acid peptide of V-CATH, when fused to the GFP N terminus, promoted the ER entry of GFP, which normally has a diffuse distribution throughout the cell. In all cases, cells were coinfected with the chiA/ER-RFP virus to visualize the ER and to help compare the fluorescence patterns of control GFPs with those from virus-expressed v-cath-gfp fusions. The GFP fluorescence patterns of the two control viruses, chiA/ER-GFP and chiA/GFP (expressing nontargeted GFP), were compared to those of the V-CATH N-terminal 12-amino-acid (N12-GFP) or 22-amino-acid (N22-GFP) peptide-GFP fusion at 40 hpi.
Figure 3 shows the schematics of the viral constructs used for this experiment and the resulting fluorescence patterns in Hi5 cells at 40 hpi. The fluorescence from the chiA/ER-RFP virus overlapped precisely with that of the chiA/ER-GFP virus (Fig. 3, row 2), as expected, but not with that of the control chiA/GFP virus showing diffuse GFP fluorescence throughout the cell, including the nucleus (row 1). The GFP fluorescence of the chiA/N22-GFP virus, expressing the v-cath-encoded N-terminal 22-amino-acid peptide fused to GFP (N22-GFP), overlapped precisely with the fluorescence of the ER-RFP marker (Fig. 3, row 4), including a lack of nuclear fluorescence, and this pattern was consistent in all infected cells. In contrast, the fluorescence of the chiA/N12-GFP virus, expressing only the v-cath-encoded N-terminal 12-amino-acid peptide fused to GFP (N12-GFP), occurred throughout the cells, including the nucleus (Fig. 3, row 3), like that of the chiA/GFP control virus expressing GFP (row 1), and was consistent in all cells of the monolayer (data not shown). Thus, the v-cath-encoded N-terminal 22-amino-acid peptide, but not the 12-amino-acid peptide, was sufficient for the ER localization of GFP.
mRFP-based BiFC.
A bimolecular fluorescence complementation (BiFC) system (16), which utilizes a split monomeric RFP (mRFP1) with enhanced fluorescence due to a Q66T amino acid substitution (17), was used to assess any molecular interaction between AcMNPV CHIA and proV-CATH during infection. In all cases the ER was labeled by coinfecting cells with the chiA/ER-GFP virus. No red fluorescence was detected when cells were infected with the chiA/ER-GFP virus alone. Control experiments, in which cells were infected with a virus expressing only one of the split mRFP-fused proteins (either CH-mRFPC/v-cath or chiA/CA-mRFPN) also produced no detectable red fluorescence (data not shown). To ensure that any BiFC pattern detected was indeed specific to interaction between CHIA and proV-CATH, coinfection with two control viruses, CH-mRFPC/v-cath, which expresses CH-mRFPC, and MYC-mRFPN-KDEL/v-cath, which expresses ER-targeted, unfused MYC-mRFPN-KDEL (to mimic the expected localization of CH-mRFPC), was monitored (Fig. 4a). No red fluorescence was detected in this control assay, even though CH-mRFPC and MYC-mRFPN-KDEL were detectable in the cell lysate by immunoblotting with commercial anti-FLAG or anti-MYC (data not shown). We also detected MYC-mRFPN-KDEL by anti-MYC-based immunofluorescent labeling, which produced a fluorescence pattern that overlapped that of the ER-GFP marker (data not shown). In contrast to all our negative-control assays, we detected red fluorescence for the CH-mRFPC/CA-mRFPN virus, suggestive of complementation due to a direct interaction between the split mRFP-tagged chitinase and cathepsin (Fig. 4b). Most cells infected with the CH-mRFPC/CA-mRFPN virus produced BiFC fluorescence, indicating that the interaction between CH-mRFPC and CA-mRFPN occurred in infected cells. Some cells expressing the ER-GFP marker did not show evidence of BiFC, perhaps because these cells were not infected with the BiFC virus. The facts that the red BiFC fluorescence pattern observed in infected cells resembled the fluorescence pattern of the green ER-GFP marker protein expressed by a coinfecting virus and that these patterns were coincident in the merged image indicated not only that there is a molecular interaction between CH-mRFPC and CA-mRFPN but also that the interaction occurred in the ER of infected cells (Fig. 4b).
Fig. 4.
