Abstract
The tripartite motif (TRIM) protein family comprises more than 60 members that have diverse functions in various biological processes. Although a small number of TRIM proteins have been shown to regulate innate immunity, much remains to be learned about the functions of the majority of the TRIM proteins. Here we identify TRIM56 as a cellular protein associated with the N-terminal protease (Npro) of bovine viral diarrhea virus (BVDV), a pestiviral interferon antagonist which degrades interferon regulatory factor 3 (IRF3) through the proteasome. We found that TRIM56 was constitutively expressed in most tissues, and its abundance was further upregulated moderately by interferon or virus. The manipulation of TRIM56 abundance did not affect the protein turnover of Npro and IRF3. Rather, ectopic expression of TRIM56 substantially impaired, while knockdown of TRIM56 expression greatly enhanced, BVDV replication in cell culture. The antiviral activity of TRIM56 depended on its E3 ubiquitin ligase activity as well as the integrity of its C-terminal region but was not attributed to a general augmentation of the interferon antiviral response. Overexpression of TRIM56 did not inhibit the replication of vesicular stomatitis virus or hepatitis C virus, a virus closely related to BVDV. Together, our data demonstrate that TRIM56 is a novel antiviral host factor that restricts pestivirus infection.
INTRODUCTION
The tripartite motif (TRIM) protein family consists of more than 60 members that have diverse functions in a broad range of biological processes, including, but not limited to, cell proliferation, differentiation, development, apoptosis, and immunity (19, 21). Proteins of the TRIM family share the characteristic tripartite (also known as RBCC) motif that comprises a RING finger, one or two B boxes, and a coiled-coil domain at the N-terminal region. The C-terminal half, however, is variable among different TRIM proteins and can have or not have one or more specific domains that determine the function specificity of some TRIM members. Based on the C-terminal domain composition of the N-terminal RBCC motif, the human TRIMs are classified into 11 subfamilies, subfamilies C-I to C-XI, with the C-IV group having the largest number of members, all of which contain the SPRY/B30.2-like domain, a conserved region whose function remains unclear but is thought to be involved in protein-protein interactions or RNA binding (21, 22, 29).
The functions of the majority of the TRIM proteins are poorly understood. Based on the presence of the RING domain, it was proposed that the TRIM proteins represent a novel class of single-RING-finger E3 ubiquitin (Ub) ligases that mediate the posttranslational modification of different substrates (18). By self-association via the coiled-coil domains, the TRIM proteins oligomerize and act as a scaffold for the assembly of multiprotein complexes that occupy specific cellular compartments (24). TRIM proteins have been shown to be involved in neurological diseases, genetic disorders, and carcinogenesis (18) and have recently demonstrated emerging roles in regulating innate immunity to viral infections, which were uncovered mainly through research on host factors that restrict retroviruses (20). For example, TRIM5α from rhesus monkey targets the incoming HIV capsid early postinfection and affects the uncoating of the preintegration complex, a process that depends on the C-terminal B30.2 domain (30). Human TRIM15, however, interferes with murine leukemia virus release by interacting with viral Gag through its B-box domain independent of the E3 ligase activity and C-terminal B30.2 domain (34). More recently, several TRIMs have been shown to positively or negatively regulate innate immune signaling pathways. TRIM25 interacts with retinoic acid-inducible gene I (RIG-I), a cytoplasmic viral pathogen recognition receptor, through its B30.2 domain and promotes the ubiquitination of RIG-I, which is essential for downstream signaling for the induction of type I interferons (IFNs) (9). Interestingly, the NS1 protein of influenza virus, a well-known IFN antagonist, associates with TRIM25 and inhibits its E3 ligase activity toward RIG-I ubiquitination (7), demonstrating the importance of TRIM25 in antiviral innate immunity. On the other hand, mouse TRIM30α associates with TAK1 and promotes the degradation of TAB2 and TAB3, leading to the inhibition of TRAF6 ubiquitination and subsequent termination of Toll-like receptor signaling (28). TRIM21 was reported previously to interact with and ubiquitinate IFN regulatory factor 8 (IRF8) and IRF3, which regulates the transcriptional activity of IRF8 and the stability of IRF3, respectively (10, 14). Despite recent advances in our understanding of the function of TRIMs and their emerging roles in innate immunity, only a small number of TRIMs have been shown to fend off viruses, and the precise mechanisms of action for most known antiviral TRIMs are still unclear (21). It is expected that the number of TRIMs having antiviral activities will continue to grow.
Bovine viral diarrhea virus (BVDV) along with classical swine fever virus of pigs and border disease virus of sheep comprise the genus Pestivirus within the family Flaviviridae. Pestiviruses cause respiratory, digestive, and reproductive problems in domestic ruminants worldwide that result in high economic losses. As with other flaviviruses, pestiviruses are small enveloped viruses that possess a positive-polarity, single-stranded RNA genome. Upon the infection of host cells, the pestivirus genomic RNA is translated into a single polyprotein, which is subsequently processed into 11 to 12 structural and nonstructural (NS) proteins. Similarly to the closely related hepatitis C virus (HCV) and other classical flaviviruses, the pestivirus NS proteins are responsible for the assembly of viral replication complexes on cytoplasmic membranes, while the structural proteins make up the virions. Unique to pestivirus-encoded proteins is a small N-terminal protease (Npro), a papain-like cysteine protease that directs its self-cleavage off the viral polyprotein (25). Npro is dispensable for pestivirus replication in vitro, but it is critical for the pestivirus counteraction of the IFN antiviral response. We and others have shown that BVDV Npro targets IRF3, a transcription factor pivotal for the control of type I IFN synthesis, for proteasomal degradation (5, 11). Although the underlying mechanism remains elusive, the Npro-induced degradation of IRF3 requires the cellular ubiquitin conjugation system, as the inactivation of the E1 ubiquitin-activating enzyme prevents Npro-induced IRF3 loss (5). In our efforts to understand how BVDV Npro promotes polyubiquitination and subsequent degradation of IRF3, we have identified TRIM56 as an interaction partner of Npro. Initially speculating that TRIM56 may be the E3 ubiquitin ligase involved in the Npro-IRF3 interplay, we have unexpectedly found that TRIM56 is an antiviral host factor against BVDV. The antiviral effect is specific for BVDV, as we found that TRIM56 had no effect on the replication of vesicular stomatitis virus (VSV) and HCV. We report that TRIM56 inhibits BVDV propagation by acting on intracellular viral RNA replication. Furthermore, we provide evidence that the antiviral function of TRIM56 depends both on its E3 ubiquitin ligase activity and on the integrity of its C-terminal region but is not attributed to a general augmentation of the IFN response. Our study adds TRIM56 to the list of antiviral TRIMs and provides novel insights for better understanding of the antiviral mechanisms of TRIM56.
MATERIALS AND METHODS
Cells.
HEK293, 293FT, human hepatoma Huh7 and Huh7.5, human monocytic leukemia THP1, and African green monkey kidney Vero cells were cultured by conventional techniques. MDBK cells were cultured similarly except that horse serum was used in place of fetal bovine serum (FBS). HeLa-FHNpro cells stably expressing Flag- and hemagglutinin (HA)-tandem-tagged BVDV Npro were cultured as described previously (36). The HeLaNpro-25 cell line is a Tet-regulated cell line that conditionally expresses Myc-tagged Npro (5). 293-Npro cells were generated by the stable transduction of HEK293 cells with a replication-incompetent retroviral vector encoding Flag-tagged Npro (pCX4pur-FlagNpro), followed by selection with puromycin. MDBK and Huh7 cells stably expressing wild-type (WT) and various mutant forms of Flag-tagged TRIM56 were created by infection with replication-incompetent retroviral vectors that encode WT or mutant TRIM56 in the pCX4bsr backbone (see details below), followed by stable selection with blasticidin. MDBK cells stably expressing WT IRF3 (BK-F3) and a mutant IRF3 with the N-terminal 133 amino acids (aa) deleted (BK-F3DN133) were derived from MDBK cells infected with replication-incompetent retroviral vectors that encode Flag- and HA-tandem-tagged human IRF3 or IRF3DN133 in the pCX4pur backbone, followed by stable selection with puromycin.
Viruses, replicons, and viral replication assays.
VSV (Indiana strain) and cytopathic (cp) BVDV (NADL strain) stocks were propagated in Vero and MDBK cells, respectively. Genotype 2a HCV JFH1 was produced and propagated in Huh7.5 cells as described previously (35, 37). For virus infection, the indicated viral inoculum was incubated with cells in Dulbecco's modified Eagle's medium (DMEM) plus 2% FBS at the indicated multiplicity of infection (MOI) in a humidified 37°C CO2 incubator. One hour later (for VSV and BVDV) or 6 h later (for HCV), the viral inoculum was removed, and infected cells were washed, refed with complete culture medium, and allowed to return to culture. At various time points postinfection, cell-free culture supernatants were removed and stored at −80°C until they were evaluated for infectious virus titers by standard plaque (for VSV, in Vero cells) and 50% tissue culture infective dose (TCID50) (for BVDV and HCV, in MDBK and Huh7.5 cells, respectively) assays, as previously described (17, 37, 41). Viral titers were expressed as PFU/ml (for VSV) and TCID50/ml (for BVDV and HCV).