Bimolecular fluorescence complementation (BiFC) assay. Virus constructs are shown above the corresponding fluorescence images. (a) Negative control for the BiFC assay. Cells were coinfected at an MOI of ∼100 with CH-mRFPC/v-cath, which expresses the ER-targeted CH-mRFPC fusion protein, and another virus, MYC-mRFPN-KDEL/v-cath, which expresses the ER-targeted MYC-mRFPN-KDEL protein. The MYC-mRFPN-KDEL protein was targeted to the ER by fusion to the chiA signal peptide and was modified by the addition of a C-terminal KDEL motif so that it accumulates in the ER of infected cells. Both of the BiFC negative-control viruses contain unmodified v-cath. (b) Cells were infected at an MOI of ∼100 with the CH-mRFPC/CA-mRFPN BiFC virus, which coexpresses the split-mRFP fusion proteins CH-mRFPC and CA-mRFPN, along with the ER-GFP virus to identify the ER. Virus-infected Hi5 cells were photographed at 48 hpi using confocal laser scanning microscopy. SP, chitinase signal peptide sequence.
Ni affinity copurification of CHIA and proV-CATH.
Protein copurification, utilizing Ni-agarose/6× histidine tag (His) affinity, was used to provide biochemical evidence for the interaction between CHIA and proV-CATH. To detect CHIA and proV-CATH in the different samples by immunoblotting, chiA was tagged with FLAG (CH) and v-cath with HA (CA) (Fig. 5a) as described above. The control CH/CA virus had FLAG-tagged chiA and HA-tagged v-cath, but neither chiA nor v-cath contained a 6× His tag. To demonstrate interaction between CH and CA, reciprocal copurifications were carried out using two experimental viruses, one coexpressing CH-HIS and CA (CH-HIS/CA) and the other coexpressing CH and CA-HIS (CH/CA-HIS). CH was detected in cell lysates from all three viruses by immunoblotting using anti-FLAG (Fig. 5b, lanes 1, 2, and 3; 58-kDa CHIA bands). Similarly, CA was also detectable in all three of the lysate samples when blots were probed with anti-HA (Fig. 5b, lanes 1, 2, and 3; 36-kDa proV-CATH bands).
As expected, CH-HIS was purified from lysates of cells infected with CH-HIS/CA by Ni affinity to the His tag (Fig. 5b, lane 4; 58-kDa CHIA band). CA (with no His tag) was copurified in the same purified CH-HIS sample (Fig. 5b, lane 4; 36-kDa proV-CATH band). As expected, CA-HIS was purified by Ni affinity to the His tag from the lysate of cells infected with CH/CA-HIS (Fig. 5b, lane 6; 36-kDa proV-CATH band). CH (with no His tag) was copurified (Fig. 5b, lane 6; 58-kDa CHIA band) with CA-HIS. From the CH/CA negative-control lysate (in which neither CH nor CA was His tagged), neither CH nor CA, though both were present in the lysate (Fig. 5b, lane 2), was retained on the Ni-agarose, demonstrating that the Ni affinity copurification of each protein was due to the presence of the respective His tag (Fig. 5b, lane 5). Thus, when CH-HIS was pulled down with Ni-agarose, CA was copurified, and likewise, when CA-HIS was pulled down with Ni-agarose, CH was copurified. These reciprocal copurification data corroborate the mRFP-based BiFC results, both suggestive of a direct binding interaction between CHIA and proV-CATH.
Effects of CHIA KDEL deletion on subcellular cotrafficking of CHIA and proV-CATH.
When the C-terminal KDEL motif of AcMNPV CHIA was deleted (24, 25), the CHIAΔKDEL protein was secreted into the cell culture medium and the hemolymph of infected insects. If proV-CATH associates with CHIA, then cotrafficking of these proteins should be observed. To determine if the CHIA KDEL sequence influences the trafficking of proV-CATH in concert with CHIA, we deleted the native chiA-encoded KDEL such that the adjacent (ΔKDEL)CH and CA were coexpressed from their native promoters in the bacmid polh locus of the (ΔKDEL)CH/CA virus as in the control CH/CA virus (Fig. 6a). Two AcMNPV-permissive cell lines, each derived from a different insect species (Sf21 from Spodoptera frugiperda and Hi5 from Trichoplusia ni) were compared for their CH and CA retention and secretion profiles. Corresponding temporal Western blot analyses were carried out on the soluble intracellular and extracellular proteins from the two cell lines infected with the control CH/CA virus or the (ΔKDEL)CH/CA virus.