The genomic J6/JFH1 HCV RNA replicon encoding Renilla luciferase (pFL-J6/JFH-5′C19Rluc2AUbi, here referred to as J6/JFH1-RL) and the subgenomic noncytopathic (ncp) BVDV (ncpBVDV) replicon encoding firefly luciferase (ncpNADL Jiv90-deltaS-luc, here referred to as BVD39) were generous gifts from Charles Rice (15, 32). We generated the BVD39-NS2 replicon construct by deleting the Npro gene, 10 N-terminal residues of capsid, an extra Leu codon, 18 C-terminal residues of capsid, 9 N-terminal residues of Erns, and 48 C-terminal residues of E2-encoding sequences from the BVD39 replicon plasmid using standard mutagenesis techniques. HCV and BVDV replicon RNAs were synthesized from linearized replicon plasmid templates by T7 RNA polymerase-mediated in vitro transcription as described previously (15, 32). For measurements of BVDV and HCV RNA replication, 5 million cells were transfected with 6 μg of in vitro-transcribed ncpBVDV or J6/JFH1-RL HCV replicon RNAs by electroporation on a Bio-Rad GenePulser XCell apparatus. The parameters used were 140 V, 500 μF, and maximum resistance (exponential decay mode). After electroporation, cells were cultured in 12-well plates for the indicated time periods prior to cell lysis (from duplicated wells) in 1× passive lysis buffer (Promega) and for measurements of Renilla (for J6/JFH1-RL RNA-transfected cells) or firefly (for ncpBVDV replicon RNA-transfected cells) luciferase activities in cell lysates. The luciferase activity at 6 h posttransfection (which represents the direct translation of input replicon RNA) was set as the baseline for the calculation of fold changes in luciferase activities at later time points, which were used to compare the rates of viral RNA replication.
Plasmids.
Plasmids were generated by conventional PCR techniques, and their identities were validated by DNA sequencing. To construct the human TRIM56 (here referred to as TRIM56) expression vector pcDNA3.1-TRIM56-V5, cDNA encoding human TRIM56 was amplified by PCR from HeLa cDNA using primers 5′-cgcggatccATGGTTTCCCACGGGTCCT-3′ (forward) and 5′-gacggtatcgatACTGTCCGGAGAACGGAC-3′ (reverse) and subsequently ligated into pcDNA3.1/V5-His-TOPO (Invitrogen). To construct the bovine TRIM56 (here referred to as boTRIM56) expression vector pEF6-boTRIM56-V5, cDNA encoding boTRIM56 was amplified by PCR from MDBK cDNA using primers 5′-cgcggatccATGGTTTCCCAGGGCTCCT-3′ (forward) and 5′-gacggtatcgatACTGTCAAGAGGGCGGAC-3′ (reverse) and subsequently ligated into pEF6/V5-His-TOPO (Invitrogen). Uppercase letters indicate TRIM56 sequences. The TRIM56 RING mutants (C24A and CC21/24AA) and various deletion mutants were generated from pcDNA3.1-TRIM56-V5 by QuikChange site-directed mutagenesis (Stratagene). The mutagenesis primers used are listed in Table 1. N-terminally and C-terminally green fluorescent protein (GFP)-tagged TRIM56 constructs (GFP-TRIM56 and TRIM56-GFP) were constructed in the pEGFP-N1 and pEGFP-C2 (both from Clontech) backbones, respectively. To generate retroviral expression vectors for WT boTRIM56 and WT and various mutant forms of TRIM56, the boTRIM56 and various TRIM56 cDNA fragments were excised from pEF6-boTRIM56-V5 and pcDNA3.1-derived TRIM56-V5 plasmids and ligated into pCX4bsr in a way that they would be expressed as proteins fused to a C-terminal Flag tag.
Table 1.
Primers used for mutagenesis of human TRIM56
| Mutation | Primer | Primer sequence |
|---|---|---|
| A (ΔRING) | del2-71F | 5′-CCTTCGCGGATCCATGTCCTTCAAGACCAACT-3′ |
| del2-71R | 5′-AGTTGGTCTTGAAGGACATGGATCCGCGAAGG-3′ | |
| B (ΔB box) | del161-205F | 5′-GTGGTATGATGAGGAGGCTGAAGCTGTGCGTG-3′ |
| del161-205R | 5′-CACGCACAGCTTCAGCCTCCTCATCATACCAC-3′ | |
| C (Δcoiled coil) | del211-363F | 5′-GCTGAAGCTGTGCGTCACCTTCTTCGGCTG-3′ |
| del211-363R | 5′-CAGCCGAAGAAGGTGACGCACAGCTTCAGC-3′ | |
| D (Δ369-742) | del369-742F | 5′-CACCTTCTTCGGCTGGGGACCATCCACATC-3′ |
| del369-742R | 5′-GATGTGGATGGTCCCCAGCCGAAGAAGGTG-3′ | |
| E (Δ77-742) | del77-742F | 5′-CTCCTTCAAGACCAACGGGACCATCCACATCT-3′ |
| del77-742R | 5′-AGATGTGGATGGTCCCGTTGGTCTTGAAGGAG-3′ | |
| F (ΔN363) | del2-363F | 5′-CTTCGCGGATCCATGCACCTTCTTCGGCTG-3′ |
| del2-363R | 5′-CAGCCGAAGAAGGTGCATGGATCCGCGAAG-3′ | |
| G (ΔRING + B box) | del2-205F | 5′-CTTCGCGGATCCATGGCTGAAGCTGTGCGT-3′ |
| del2-205R | 5′-ACGCACAGCTTCAGCCATGGATCCGCGAAG-3′ | |
| H (Δ211-284) | del211-284F | 5′-GCTGAAGCTGTGCGTGAAGAAGCTGCTCGG-3′ |
| del211-284R | 5′-CCGAGCAGCTTCTTCACGCACAGCTTCAGC-3′ | |
| I (Δ290-353) | del290-353F | 5′-GAAGAAGCTGCTCGGCTCCATCCTGGGCTC-3′ |
| del290-353R | 5′-GAGCCCAGGATGGAGCCGAGCAGCTTCTTC-3′ | |
| J (Δ355-433) | del355-433F | 5′-CCACAGCTGGAGCTCACCTTGGAGGAGGAC-3′ |
| del355-433R | 5′-GTCCTCCTCCAAGGTGAGCTCCAGCTGTGG-3′ | |
| K (Δ431-519) | del431-519F | 5′-ACAACCCGAGAAGAGGAGATCCTGGTGGCG-3′ |
| del431-519R | 5′-CGCCACCAGGATCTCCTCTTCTCGGGTTGT-3′ | |
| L (Δ515-610) | del515-610F | 5′-GATCACCGGGCTCTGTGTGGAGGTGTACAATA-3′ |
| del515-610R | 5′-TATTGTACACCTCCACACAGAGCCCGGTGATC-3′ | |
| M (Δ621-695) | del621-695F | 5′-AATATGGAAGGCAGCCTGCTTCGAGAAGTCAACAAG-3′ |
| del621-695R | 5′-CTTGTTGACTTCTCGAAGCAGGCTGCCTTCCATATT-3′ | |
| N (Δ693-750) | del693-750F | 5′-GATAAGAAGGGCTACATCCGTTCTCCGGACAGTATC-3′ |
| del693-750R | 5′-GATACTGTCCGGAGAACGGATGTAGCCCTTCTTATC-3′ | |
| C24A | TM56 C24A+ | 5′-ACTTCCTGGCCTGTAAAATCGCCCTGGAGCAGCTGCG-3′ |
| TM56 C24A− | 5′-CGCAGCTGCTCCAGGGCGATTTTACAGGCCAGGAAGT-3′ | |
| CC21/24AA | TM56 CC21/24AA+ | 5′-AGCGACTTCCTGGCCGCTAAAATCGCCCTGGAGCAGCTGCG-3′ |
| TM56 CC21/24AA− | 5′-CGCAGCTGCTCCAGGGCGATTTTAGCGGCCAGGAAGTCGCT-3′ |
34AIRESneo encoding N-terminally Flag-tagged HCV NS3/4A was constructed from HCV-N (2) sequenced in the pBICEP-CMV-1 backbone (Sigma). cDNA expression plasmids for BVDV NS4A, NS4B, and NS5B were constructed from the BVD39 replicon sequence and ligated in frame with downstream Myc-6×His tags into pEF1/Myc-His (Invitrogen). Myc-tagged BVDV NS3, NS3/4A, NS2-3, and NS5A constructs (ncp Nose strain) were generous gifts from Hiroomi Akashi (38). pIRES-V5-TRIM25 (8) and p3XFLAG-Ub (6) were gifts from Jae Jung and Susan Baker, respectively. pcDNA6-mycNpro (the WT and L8P and C69A mutants) were described previously (5).
RNA interference (RNAi).
A short hairpin RNA (shRNA) construct targeting boTRIM56 (pLKO.1pur-boT56-097shRNA) was constructed in the pLKO.1-TRC cloning vector (Addgene) by using the following oligonucleotides: 5′-ccggCGCGCGGCTCTATCTCATCAActcgagTTGATGAGATAGAGCCGCGCGtttttg-3′ (forward) (the boTRIM56 sequence is in uppercase type) and 5′-aattcaaaaaCGCGCGGCTCTATCTCATCAActcgagTTGATGAGATAGAGCCGCGCG-3′ (reverse). The TRIM56 shRNA in the pLKO.1puro vector (pLKO.1puro-T56-097shRNA) was purchased from Openbiosystems (catalog number TRCN0000073097), in which the TRIM56 target sequence was CGCACGGCTCTATCTCATCAA. To generate the TRIM56 shRNA vector (pRevTRE-T56-097shRNA) that allows the selection of transfected cells with hygromycin, the U6 promoter-huTRM56 shRNA cDNA cassette was excised from pLKO.1puro-T56-097shRNA by EcoRI (polished to a blunt end) and ClaI digestion and ligated into pRevTRE (Clontech), which was digested with XhoI (polished to a blunt end) and ClaI.