In both cell lines infected with the native control CH/CA virus, the intracellular levels of both CH and CA were much higher than the corresponding extracellular levels throughout the 48-h time course (Fig. 6). Moreover, intracellular CH and CA were detected earlier than extracellular CH and CA, respectively. For example, in Hi5 cells infected with the CH/CA virus, intracellular CH and CA were first detected by 24 and 36 hpi, while the detection of extracellular CH and CA was delayed to 42 and 48 hpi, respectively (Fig. 6b). In Sf21 cells infected with the CH/CA virus, intracellular CH and CA were first detected by 18 hpi and 24 hpi, respectively (Fig. 6c). In comparison, the first detection of extracellular CH was delayed to 30 hpi, while extracellular CA was only barely detected as late as 48 hpi in Sf21 cells. Thus, for both Hi5 and Sf21 cells infected with the control virus, both proteins remained largely intracellular and were secreted only about 12 to 24 h after initial intracellular detection.
In contrast, for both cell lines infected with the (ΔKDEL)CH/CA virus, which lacks the KDEL ER retention motif in CHIA, the levels of extracellular (ΔKDEL)CH and CA throughout the 48-h time course exceeded intracellular levels. Both extracellular proteins were also detected earlier than extracellular proteins for the control virus. In Hi5 cells infected with the (ΔKDEL)CH/CA virus, extracellular (ΔKDEL)CH and CA were first detected much earlier, at 24 and 36 hpi, respectively, than intracellular (ΔKDEL)CH and CA, detection of which was delayed until 30 and 48 hpi, respectively (Fig. 6b). Similar results were obtained for Sf21 cells infected with the (ΔKDEL)CH/CA virus (Fig. 6c). The overall extracellular levels of both (ΔKDEL)CH and CA were higher than the intracellular levels over the 48-h time course. While detection of extracellular (ΔKDEL)CH and CA was somewhat delayed relative to the first detection of intracellular proteins, it was not delayed as much as for the control virus. Thus, in both Hi5 and Sf21 cells infected with the (ΔKDEL)CH/CA virus, both (ΔKDEL)CH and CA were secreted to the extracellular medium much earlier and at higher levels than in cells infected with the control virus.
Irrespective of the cell line or virus construct used, CA was always detected after CH. Depending on the virus and cells used, the timing of detection of CA ranged from 6 to 18 h later than that for CH in the corresponding cell/virus system. For example, intracellular CA was not detected until 6 h after the detection of intracellular CH following infection of Sf21 cells with either virus. Similarly, the intracellular detection of CA was delayed by 12 or 18 h compared to that of CH in Hi5 cells infected with the CH/CA or (ΔKDEL)CH/CA virus, respectively.
In all cases the localization, secretion, and relative levels of CA matched those of CH or (ΔKDEL)CH. For the CH construct with KDEL (control), both CH and CA remained largely intracellular throughout the replication cycle, while for the CH construct lacking the KDEL ER retention motif, both (ΔKDEL)CH and CA were secreted earlier and at higher levels than those for the control CH/CA virus. This suggests that the CH KDEL motif is responsible for the intracellular retention of both CH and CA and that a lack of the CH KDEL allows for the secretion of both CH and CA. This KDEL-dependent coretention of CH and CA could be explained by a direct interaction between these two proteins.
To be certain that deletion of the CHIA KDEL motif did not inadvertently reduce cell viability or otherwise cause premature cell lysis, which could have instead disrupted the cellular retention of (ΔKDEL)CH and CA relative to that with the control virus, we performed a simple comparative infected-cell viability assay based on trypan blue exclusion. At 36 hpi and 48 hpi, times at which there were marked differences in the CH and CA distributions between control CH/CA and (ΔKDEL)CH/CA cultures, we similarly found 95% and 90% cell viabilities for both virus-infected cultures. The results of this viability assay therefore suggested that the CHIA KDEL deletion did not affect the viability or integrity of the (ΔKDEL)CH/CA-infected cells relative to that of control CH/CA virus-infected cells.
DISCUSSION
This study was designed to better define the mechanism(s) for the subcellular trafficking, cellular retention, and possible protein-protein interactions of AcMNPV proV-CATH, the inactive progenitor of the V-CATH cysteine protease. Our studies utilized bacmid-derived viruses that had their native chiA-v-cath locus deleted. Instead of being in the native locus, our chiA-v-cath coexpression constructs, maintaining their native intergenic promoters and genomic organization, were relocalized to the polh locus of the chiA-v-cath deletion bacmids. We determined by Northern and Western blotting that the CH and CA constructs were expressed in a manner similar to that of native AcMNPV chiA and v-cath. In particular, we noticed that neither the inherent timing of simultaneous transcription of chiA and v-cath RNA (from 9 hpi) or the lag in proV-CATH protein detection relative to that for CHIA, as previously described (12), changed for the bacmid polh locus-expressed CH and CA genes. Thus, the simultaneous expression kinetics of the adjacent chiA and v-cath isoform genes were conserved with that of the native-locus-expressed genes in all of our viral constructs.
We used GFP fused to CHIA (CH-GFP) and mRFP fused to proV-CATH (CA-RFP) to enable visualization of the fluorescence patterns produced by both fusion proteins in live virus-infected cells. When CH-GFP and CA-RFP were each expressed by separate viruses, the fluorescence pattern of each matched that of the corresponding ER marker protein, ER-RFP or ER-GFP. When CH-GFP and CA-RFP were coexpressed by a single virus, the CH-GFP and CA-RFP patterns were identical to each other and to those found when each fluorescent fusion was coexpressed with adjacent unmodified v-cath or chiA genes. The fact that CA-RFP was detectable when coexpressed with CH-GFP suggests that the GFP fusion with chiA did not affect the postulated chaperone activity of CHIA, which is needed for the proper folding and trafficking of proV-CATH. It also suggests that the solubility of CA-RFP is not affected due to its RFP fusion and, furthermore, that the putative interaction between CHIA and proV-CATH is not changed.
To determine the N-terminal portion of the v-cath-expressed protein that enables its ER entry, we fused either the first 12 or the 22 codons of v-cath to gfp. In the N12-GFP protein, the native preproV-CATH signal cleavage site (6) (after Ala18) was absent, but it was present in N22-GFP. The 22 codons, but not the 12 codons, encoding the N terminus of V-CATH enabled efficient ER translocation of normally diffuse GFP, demonstrating that the N-terminal 22-amino-acid sequence containing the preproV-CATH signal cleavage site was sufficient for the initial ER localization of proV-CATH.
We noted reconstituted mRFP fluorescence due to CH-mRFPC-CA-mRFPN interaction in our bimolecular fluorescence complementation (BiFC) assay. Using this assay we detected fluorescence, signifying that CHIA and proV-CATH do interact during replication within virus-infected cells. The putative CHIA–proV-CATH interaction is likely to have occurred in the ER of infected cells, where CHIA KDEL directs CHIA accumulation and perhaps assists in proV-CATH folding. The ER colocalization of the reconstituted mRFP fluorescence pattern with the ER-GFP marker agrees with our previous data for proV-CATH-DsRED accumulation in the ER of infected cells (13) and with the CH-GFP/CA-RFP colocalization data reported here. The CH-mRFPC-CA-mRFPN interaction in the ER is consistent with the notion that CHIA assists in the folding of proV-CATH and agrees with the several suggestions in the literature that CHIA and proV-CATH bind to each other (3–5, 14, 15, 19). In addition to documenting CHIA–proV-CATH interactions in vivo by mRFP-based BiFC, we demonstrated the interaction biochemically in vitro by (reciprocal) His tag-dependent copurification of CHIA and proV-CATH. We report that when CH-HIS was purified by Ni affinity, CA was copurified with it. Similarly, in the reciprocal experiment, when CA-HIS was purified by Ni affinity, CH was copurified with it. This finding further corroborates our colocalization and BiFC data, and it strongly suggests that there is a specific interaction between these two viral proteins.
Knowing that CHIA has an ER retention mechanism based on its native C-terminal KDEL motif, and with evidence for a CHIA–proV-CATH interaction in the ER of infected cells, we wanted to test whether the interaction of proV-CATH with ER-retained CHIA might provide a mechanism for proV-CATH cellular retention as well. When we deleted CHIA KDEL, we found synchronous, premature cosecretion and reduced intracellular accumulation of both CA and (ΔKDEL)CH relative to those for a control virus with intact CH KDEL. Thus, the intracellular (ER-retained) or extracellular (secreted) localization of proV-CATH mimicked that of CHIA with or without the KDEL, respectively. The fact that the same native coretention and ΔKDEL cosecretion was observed in cell lines derived from different tissues of different host organisms (Sf21, from ovaries of the fall armyworm, and Hi5, from embryonic T. ni tissue) suggests that the proV-CATH retention process is not host cell dependent.