For the stable knockdown of boTRIM56, MDBK cells were transfected with the linearized pLKO.1pur-boT56-097shRNA vector and selected in medium containing puromycin (5 μg/ml). For the stable knockdown of TRIM56 in HeLa-FHNpro cells, cells were transfected with the linearized pRevTRE-T56-097shRNA construct and selected with hygromycin (200 μg/ml). After 2 to 3 weeks, individual cell colonies were picked, expanded, and screened for boTRIM56 and TRIM56 expression by quantitative real-time PCR (for MBDK boTRIM56 knockdown clones) and/or Western blotting (for HeLa-FHNpro TRIM56 knockdown clones).
For the transient knockdown of TRIM56 in 293-Npro cells, two synthetic Stealth small interfering RNAs (siRNAs) (Invitrogen) were used. The siRNA target sequences were AGTTCAAAGGCAGGCTCAAGTCAAT (siRNA 1) and CCACGTGGAGGTGTACAATATGGAA (siRNA 2).
RT-PCR.
Total cellular RNA was extracted from cells grown in 60-mm culture dishes following the indicated treatments by using TRIzol (Invitrogen), treated with DNase I to remove potential genomic DNA contamination, and reverse transcribed to cDNA by using Moloney murine leukemia virus (MMLV) reverse transcriptase (RT) (Promega) according to the manufacturer's recommendations.
For semiquantitative RT-PCR, the following primers were used: human/bovine TRIM56 (5′-GCTCTATCTCATCAACC-3′ [forward] and 5′-GTGGATGGTSCCGTTACTGAG-3′ [reverse]), human RIG-I (16), human glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (Clontech), and boGAPDH (5′-CAAGTTCAACGGCACAGTCAA-3′ [forward] and 5′-TGGTCATAAGTCCCTCCACGAT-3′ [reverse]).
Quantitative real-time PCR was performed by using iQ SYBR green Supermix (Bio-Rad) with an iCycler IQ5 real-time PCR machine (Bio-Rad). The following primers were used to detect human/bovine TRIM56: 5′-CTGCTTGGMGACTTCCTGAC-3′ (forward) and 5′-GTGGATGGTSCCGTTACTGAG-3′ (reverse). The relative abundance of each target was obtained by normalization with endogenous 28S rRNA or β-actin (4).
IFN treatment, virus infections, and IFN-β promoter assay.
Where indicated, cells were transfected with poly(I:C) or poly(dA:dT) (both from GE Health) and incubated with 500 U/ml of recombinant human IFN-α in culture medium, infected with 50 to 100 hemagglutinin units (HAU)/ml of Sendai virus (SeV) (Cantell strain; Charles River Laboratories) for 16 h, or infected with VSV or cytopathic BVDV NADL at the indicated MOIs for the indicated time periods (5, 37, 41). IFN-β promoter activities were determined by cotransfecting cells with IFN-β-Luc and pRL-TK (internal control for the normalization of transfection efficiency), followed by a dual luciferase assay, as described previously (5, 36, 37).
Affinity purification of Npro-associated cellular proteins.
Npro-associated cellular proteins were purified from HeLa-FHNpro cells that stably express N-terminal Flag-HA-tandem-tagged BVDV Npro (36). Cells grown in 35 150-mm dishes were lysed in a buffer containing 50 mM HEPES (pH 7.4), 1.5 mM EDTA, 150 mM NaCl, 10% glycerol, 10 mM NaF, 1 mM Na3VO4, 0.5 mM dithiothreitol (DTT), 1% Triton X-100, and protease inhibitor cocktail (Sigma); clarified by centrifugation; and immunoprecipitated with anti-Flag M2 agarose (Sigma). The bound proteins were eluted with Flag peptide, fractionated on a 4 to 12% SDS-PAGE gel, and stained with Coomassie blue using a colloidal blue staining kit (Invitrogen). Protein bands were excised from the gel and analyzed on a matrix-assisted laser desorption ionization-tandem time of flight (MALDI-TOF/TOF) mass spectrometer.
Immunoprecipitation and immunoblots.
Cellular extracts were prepared and subjected to immunoprecipitation (IP) and/or immunoblot analysis as previously described (5). The following polyclonal antibodies (pAbs) and monoclonal antibodies (MAbs) were used: rabbit anti-TRIM56 (Bethyl Labs); rabbit anti-TRIM56 937-2 (generated by immunizing rabbits with a keyhole limpet hemocyanin [KLH]-coupled peptide, RAGWYDEEARERQAAQ); anti-Myc tag 9B11 MAb (Cell Signaling Tech); anti-V5 MAb (Invitrogen); rabbit anti-FLAG pAb, anti-FLAG M2, and anti-actin MAbs (Sigma); and peroxidase-conjugated secondary anti-rabbit and anti-mouse pAbs (Southern Biotech). Protein bands were visualized by using enhanced chemiluminescence (Millipore), followed by exposure to X-ray films.
Confocal imaging.
HeLaNpro-25 cells induced for Myc-Npro expression were cultured on LabTek chamber slides and transiently transfected with the GFP-TRIM56 or TRIM56-GFP vector. Two days later, cells were fixed in 4% paraformaldehyde in phosphate-buffered saline (PBS) for 20 min, permeabilized with 0.25% Triton X-100 for 10 min, and subsequently incubated with anti-Myc MAb and washed in PBS, followed by incubation with Texas Red-conjugated anti-mouse IgG (Southern Biotech). Slides were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and examined under a Zeiss LSM-510 laser scanning confocal microscope within the Department of Microbiology, Immunology and Biochemistry, University of Tennessee Health Science Center.
RESULTS
Identification of TRIM56 as a cellular protein that associates with BVDV Npro.
To explore the mechanism of the pestivirus Npro-mediated proteasomal degradation of IRF3, we studied cellular proteins associated with BVDV Npro in HeLa-FHNpro cells that stably express Flag- and HA-tandem-tagged BVDV Npro (FH-Npro) (36). As described previously (36), these cells expressed high levels of FH-Npro and had no detectable IRF3 protein compared with control HeLa cells (Fig. 1A, compare lanes 1 and 3). Treatment with epoxomicin, a potent inhibitor of the proteasome, stabilized the IRF3 protein in HeLa-FHNpro cells (Fig. 1A, compare lanes 2 and 1). We purified Npro-containing complexes by immunoprecipitation (IP) with anti-Flag M2 monoclonal antibody from HeLa-FHNpro cells and, as a negative control, from parental HeLa cells. The immunoprecipitated materials were eluted from the matrix with the Flag peptide, fractionated on an SDS-PAGE gel, and visualized by Coomassie blue staining (Fig. 1B). Proteins specifically associated with FH-Npro (Fig. 1B, lane 3) were excised, and their identities were determined by mass spectrometry (see Table S1 in the supplemental material). In parallel, only a few polypeptides were purified from control HeLa cells by anti-Flag IP (Fig. 1B, lane 2), and none were found to overlap with those proteins coimmunoprecipitated with FH-Npro, as determined by mass spectrometry (data not shown). The cellular proteins associated with FH-Npro included TRIM56, cytoskeleton proteins (β-actin and β-tubulin), chaperone proteins (hsp27 and hsc70), KIAA0913, and DNA-PK, among many others (Fig. 1B and Table S1). Because the TRIM family proteins were proposed to represent a class of “single-protein RING finger” E3 ubiquitin ligases (18), we hypothesized that TRIM56 may be involved in the interplay between Npro and IRF3, and we thus focused our studies on characterizing the functions of TRIM56.
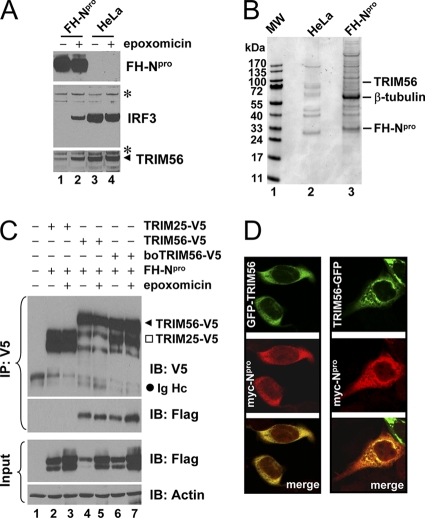
Fig. 1.
Identification of TRIM56 as a protein interaction partner of BVDV Npro. (A) Immunoblot analysis of Flag- and HA-tandem-tagged Npro (FHNpro) (using anti-Flag antibody), IRF3, and TRIM56 expression in HeLa-FHNpro (lanes 1 and 2) and parental HeLa (lanes 3 and 4) cells. Where indicated, cells were treated with epoxomicin (100 nM) overnight prior to cell lysis. Asterisks indicate nonspecific bands, which demonstrate equal sample loading. (B) Protein complexes were purified from control HeLa and HeLa-FHNpro cells by anti-Flag affinity purification and fractionated on SDS-PAGE gels, followed by Coomassie blue staining. The protein bands for TRIM56, β-tubulin, and FH-Npro (identified later by mass spectrometry) are indicated on the right. (C) Co-IP analysis of the interaction of FH-Npro with V5-tagged human TRIM56 (TRIM56) and bovine TRIM56 (boTRIM56) in cotransfected 293FT cells. TRIM25-V5 was used as a negative control for the specificity of the Npro-TRIM56 interaction. Where indicated, cells were treated with epoxomicin overnight prior to cell lysis. Note that epoxomicin had no effect on the Npro-TRIM56/boTRIM56 associations. The input panel shows the immunoblotting of 1/10 of the whole-cell lysates used for IP. Note that ectopically expressed FH-Npro was detected as a doublet in immunoblot analyses of the whole-cell lysates. The upper band may represent monoubiquitinated FH-Npro. IB, immunoblot; Ig Hc, IgG heavy chain. (D) HeLaNpro-25 cells induced for Npro expression were transiently transfected with a vector encoding GFP-TRIM56 (left) or TRIM56-GFP (right) and subsequently fixed for immunostaining of Npro (using anti-Myc antibody). The subcellular localizations of GFP-tagged TRIM56 and Myc-Npro were examined by confocal microscopy.