When a lepidopteran insect is infected by a native chiA–v-cath-expressing baculovirus (such as AcMNPV), the timing, enzymatic maturation, and cellular release of the V-CATH and CHIA enzymes are regulated in order to allow liquefaction only after optimal viral replication and progeny OB formation have occurred. That is, chiA and v-cath are regulated codependently and coordinately, both temporally and spatially. The genetic organization of these two AcMNPV genes, in a contiguous antiparallel manner and with intergenic late transcription initiation sites, is conserved in the group 1 alphabaculoviruses and in some granuloviruses (27), ensuring such coregulation of transcription. Regardless of whether chiA and v-cath are encoded in this conserved antiparallel manner, most alpha- and betabaculoviruses, which infect lepidopteran larvae, encode homologues of both. Infection by gamma- and deltabaculoviruses, all of which lack chiA and v-cath, of hymenopteran and dipteran larvae, respectively, is restricted to the host gastrointestinal tract. It is likely that the gamma- and deltabaculoviruses do not need to encode homologues of chiA or v-cath because, unlike the alpha- and betabaculoviruses, they do not require terminal host liquefaction in order to disseminate their progeny viral OBs effectively. Rather, viral OB dissemination of these viruses occurs due to the sloughing off and release of OBs and infected midgut epithelial cells into the surroundings (1, 20).
Although chiA and v-cath have similar temporal transcription profiles, there is a delay in the translation of proV-CATH relative to that of CHIA. Presumably, enzymatically active CHIA is retained in the ER of cells due to its KDEL motif, preventing it from prematurely degrading host cuticular chitin. Since CHIA accumulates in the ER several hours prior to the time when proV-CATH is translated, sufficient CHIA would be available to bind nascent proV-CATH when it is translated later, possibly upon its cotranslational import into the ER. This timing would allow CHIA to assist in the folding of proV-CATH. Thus, the CHIA–proV-CATH interaction could retain both proteins in the ER of cells, under the direction of CHIA KDEL, until cell lysis occurs, when both enzymes are released and activated. In addition, perhaps the binding of proV-CATH to CHIA allows the retention of proV-CATH in a soluble proenzyme form that remains competent for proteolytic maturation into the active V-CATH enzyme. The maintenance of proV-CATH in a soluble form is dependent on CHIA, since proV-CATH in a ΔCHIA virus is rendered insoluble and is not activated (15). The fact that CHIA is required to promote proV-CATH maturation to active V-CATH during cell death implicates CHIA as a key factor in the control of CHIA–V-CATH-codependent host liquefaction. However, the fact that V-CATH maturation occurs only upon the lysis of infected cells (ultimately resulting in the cellular release and activation of the two enzymes), which commences the dissolution of host tissues, illustrates the importance of the molecular interaction between CHIA and proV-CATH and thus their coretention in the ER until cell lysis occurs.
If this regulation in time and localization is compromised, for example, by the secretion of native or foreign protease or chitinase (lacking a KDEL), then the timing of liquefaction in relation to optimal OB production should similarly be compromised. For example, an AcMNPV bacmid engineered to express and secrete a heterologous papain-like cathepsin (ScathL) derived from Sarcophaga peregrina killed Heliothis virescens larvae 30% faster than control AcMNPV but exhibited a 0.5 log reduction in OB production (8). Also, when T. ni was infected with a mutant AcMNPV expressing CHIAΔKDEL, CHIA (and presumably proV-CATH) was secreted constitutively during replication, and host insects died and were liquefied earlier (about 8 h) than when they were infected with control AcMNPV (24). The decrease in host survival time was even further exacerbated (24 h less than that with the control virus) when CHIAΔKDEL was overexpressed from the strong polh promoter (25).
When Cydia pomonella granulovirus (CpGV) chiA, which lacks a KDEL or equivalent motif and is therefore constitutively secreted from infected cells, was expressed by a BmNPV chiA deletion mutant, BmNPV proV-CATH was expressed, folded, and catalytically activated to V-CATH (4). This suggests that CpGV CHIA and BmNPV proV-CATH can be coregulated and function together. However, Daimon et al. (4) also noted that the level of host liquefaction was lower for the CpGV CHIA-expressing virus than for native BmNPV, highlighting the importance of the KDEL motif in CHIA for the efficiency of host liquefaction. This finding demonstrates that the differential intracellular regulation of CHIA and proV-CATH of different baculoviruses reflects each virus's innate dependence on these two baculoviral proteins for the promotion of host liquefaction. This also raises the possibility that complex and multifaceted CHIA and proV-CATH regulation at both the temporal and spatial levels might have evolved to coordinate intracellular CHIA and proV-CATH regulation/retention because of its ecological importance to optimal virus production, virus spread, and thus baculovirus success in nature.