To confirm the TRIM56-Npro interaction, we performed co-IP experiments in 293FT cells transiently coexpressing V5-tagged TRIM56 and FH-Npro. Cells coexpressing V5-tagged TRIM25 and FH-Npro were used as a control for specificity (Fig. 1C). After IP with anti-V5, FH-Npro was found to form a complex with TRIM56-V5 (regardless of whether TRIM56 was of human or bovine origin) but not with TRIM25-V5. The inhibition of the proteasome had little effect on the association of Npro with TRIM56. To examine whether Npro colocalizes with TRIM56, we transiently transfected GFP-TRIM56 or TRIM56-GFP constructs (in which GFP was tagged at the N and C termini of TRIM56, respectively) into HeLaNpro-25 cells stably expressing Myc-tagged Npro (5) and examined the subcellular localizations of Npro and TRIM56 by fluorescence confocal microscopy (Fig. 1D). While Myc-Npro resided in both the cytoplasm and nucleus, GFP-tagged TRIM56 was distributed exclusively in the cytoplasm. Importantly, Myc-Npro colocalized with GFP-TRIM56 and TRIM56-GFP extensively in the cytoplasm. Collectively, these data corroborate our earlier finding that TRIM56 is an interaction partner of BVDV Npro.
The C-terminal portion of the TRIM56 protein mediates the interaction with Npro.
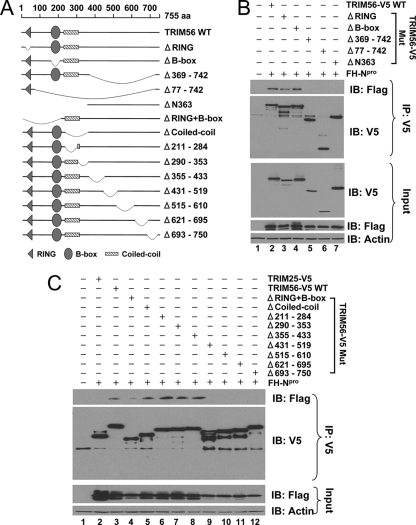
Both the human and bovine TRIM56 proteins comprise 755 amino acids (aa), with 87% amino acid identity between them. Like other TRIM family proteins, TRIM56 from either species has the N-terminal signature tripartite motifs, i.e., the RING, B-box, and coiled-coil domains. However, neither human nor bovine TRIM56 has sequence homology to any known domain structures in its C-terminal portion (Fig. 2A, and see Fig. S1 in the supplemental material). Thus, TRIM56 belongs to the C-V subfamily of TRIMs (21, 29). To determine which part of TRIM56 mediates its interaction with Npro, we generated an array of human TRIM56 deletion mutants (Fig. 2A) and determined their abilities to interact with Npro by co-IP (Fig. 2B and C). We found that the deletion of the C-terminal half of TRIM56 (Δ369-742) (Fig. 2B, lane 5) abrogated the ability of TRIM56 to interact with Npro, while the deletion of RING (Fig. 2B, lane 3), the B box (Fig. 2B, lane 4), or both (Fig. 2C, lane 4) or the coiled-coil domain (Fig. 2C, lanes 5 and 6) failed to do so. To determine which part of the C-terminal sequence of TRIM56 is critical for its association with Npro, we conducted a more refined internal deletion analysis of TRIM56 starting from aa 290, producing 6 mutants, with each one removing approximately 60 to 90 aa toward the C terminus (Δ290-353, Δ355-433, Δ431-519, Δ515-610, Δ621-695, and Δ693-750) (Fig. 2A). Co-IP analysis demonstrated that the region at aa 431 to 750 of TRIM56 mediates the association with Npro, as a deletion of any part of this region of as few as 57 aa abrogated the TRIM56-Npro interaction (Fig. 2C, lanes 9 to 12). We conclude from these experiments that the integrity of the C-terminal region of TRIM56 is critical for its association with pestivirus Npro. It should be noted that a large N-terminal deletion mutant (ΔN363) (Fig. 2B, lane 7) in which the tripartite motif was completely deleted also no longer associated with Npro. We speculate that this large N-terminal deletion may affect the overall folding of the TRIM56 protein, thereby abrogating the TRIM56-Npro interaction.
Fig. 2.
The C-terminal portion of TRIM56 is important for its association with Npro. (A) Schematic representation of human TRIM56 protein domains and the individual TRIM56 deletion mutants tested in this study. (B and C) Co-IP analysis of the association of FH-Npro with V5-tagged, WT TRIM56 or the indicated TRIM56 mutant in cotransfected 293FT cells. In C, TRIM25-V5 was used as a negative control for the interaction with FH-Npro.
TRIM56 has RING-dependent E3 ubiquitin ligase activity and self-associates in cells.
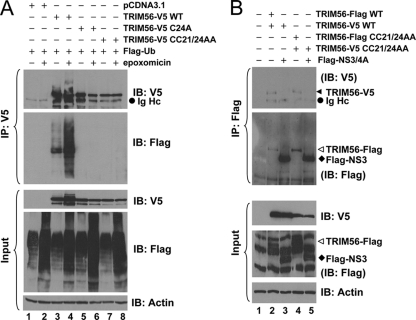
Many but not all of the TRIM proteins have intrinsic E3 ubiquitin ligase activity, which is dependent on the N-terminal RING domain and characterized by the ability to mediate the transfer of ubiquitin both to heterologous substrates and to themselves (18). When coexpressed with Flag-Ub, TRIM56-V5 was expressed as a smear of high-molecular-mass proteins that reacted with anti-Flag in addition to a product of TRIM56's predicted mass (Fig. 3A, lane 3), a phenomenon which was enhanced following the inhibition of the proteasome by epoxomicin (lane 4). This suggests that TRIM56 promotes the ubiquitination of itself. Mutations of the conserved cysteine residues within the RING domain of TRIM56 completely abrogated its ability to induce self-ubiquitination (Fig. 3A, lanes 5 and 6 [Cys 24 to Ala] and lanes 7 and 8 [both Cys 21 and Cys 24 to Ala]), confirming that the E3 ubiquitin ligase activity of TRIM56 depends on its RING domain.
Fig. 3.
TRIM56 is a RING-type E3 Ub ligase and self-associates. (A) HEK293 cells were cotransfected with Flag-Ub, pcDNA3.1, and WT or C24A or CC21/24AA mutant TRIM56-V5 and mock treated or treated with 50 nM epoxomicin. (Top) Cell lysates were subjected to IP with anti-V5 MAb, followed by immunoblot analysis with anti-V5 MAb and anti-Flag M2. (Bottom) Immunoblot analysis of whole-cell lysates (input) for WT and C24A and CC21/24AA mutant TRIM56-V5 (using MAb anti-V5 antibody), Flag-Ub (using rabbit anti-Flag), and actin. (B) Co-IP analysis of the interactions of TRIM56-Flag with TRIM56-V5 (lane 2), TRIM56-V5 with Flag-NS3/4A (lane 3, as a negative control), CC21/24AA TRIM56-Flag with CC21/24AA TRIM56-V5 (lane 4), and CC21/24AA TRIM56-V5 with Flag-NS3/4A (lane 5, as a negative control) in cotransfected 293FT cells. (Top) Cell lysates were immunoprecipitated with anti-Flag, followed by immunoblot analysis with anti-V5 MAb and anti-Flag MAb. (Bottom) Immunoblot analysis of whole-cell lysates (input) for WT and CC21/24AA mutant TRIM56-V5 (using anti-V5 MAb), WT and CC21/24AA mutant TRIM56-Flag, Flag-NS3/4A (using rabbit anti-Flag), and actin.
The TRIM proteins are known to homomultimerize through their coiled-coil domains, thereby identifying specific cellular compartments (24). We found that TRIM56-V5 associated with TRIM56-Flag in co-IP experiments regardless of whether the RING domain was intact (WT) or mutated (CC21/24AA) (Fig. 3B, lanes 2 and 4). As a control for specificity, TRIM56-V5 (WT or CC21/24AA) did not form a complex with Flag-tagged NS3/4A from HCV (Fig. 3B, lanes 3 and 5). These data indicate that like other TRIMs, TRIM56 also oligomerizes, and such activity is separated from its E3 ubiquitin ligase activity.
TRIM56 does not mediate Npro-induced IRF3 loss, nor does it target Npro for proteasomal degradation.