Supplementary Material
ACKNOWLEDGMENTS
The work described here was supported by grants to P.J.K. from the Natural Sciences and Engineering Research Council of Canada (STPGP 365213 and RGPIN 8395).
We acknowledge the excellent technical support of David Leishman. We are grateful to Monique van Oers (Wageningen University) for the contribution of the chiA–v-cath deletion bacmid (AcBACΔCC) and to Joachim F. Uhrig (Max Planck Institute for Plant Breeding Research) for the contribution of the split mRFP constructs pBatTL-smRFPN and pBatTL-smRFPC.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Becnel J. J. 2006. Transmission of viruses to mosquito larvae mediated by divalent cations. J. Invertebr. Pathol. 92:141–145 [DOI] [PubMed] [Google Scholar]
- 2. Brömme D., Okamoto K. 1995. The baculovirus cysteine protease has a cathepsin B-like S2-subsite specificity. Biol. Chem. Hoppe-Seyler 376:611–615 [DOI] [PubMed] [Google Scholar]
- 3. Daimon T., Katsuma S., Kang W., Shimada T. 2006. Comparative studies of Bombyx mori nucleopolyhedrovirus chitinase and its host ortholog, BmChi-h. Biochem. Biophys. Res. Commun. 345:825–833 [DOI] [PubMed] [Google Scholar]
- 4. Daimon T., Katsuma S., Kang W. K., Shimada T. 2007. Functional characterization of chitinase from Cydia pomonella granulovirus. Arch. Virol. 152:1655–1664 [DOI] [PubMed] [Google Scholar]
- 5. Daimon T., Katsuma S., Shimada T. 2007. Mutational analysis of active site residues of chitinase from Bombyx mori nucleopolyhedrovirus. Virus Res. 124:168–175 [DOI] [PubMed] [Google Scholar]
- 6. Gotoh T., Awa H., Kikuchi K., Nirasawa S., Takahashi S. 2010. Prorenin processing enzyme (PPE) produced by baculovirus-infected Sf-9 insect cells: PPE is the cysteine protease encoded in the acMNPV gene. Biosci. Biotechnol. Biochem. 74:370–374 [DOI] [PubMed] [Google Scholar]
- 7. Granados R. R., Lawler K. A. 1981. In vivo pathway of Autographa californica baculovirus invasion and infection. Virology 108:297–308 [DOI] [PubMed] [Google Scholar]
- 8. Harrison R. L., Bonning B. C. 2001. Use of proteases to improve the insecticidal activity of baculoviruses. Biol. Control 20:199–209 [Google Scholar]
- 9. Haseloff J., Siemering K. R., Prasher D. C., Hodge S. 1997. Removal of a cryptic intron and subcellular localization of green fluorescent protein are required to mark transgenic Arabidopsis plants brightly. Proc. Natl. Acad. Sci. U. S. A. 94:2122–2127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Hawtin R. E., et al. 1995. Identification and preliminary characterization of a chitinase gene in the Autographa californica nuclear polyhedrosis virus genome. Virology 212:673–685 [DOI] [PubMed] [Google Scholar]
- 11. Hawtin R. E., et al. 1997. Liquefaction of Autographa californica nucleopolyhedrovirus-infected insects is dependent on the integrity of virus-encoded chitinase and cathepsin genes. Virology 238:243–253 [DOI] [PubMed] [Google Scholar]
- 12. Hodgson J. J., Arif B. M., Krell P. J. 2007. Reprogramming the chiA expression profile of Autographa californica multiple nucleopolyhedrovirus. J. Gen. Virol. 88:2479–2487 [DOI] [PubMed] [Google Scholar]
- 13. Hodgson J. J., Arif B. M., Krell P. J. 2009. Autographa californica multiple nucleopolyhedrovirus and Choristoneura fumiferana multiple nucleopolyhedrovirus v-cath genes are expressed as pre-proenzymes. J. Gen. Virol. 90:995–1000 [DOI] [PubMed] [Google Scholar]
- 14. Hom L. G., Ohkawa T., Trudeau D., Volkman L. E. 2002. Autographa californica M nucleopolyhedrovirus proV-CATH is activated during infected cell death. Virology 296:212–218 [DOI] [PubMed] [Google Scholar]