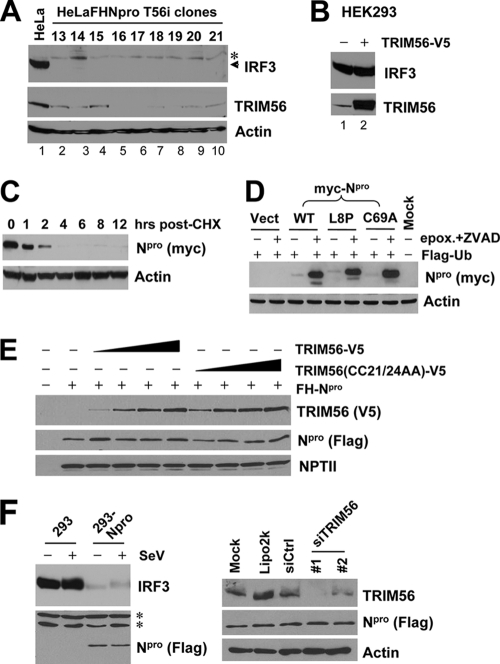
Because TRIM56 is a RING-type E3 ligase associated with Npro, we set out to determine whether TRIM56 is involved in the Npro-mediated proteasomal degradation of IRF3. To test this, we stably transfected a TRIM56 shRNA into in HeLa-FHNpro cells and analyzed the IRF3 protein abundance in a number of cell clones in which the expression of endogenous TRIM56 was downregulated to various extents (Fig. 4A, lanes 2 to 10). Regardless of the TRIM56 protein levels (low to undetectable) in these cell clones, the IRF3 protein remained undetectable (Fig. 4A), suggesting that TRIM56 is not essential for Npro-induced IRF3 degradation. Consistent with this, overexpression of TRIM56 in HEK293 cells did not reduce the IRF3 protein abundance (Fig. 4B).
Fig. 4.
TRIM56 does not regulate the protein abundances of IRF3 and BVDV Npro. (A) Knockdown of TRIM56 does not reverse Npro-induced IRF3 degradation. Data shown are for immunoblot analyses of IRF3, TRIM56, and actin in control HeLa cells (lane 1) and HeLa-FHNpro-derived cell clones stably transfected with a TRIM56 shRNA (lanes 2 to 10). The asterisk indicates a nonspecific band. (B) Overexpression of TRIM56 does not reduce IRF3 protein levels in transiently transfected HEK293 cells. (C) Immunoblot analysis of Npro and actin in HeLaNpro-25 cells induced for Npro expression following treatment with cycloheximide (CHX) (75 μg/ml) for the indicated times. (D) Immunoblot analysis of Npro and actin expression in 293FT cells cotransfected with Flag-Ub and Myc-tagged WT or L8P or C69A mutant Npro. Where indicated, cells were treated with epoxomicin (50 nM) plus a pancaspase inhibitor, ZVAD (40 μM). (E) Overexpression of WT or mutant TRIM56 does not regulate the protein level of cotransfected FH-Npro in HEK293 cells. The empty pcDNA3.1 vector was supplemented to keep the total DNA transfected constant. An immunoblot of neomycin phosphotransferase II (NPTII) is shown to demonstrate equal transfection efficiency and sample loading. (F) Knockdown of endogenous TRIM56 does not regulate Npro protein abundance in HEK293 cells stably expressing Flag-Npro (293-Npro). (Left) Immunoblot analysis of IRF3 and Npro in control HEK293 and 293-Npro cells (mock infected or infected with SeV). (Right) Immunoblot analysis of TRIM56, Npro, and actin expression in 293-Npro cells mock transfected or transfected with Lipofectamine alone or Lipofectamine complexed with a negative-control siRNA or with individual siRNAs (siRNAs 1 and 2) specifically targeting TRIM56.
BVDV Npro is a short-lived protein with a half-life of less than 1 h (Fig. 4C), a phenomenon similar to what was recently described for Npro from classical swine fever virus (27). The inhibition of the proteasome by epoxomicin substantially stabilized the ectopically expressed Npro protein regardless of whether Npro was of the WT sequence or bore mutations that abrogated the ability of Npro to degrade IRF3 (L8P and C69A) (5) (Fig. 4D). We considered the possibility that TRIM56 may interact with and promote Npro degradation through the proteasome. However, overexpression of TRIM56 did not reduce the protein abundance of coexpressed Npro (Fig. 4E), nor did it enhance Npro polyubiquitination (data not shown). Furthermore, silencing of endogenous TRIM56 expression did not upregulate the Npro protein level in HEK293 cells stably expressing Flag-tagged Npro (293-Npro) (Fig. 4F). Collectively, we conclude that TRIM56 is not the E3 ubiquitin ligase recruited by Npro to degrade IRF3, nor does it contribute to Npro protein turnover by the proteasome.
Ectopically expressed TRIM56 exerts antiviral activity against BVDV replication by means of its E3 ligase activity.
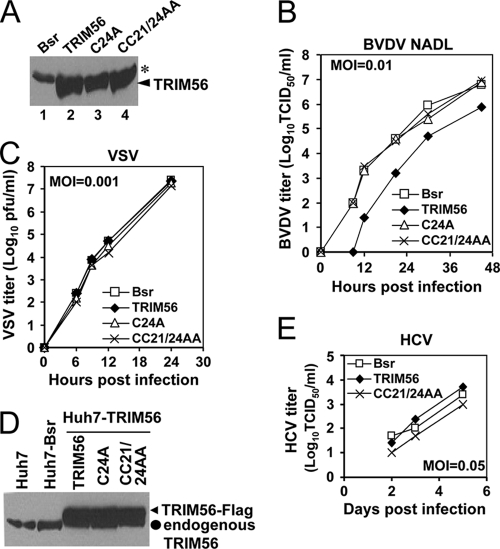
Although we did not find TRIM56 to be involved in regulating the protein abundances of IRF3 and Npro, its association with a BVDV protein (Npro) prompted us to determine whether TRIM56 has any effect on BVDV replication. To this end, we generated MDBK cells stably expressing WT or the E3 ligase-deficient RING mutant form (C24A and CC21/24AA) of human TRIM56 or the empty vector (Bsr). Immunoblot analysis demonstrated that the WT and mutant TRIM56 proteins in these cells were expressed at comparable levels (Fig. 5A, lanes 2 to 4). When infected with the cytopathic (cp) NADL strain of BVDV (MOI of 0.01), cells expressing WT TRIM56 consistently released 1 to 1.5 logs fewer infectious progeny virus into culture medium than did Bsr cells during the 48-h observation period, while the virus yields were undistinguishable between Bsr cells and cells expressing the C24A or CC21/24AA mutant TRIM56 protein (Fig. 5B). In agreement with this, WT TRIM56-expressing MDBK cells consistently had substantially fewer cytopathic effects (CPE) and better cell survival than did Bsr cells upon infection with cp BVDV NADL at increasing MOIs (see Fig. 8C). These data suggest that TRIM56 is an antiviral cellular factor against BVDV infection and that the anti-BVDV activity of TRIM56 depends on its E3 ligase activity.
Fig. 5.
Ectopically expressed TRIM56 restricts BVDV replication in bovine kidney (MDBK) cells by means of its E3 Ub ligase activity. (A) Immunoblot analysis of ectopically expressed TRIM56 in MDBK cells stably expressing a control vector (Bsr), WT TRIM56, and RING mutant TRIM56 (C24A and CC21/24AA). The asterisk indicates a nonspecific band that demonstrates equal sample loading. (B) Progeny virus production in culture supernatants of MDBK-Bsr, -TRIM56, -C24A, and -CC21/24AA cells at various time points postinfection with BVDV NADL (MOI of 0.01). Data shown are representative of three independently conducted experiments. (C) Progeny virus production in culture supernatants of MDBK-Bsr, -TRIM56, -C24A, and -CC21/24AA cells at various time points postinfection with VSV (MOI of 0.001). Data shown are representative of two independently conducted experiments. (D) Immunoblot analysis of TRIM56 expression (using anti-TRIM56) in parental Huh7 cells and Huh7 cells stably transduced with the control vector (Bsr), WT TRIM56 (TRIM56), or the TRIM56 RING mutants (C24A or CC21/24AA). (E) Progeny virus production in culture supernatants of Huh7-Bsr, -TRIM56, and -CC21/24AA cells at various time points postinfection with HCV JFH1 (MOI of 0.05). At day 1, the infectious progeny virus titers were all below the detection limit (5 TCID50/ml). Data shown are representative of two independently conducted experiments.
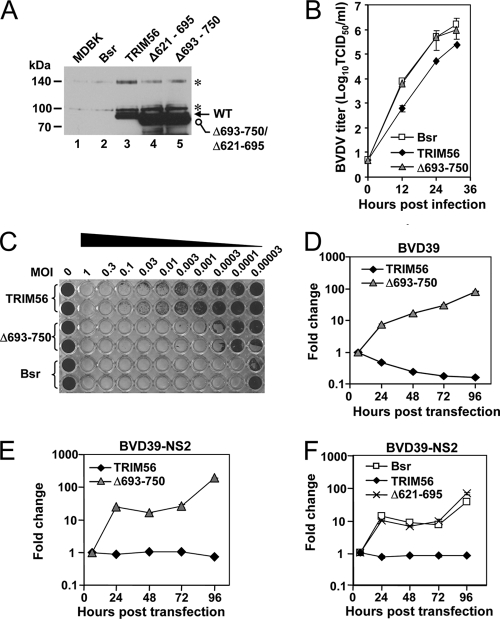
Fig. 8.
The C-terminal structural integrity is important for TRIM56's antiviral function against BVDV. (A) Immunoblot detection of ectopically expressed TRIM56 in parental MDBK cells (lane 1) and MDBK cells stably expressing the Bsr vector (lane 2), WT TRIM56 (lane 3), or the Δ621-695 (lane 4) and Δ693-750 (lane 5) mutant TRIM56 proteins. Asterisks indicate nonspecific bands. (B) Progeny BVDV titers in culture supernatants of MDBK-Bsr, MDBK-TRIM56, and MDBK-Δ693-750 mutant cells at various time points postinfection with cp BVDV NADL (MOI of 0.005). Data shown are representative of three independent experiments. (C) Duplicate wells of MDBK-TRIM56, MDBK-Δ693-750 mutant, and MDBK-Bsr cells grown in a 96-well plate were infected with increasing MOIs (from right to left, MOIs of 0.00003 through 1) of the cp BVDV NADL for 70 h prior to cell fixation and crystal violet staining. The very left column shows uninfected cells. The image shown is representative of two independent experiments. (D and E) Replication of the BVD39 (D) and BVD39-NS2 (E) replicons in MDBK-TRIM56 and MDBK-Δ693-750 mutant cells. Data shown are representative of three independent experiments. (F) Replication of the BVD39-NS2 replicon in MDBK-Bsr, MDBK-TRIM56, and MDBK-Δ621-695 mutant cells. Data shown are representative of three independent experiments.
TRIM25 positively regulates RIG-I signaling via the ubiquitination of the CARD domain of RIG-I (9), thereby enhancing the type I IFN antiviral response. We considered the possibility that the antiviral effect of TRIM56 may result from a general augmentation of the IFN response. To investigate this possibility, we studied the effects of TRIM56 overexpression on viral activation of the IFN-β promoter and on the replication of VSV, a rhabdovirus extremely sensitive to IFN′s antiviral action. We found that Sendai virus (SeV) infection strongly activated the IFN-β promoter in HEK293 cells overexpressing WT TRIM56, as it did to a similar extent in control HEK293 cells and in cells overexpressing the E3 ligase-null CC21/24AA mutant TRIM56 protein (data not shown). VSV replicated with similar efficiencies among MDBK cells expressing WT, C24A, and CC21/24AA TRIM56 proteins and the Bsr vector (Fig. 5C), in contrast to the compromised BVDV propagation selectively in cells expressing WT TRIM56 (Fig. 5B). We further studied the effect of TRIM56 overexpression on the replication of HCV, a flavivirus that is closely related to BVDV. The stable, ectopic expression of the WT or the CC21/24AA RING mutant form of TRIM56 in HCV-permissive hepatoma Huh7 cells (Fig. 5D) had little effect on the propagation of cell culture-derived HCV JFH1 (Fig. 5E), indicating that TRIM56 does not restrict HCV replication. Taken collectively, the TRIM56 E3 ubiquitin ligase specifically inhibits BVDV infection, and such an antiviral effect is not attributed to a general augmentation of the IFN antiviral response.
TRIM56 inhibits BVDV replication by targeting an intracellular viral RNA replication step.
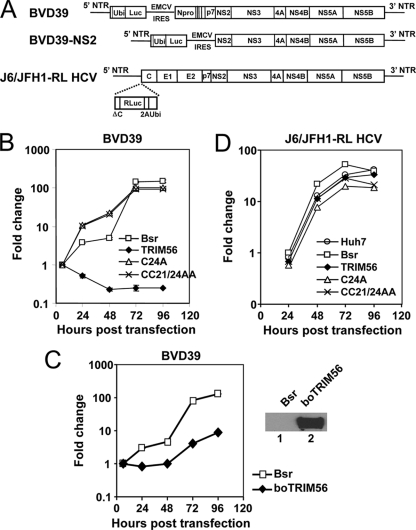
To determine how TRIM56 restricts BVDV infection, we investigated the impact of TRIM56 on the replication of a bicistronic, subgenomic BVDV RNA replicon, BVD39 (Fig. 6A, top). This replicon RNA self-replicates continuously in the cytoplasm upon transfection into MDBK cells, expressing luciferase as a measure of intracellular BVDV RNA replication (15). The BVD39 replicon replicated efficiently in control MDBK-Bsr cells as well as in MBDK cells stably expressing the C24A or CC21/24AA mutant TRIM56 protein, resulting in an increase in luciferase activity over 72 h, followed by a plateau later. In contrast, the replication of this BVDV RNA was blunted in cells expressing WT TRIM56 (Fig. 6B). The stable, ectopic expression of boTRIM56 also substantially reduced cellular permissiveness for the BVD39 replicon compared with MDBK-Bsr cells (Fig. 6C), confirming that boTRIM56, like its human homolog, also suppresses BVDV RNA replication when ectopically expressed. In contrast, ectopically expressed TRIM56 (WT or the C24A and C21/24AA mutants) had no demonstrable effect on the replication of a genome-length J6/JFH1 HCV RNA replicon encoding Renilla luciferase (J6/JFH1-RL) (Fig. 6A) in HCV-permissive Huh7 cells (Fig. 6D). These results suggest that TRIM56 specifically restricts BVDV infection by targeting an intracellular BVDV RNA replication step.
Fig. 6.
TRIM56 inhibits BVDV infection by targeting intracellular viral RNA replication. (A) Schematic representation of the subgenomic ncpBVDV RNA replicons encoding firefly luciferase, BVD39 (which encodes Npro) (15) and BVD39-NS2 (which does not encode Npro), and the genome-length J6/JFH1-RL HCV replicon encoding Renilla luciferase (32). NTR, nontranslated region. (B) Replication of the BVD39 replicon in transfected MDBK-Bsr, -TRIM56, -C24A, and -CC21/24AA cells. Data shown represent the fold increases in luciferase activity over that present at 6 h (which represents the direct translation of input replicon RNA) in each cell type and are representative of two independently conducted experiments. (C, left) Replication of the BVD39 replicon in MDBK-Bsr cells and MDBK-boTRIM56 cells stably overexpressing boTRIM56. Data shown are representative of two independent experiments. (Right) Immunoblot analysis of ectopically expressed boTRIM56 (using anti-Flag antibody) in MDBK-boTRIM56 cells (lane 2). MDBK-Bsr cells (lane 1) served as a negative control. (D) Replication of the J6/JFH1-RL HCV replicon in transfected Huh7 and Huh7-Bsr, -TRIM56, -C24A, and -CC21/24AA cells. Data shown represent the fold increases in luciferase activity over that present at 6 h in each cell type and are representative of two independent experiments.
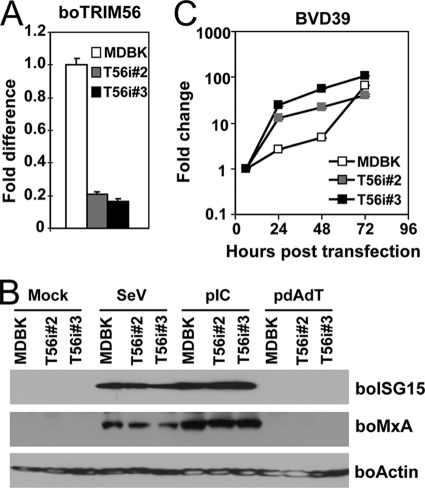
The endogenous bovine TRIM56 restricts BVDV replication.
Next, we determined whether endogenous boTRIM56, expressed at physiological levels, also restricts BVDV replication. We stably transfected MDBK cells with an shRNA construct that targets specifically boTRIM56. Because neither the commercially available TRIM56 antibodies nor our custom-made one could detect the endogenous boTRIM56 protein (data not shown), we utilized real-time RT-PCR to screen MDBK cell clones with the efficient knockdown of boTRIM56 mRNA. Two individual clones, designated T56i#2 and T56i#3, were selected for further analysis because they expressed only ∼20% TRIM56 mRNA compared with the parental MDBK cells (Fig. 7A). Both T56i cell clones responded similarly to SeV infection or transfected poly(I:C) in the induction of two well-known IFN-stimulated genes (ISGs), ISG15 and MxA, compared with control MDBK cells (Fig. 7B), indicating that the knockdown of TRIM56 did not impair the type I IFN response in these cells. We found that both T56i#2 and T56i#3 cells were substantially more permissive than control MDBK cells in supporting the replication of the BVD39 replicon, allowing earlier, more efficient BVDV RNA replication than the latter (Fig. 7C). Taken together, these data confirm the biological relevance of TRIM56's antiviral activity against BVDV.
Fig. 7.
The endogenous bovine TRIM56 restricts BVDV replication in MDBK cells. (A) Real-time RT-PCR analysis of boTRIM56 mRNA expression in parental MDBK cells and two MDBK cell clones stably transfected with a boTRIM56 shRNA (T56i#2 and T56i#3). (B) Effect of the stable knockdown of boTRIM56 in MDBK cells on viral induction of the type I IFN response. Parental MDBK and MDBK-T56i #2 and -T56i#3 cells were mock treated, infected with SeV, or transfected with poly(I:C) (pIC) or poly(dA:dT) (pdAdT). Cells were lysed for immunoblot analysis of bovine ISG15, MxA, and actin expression. Note that MDBK cells did not respond to transfected poly(dA:dT) to induce ISG15 and MxA expression. (C) Replication of the BVD39 replicon in MDBK and MDBK-T56i #2 and -T56i#3 cells. Data shown are representative of two independent experiments.
The integrity of the C-terminal region of TRIM56, but not the TRIM56-Npro interaction, is critical for TRIM56's antiviral function.