- 15. Hom L. G., Volkman L. E. 2000. Autographa californica M nucleopolyhedrovirus chiA is required for processing of V-CATH. Virology 277:178–183 [DOI] [PubMed] [Google Scholar]
- 16. Hu C. D., Grinberg A. V., Kerppola T. K. 2006. Visualization of protein interactions in living cells using bimolecular fluorescence complementation (BiFC) analysis. Curr. Protoc. Cell Biol. Chapter 21, Unit 21.3. doi: 10.1002/0471143030.cb2103s29 [DOI] [PubMed] [Google Scholar]
- 17. Jach G., Pesch M., Richter K., Frings S., Uhrig J. F. 2006. An improved mRFP1 adds red to bimolecular fluorescence complementation. Nat. Methods 3:597–600 [DOI] [PubMed] [Google Scholar]
- 18. Kaba S. A., Salcedo A. M., Wafula P. O., Vlak J. M., van Oers M. M. 2004. Development of a chitinase and v-cathepsin negative bacmid for improved integrity of secreted recombinant proteins. J. Virol. Methods 122:113–118 [DOI] [PubMed] [Google Scholar]
- 19. Katsuma S., Nakanishi T., Daimon T., Shimada T. 2009. N-linked glycans located in the pro-region of Bombyx mori nucleopolyhedrovirus V-CATH are essential for the proper folding of V-CATH and V-CHIA. J. Gen. Virol. 90:170–176 [DOI] [PubMed] [Google Scholar]
- 20. Lauzon H. A., et al. 2004. Sequence and organization of the Neodiprion lecontei nucleopolyhedrovirus genome. J. Virol. 78:7023–7035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Ohkawa T., Majima K., Maeda S. 1994. A cysteine protease encoded by the baculovirus Bombyx mori nuclear polyhedrosis virus. J. Virol. 68:6619–6625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. O'Reilly D. R., Miller L. K., Luckow V. A. 1992. Baculovirus expression vectors: a laboratory manual. W. H. Freeman, New York, NY [Google Scholar]
- 23. Rebelo A. R., Niewiadomski S., Prosser S. W., Krell P., Meng B. 2008. Subcellular localization of the triple gene block proteins encoded by a foveavirus infecting grapevines. Virus Res. 138:57–69 [DOI] [PubMed] [Google Scholar]
- 24. Saville G. P., Patmanidi A. L., Possee R. D., King L. A. 2004. Deletion of the Autographa californica nucleopolyhedrovirus chitinase KDEL motif and in vitro and in vivo analysis of the modified virus. J. Gen. Virol. 85:821–831 [DOI] [PubMed] [Google Scholar]
- 25. Saville G. P., Thomas C. J., Possee R. D., King L. A. 2002. Partial redistribution of the Autographa californica nucleopolyhedrovirus chitinase in virus-infected cells accompanies mutation of the carboxy-terminal KDEL ER-retention motif. J. Gen. Virol. 83:685–694 [DOI] [PubMed] [Google Scholar]
- 26. Slack J. M., Kuzio J., Faulkner P. 1995. Characterization of v-cath, a cathepsin L-like proteinase expressed by the baculovirus Autographa californica multiple nuclear polyhedrosis virus. J. Gen. Virol. 76(Pt 5):1091–1098 [DOI] [PubMed] [Google Scholar]
- 27. Slack J. M., Ribeiro B. M., de Souza M. L. 2004. The gp64 locus of Anticarsia gemmatalis multicapsid nucleopolyhedrovirus contains a 3′ repair exonuclease homologue and lacks v-cath and ChiA genes. J. Gen. Virol. 85:211–219 [DOI] [PubMed] [Google Scholar]
- 28. Thomas C. J., et al. 1998. Localization of a baculovirus-induced chitinase in the insect cell endoplasmic reticulum. J. Virol. 72:10207–10212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Wang F., Zhang C. X., Kumar V. S., Wu X. F. 2005. Influences of chitinase gene deletion from BmNPV on the cell lysis and host liquefaction. Arch. Virol. 150:981–990 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.