TRIM5α depends on its C-terminal B30.2/SPRY domain for its antiviral activity against HIV (30). Although the C-terminal region of TRIM56 does not have known domain structures, it mediates the interaction of TRIM56 with Npro (Fig. 2), suggesting that this region is critical for TRIM56's ability to interact with viral and, possibly, cellular proteins. We determined whether the C-terminal portion of TRIM56 is also critical for its antiviral function. To this end, we generated MDBK cells stably expressing a mutant TRIM56 protein lacking aa 693 to 750 at the C terminus (Fig. 8A, lane 5). Although its expression level was higher than that of WT TRIM56, this TRIM56 mutant was no longer able to inhibit BVDV propagation (Fig. 8B) or the replication of the BVD39 RNA replicon (Fig. 8D). It also failed to protect cells from BVDV-induced CPE (Fig. 8C). Since the Δ693-750 mutant also lost its ability to interact with Npro (Fig. 2C, lane 12), we wondered whether the TRIM56-Npro interaction determines TRIM56's antiviral activity against BVDV. To investigate this, we determined how TRIM56 affected the replication of a BVDV RNA replicon that does not encode Npro, BVD39-NS2 (Fig. 6A, middle). We found that WT TRIM56 still restricted the replication of the BVD39-NS2 replicon, while the Δ693-750 mutant failed to do so (Fig. 8E). Similarly, we found a TRIM56 mutant lacking aa 621 to 695 (in which the deletion was close to the C terminus but upstream of the Δ693-750 deletion) also lost the antiviral activity against the BVD39-NS2 replicon when stably expressed in MDBK cells (Fig. 8F; see Fig. 8A, lane 4, for a characterization of its expression). We conclude from these experiments that the C-terminal structural integrity, but not the TRIM56-Npro interaction, is important for the antiviral activity of TRIM56 against BVDV.
TRIM56 does not promote the degradation of BVDV NS proteins required for viral RNA replication.
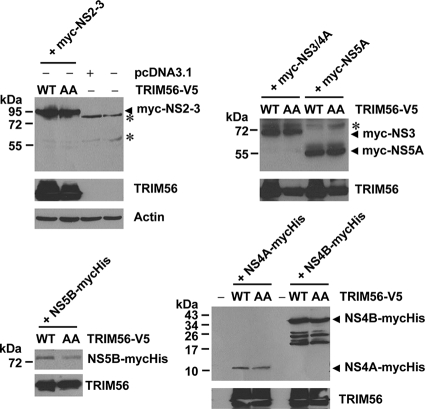
Because our data suggested that the E3 ubiquitin ligase activity of TRIM56 was required for its antiviral effect against BVDV RNA replication, we considered the possibility that TRIM56 may target one or more BVDV NS proteins required for BVDV RNA replication for degradation. To test this hypothesis, the individual BVDV NS proteins were ectopically coexpressed with excessive WT TRIM56 or the E3 ligase-deficient CC21/24AA mutant TRIM56 protein (at a ratio of 1:6) in 293FT cells, and their protein abundances were determined by immunoblot analysis (Fig. 9). We found that the BVDV NS2-3, NS3, NS4A, NS4B, NS5A, and NS5B proteins were expressed at comparable levels in cells coexpressing WT or CC21/24AA mutant TRIM56, suggesting that they are not targeted by TRIM56 E3 ligase for degradation. Therefore, another mechanism(s) of action is responsible for the antiviral effect of TRIM56 on BVDV.
Fig. 9.
TRIM56 does not promote the degradation of BVDV NS proteins. 293FT cells grown in 6-well plates were mock transfected or cotransfected with 0.25 μg of the indicated Myc- or Myc-6×His-tagged ncpBVDV NS protein construct and 1.5 μg of the V5-tagged TRIM56 (WT or the CC21/24AA mutant [AA]) vector. Cells were lysed and subjected to immunoblot analysis of the expression of BVDV NS proteins (using anti-Myc antibody) and TRIM56 (using anti-V5 antibody). Asterisks indicate nonspecific bands.
TRIM56 is a ubiquitous protein whose expression is further upregulated by IFN or virus infection.
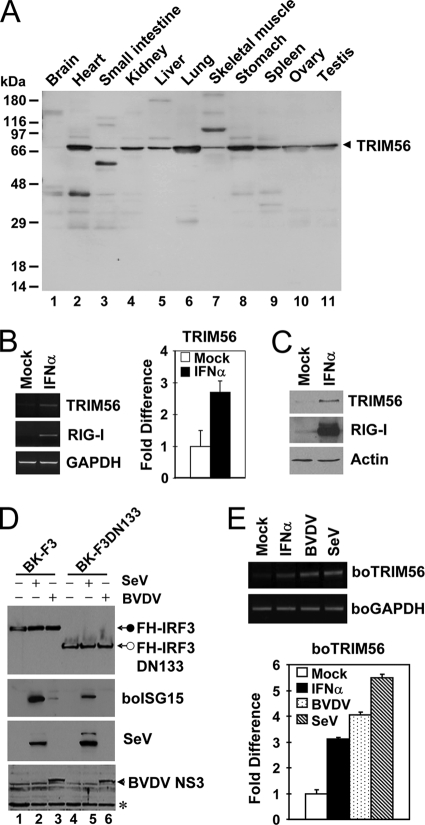
To determine the tissue distribution of TRIM56, we assessed the expression of human TRIM56 protein by multiple-tissue Western blotting (Fig. 10A). We found that TRIM56 was expressed ubiquitously in all human tissues examined, including brain, heart, small intestine, kidney, liver, lung, skeletal muscle, stomach, spleen, ovary, and testis. In all tissues, TRIM56 was expressed mainly as a protein of approximately 81 kDa, which corresponds to the predicted molecular mass of human TRIM56 (Fig. 10A, arrowhead). Of note, lung and stomach were among the organs with the highest levels of expression of TRIM56, while the brain expressed the lowest but nonetheless detectable levels of the TRIM56 protein.
Fig. 10.
TRIM56 is ubiquitously expressed, and its expression is further upregulated by virus or IFN-α. (A) Expression of the TRIM56 protein in various human tissues detected by anti-TRIM56 antibody using Human Tissues INSTA-Blot (Calbiochem). (B) Semiquantitative RT-PCR (left) and real-time PCR (right) assessments of TRIM56 mRNA abundance in HeLa cells mock treated or stimulated with 500 U/ml of IFN-α for 16 h. RIG-I was used as a positive control for IFN treatment (left). (C) Immunoblot analysis of TRIM56 and RIG-I expression in THP1 cells mock treated or stimulated with IFN-α. Similar results were obtained with HeLa cells (data not shown). (D) Stable retroviral transduction of IRF3, but not that of IRF3 with an N-terminal 133-aa deletion (DN133), renders MDBK cells able to upregulate boISG15 expression following BVDV infection. Cells were mock treated (lanes 1 and 4) or infected with SeV (lanes 2 and 5) or with BVDV (lanes 3 and 6) for 12 h prior to cell lysis and immunoblot analysis of the ectopically expressed, Flag- and HA-tandem-tagged human IRF3 (FH-IRF3) or IRF3 DN133 (FH-IRF3DN133) (using anti-Flag antibody), bovine ISG15, SeV, and BVDV NS3. The asterisk indicates a nonspecific band, which demonstrates equal protein sample loading. (E) Semiquantitative RT-PCR (top) and real-time RT-PCR (bottom) analyses of boTRIM56 mRNA abundance in BK-F3 cells mock treated, stimulated with 500 U/ml of IFN-α, or infected with cp BVDV NADL (MOI of 0.1) or SeV (50 HAU/ml) for 16 h.
Many TRIM proteins are transcriptionally regulated by IFNs (3, 23). We found that both the mRNA (Fig. 10B) and protein (Fig. 10C) levels of human TRIM56 were moderately upregulated by IFN-α (∼3-fold). IFN also upregulated boTRIM56 transcripts to a similar extent in bovine cells (Fig. 10E, solid bar). To determine whether TRIM56 is induced by viruses, in particular BVDV, we studied TRIM56 expression in MDBK cells stably overexpressing Flag- and HA-tandem-tagged human IRF3 (BK-F3) (Fig. 10D, lanes 1 to 3). In BK-F3 cells, the IRF3 protein was no longer degraded by Npro following BVDV infection because of its overexpression, allowing cells to mount an IFN response to BVDV infection, as evidenced by the induction of bovine ISG15 (Fig. 10D, lane 3). This phenotype, however, was not observed for BK-F3DN133 cells (Fig. 10D, lanes 4 to 6) stably overexpressing an IRF3 mutant lacking the N-terminal 133 aa (lane 6). We found that TRIM56 mRNA was induced by 4- and 5.5-fold upon infection with BVDV and SeV, respectively, in BK-F3 cells (Fig. 10E). Collectively, the ubiquitous tissue distribution of TRIM56 and its upregulation by IFN and viruses are consistent with the role of TRIM56 as an antiviral host factor.
DISCUSSION
The TRIM family proteins have recently been recognized to have emerging roles in innate immunity to viral infections (21). Among the over 60 TRIM proteins known to be encoded by the human and mouse genomes, only a few have been characterized for their antiviral activities, and most of them target viruses of the family Retroviridae at different stages of the viral life cycle (20, 21). In the current study we have identified TRIM56 as a novel player in antiviral innate immunity, which acts specifically to inhibit the replication of BVDV, a positive-stranded RNA virus of the genus Pestivirus within the family Flaviviridae. We demonstrate that TRIM56 possesses RING-dependent E3 ubiquitin ligase activity and self-associates (Fig. 3), features demonstrated previously for other TRIM family E3 ligases (18, 24). When ectopically expressed, TRIM56 from either bovine or human species strongly and specifically suppressed BVDV replication (Fig. 5 and 6). Importantly, the knockdown of endogenous TRIM56 rendered bovine cells substantially more permissive to BVDV replication (Fig. 7), confirming that the physiological level of TRIM56 is able to execute the antiviral function. Furthermore, we have shown that both the E3 ubiquitin ligase activity and the integrity of the C-terminal part of TRIM56 are crucial for its antiviral activity against BVDV, as the mutation of the conserved Cys residue(s) within the RING domain or the deletion of as few as 57 aa in the C-terminal region abrogated TRIM56's restriction of BVDV replication (Fig. 5, 6, and 8). To our knowledge, this is the first study that demonstrates that the TRIM56 E3 ubiquitin ligase inhibits the replication of an RNA virus.
TRIM25 was recently shown to promote IFN induction by the ubiquitination of RIG-I (9), demonstrating that TRIM proteins can participate in innate immune signaling that nonspecifically augments antiviral responses. However, several lines of evidence suggest that the antiviral activity of TRIM56 against BVDV does not result from a general enhancement of the IFN antiviral response. First, the ectopic expression of TRIM56 by itself did not enhance the SeV-induced IFN-β promoter, which activates the RIG-I pathway (39), in our experiments (data not shown). Because flaviviruses, including HCV, which is a virus closely related to BVDV, are sensed by RIG-I in host cells to trigger IFN induction (13, 26), BVDV is likely to be recognized by RIG-I. Second, although MDBK cells overexpressing TRIM56 were substantially less permissive for BVDV replication than were cells expressing the control vector or the E3 ligase-deficient TRIM56 mutants (Fig. 5B and 6B), no difference was seen among these cells with regard to their ability to support the propagation of VSV (Fig. 5C), a virus routinely used for IFN antiviral activity assays. Third, the ectopic expression of TRIM56 or its RING mutants in HCV-permissive Huh7 cells had no appreciable effect on the replication of HCV (Fig. 5E and 6D), which is sensitive to the action of IFN in cell culture (17), indicating that the antiviral activity of TRIM56 is virus specific. Fourth, the stable knockdown of endogenous boTRIM56 in MDBK cells did not apparently affect the SeV- or transfected poly(I:C)-induced expression of ISG15 and MxA (Fig. 7B), suggesting that the enhanced permissiveness for BVDV replication in the boTRIM56 knockdown MDBK cells (Fig. 7C) did not result from an impairment of the induction of the IFN response. Last, BVDV infection of MDBK cells does not induce a detectable IFN response because of the rapid degradation of IRF3 by Npro (5).
While this paper was in preparation, Tsuchida and colleagues reported the identification of mouse TRIM56 as a regulator of double-stranded DNA (dsDNA)-mediated type I IFN induction by screening a mouse cDNA library for genes that enhance intracellular dsDNA [poly(dA:dT)]-activated IFN-β promoter activity (33). Those authors showed that TRIM56 interacted with and promoted the ubiquitination of STING (also known as MITA/ERIS/MYPS), an adaptor protein essential for the dsDNA induction of IFN (12, 31, 40). In that study, the transient knockdown of TRIM56 in HEK293 cells greatly reduced dsDNA-induced IFN induction and, to a less extent, attenuated transfected poly(I:C)-induced responses. We did not observe much of an effect of the stable knockdown of TRIM56 in MDBK cells on SeV- and poly(I:C)-induced ISG induction (Fig. 7B), indicating that there may be cell type-specific differences in the role of TRIM56 in the induction of the IFN response. Interestingly, MDBK cells failed to upregulate ISG15 and MxA expression following the transfection of dsDNA, although they responded robustly to SeV and transfected poly(I:C) (Fig. 7B).
Our data indicate that TRIM56 targets BVDV propagation by suppressing intracellular viral RNA replication, as it inhibited the replication of subgenomic BVDV RNA replicons that bypass the viral entry and uncoating steps and also did not lead to the packaging and release of progeny viruses (due to the lack of viral structural genes). However, we cannot exclude the possibility that TRIM56 may also act on these early and late steps of the BVDV life cycle, as demonstrated previously for other antiviral TRIMs in restricting retroviruses (20, 21). As in the case of HCV and other flaviviruses, pestiviruses replicate their RNAs on cytoplasmic membranes (25). We found that TRIM56 is expressed exclusively in the cytoplasm (Fig. 1D), a subcellular location which is consistent with TRIM56's action on BVDV RNA replication.
Previous studies of retroviral restriction factors suggested distinct antiviral mechanisms of TRIMs in restricting different retroviruses. TRIM22 depends crucially on its E3 ligase activity for disrupting HIV particle production (1). However, TRIM5α fully relies on its C-terminal SPRY domain but only partially on the E3 ligase activity for inhibiting HIV uncoating (30). In contrast, the B box, but not the E3 ligase activity or the SPRY domain, is essential for the restriction of murine leukemia virus release by TRIM15 (34). Our data show that TRIM56 depends on both the E3 ligase activity and C-terminal sequence integrity for its antiviral function against BVDV (Fig. 5, 6, and 8). However, when coexpressed with TRIM56, none of the NS proteins essential for BVDV RNA replication (NS2 to NS5B) demonstrated any reduction in abundance (Fig. 9), indicating that they are not targeted by the TRIM56 E3 ligase for degradation. Whether the TRIM56 E3 ligase promotes posttranslational modification(s) of one or more BVDV NS proteins and/or cellular factors to impair BVDV replication will require future investigation.
What is the role of the C-terminal region in the antiviral action of TRIM56? Although this portion of TRIM56 has no known domain structures, its integrity is crucial for TRIM56's ability to interact with Npro and also to confer BVDV restriction. However, Npro itself is not essential for BVDV replication (25), and the anti-BVDV activity of TRIM56 does not rely on the TRIM56-Npro interaction, since the replication of the BVD39-NS2 RNA replicon that does not encode Npro was still blunted upon TRIM56 overexpression (Fig. 8E and F). Conceivably, the TRIM56 C-terminal region may mediate critical protein-protein interactions between TRIM56 and cellular and/or viral proteins that determine viral fitness. Thus far, we have been unable to coimmunoprecipitate TRIM56 with BVDV NS2-3, NS3, NS4A, NS4B, NS5A, or NS5B (data not shown). Alternatively, the C-terminal part of TRIM56 may bind to BVDV RNA and regulate viral RNA replication directly. Of note, the SPRY domain, which is present in the C-terminal region of many TRIMs (although not in TRIM56), was proposed previously to bind RNA (22).
Examination of a wide variety of human tissues allowed us to demonstrate the ubiquitous expression of the TRIM56 protein (Fig. 10A). With the exception of brain, in which the TRIM56 level was low, constitutive expression of TRIM56 was observed for all other tissues examined. Of note, lung and stomach expressed the highest levels of TRIM56. Because the respiratory and digestive tracts are continuously exposed to pathogenic microorganisms in the outside environment, it is reasonable to speculate that TRIM56 is evolutionarily conserved to combat other viral pathogens in addition to BVDV. The observations that TRIM56 expression is upregulated by viruses or IFN (Fig. 10) (3, 33) provide further support for the notion that TRIM56 is an antiviral host factor.
What is the biological significance of the Npro-TRIM56 interaction? Interestingly, we reproducibly observed that the protein abundance of TRIM56 was profoundly lower in HeLa-FHNpro cells than in control HeLa cells (Fig. 1A, compare lanes 1 and 3) and that the inhibition of the proteasome substantially increased the TRIM56 level (Fig. 1A, compare lanes 1 and 2) in HeLa-FHNpro cells. This suggests that Npro likely promotes proteasomal degradation of TRIM56, as it does for IRF3. Thus, not only is TRIM25 subjected to viral control (7), but TRIM56-mediated antiviral defense is also targeted for inhibition by virus-encoded proteins, and the degradation of TRIM56 by pestivirus Npro represents one such example.
Supplementary Material
ACKNOWLEDGMENTS
We are grateful to Bo Xu and Robert English for assistance with TRIM56 antibody production and mass spectrometry analysis, respectively, and to Michael Whitt for assistance with confocal imaging. We also thank Charles Rice for providing HCV and BVDV replicon constructs, Hiroomi Akashi for BVDV NS expression vectors, Jae Jung for the TRIM25 plasmid, and Susan Baker for the Flag-Ub construct.
This work was supported in part by NIH grant AI069285 to K.L.
Footnotes
Supplemental material for this article may be found at http://jvi.asm.org/.
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Barr S. D., Smiley J. R., Bushman F. D. 2008. The interferon response inhibits HIV particle production by induction of TRIM22. PLoS Pathog. 4:e1000007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Beard M. R., et al. 1999. An infectious molecular clone of a Japanese genotype 1b hepatitis C virus. Hepatology 30:316–324 [DOI] [PubMed] [Google Scholar]
- 3. Carthagena L., et al. 2009. Human TRIM gene expression in response to interferons. PLoS One 4:e4894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan M. W., et al. 2005. Hypermethylation of 18S and 28S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin. Cancer Res. 11:7376–7383 [DOI] [PubMed] [Google Scholar]
- 5. Chen Z., et al. 2007. Ubiquitination and proteasomal degradation of interferon regulatory factor-3 induced by Npro from a cytopathic bovine viral diarrhea virus. Virology 366:277–292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Clementz M. A., et al. 2010. Deubiquitinating and interferon antagonism activities of coronavirus papain-like proteases. J. Virol. 84:4619–4629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gack M. U., et al. 2009. Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5:439–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gack M. U., et al. 2008. Roles of RIG-I N-terminal tandem CARD and splice variant in TRIM25-mediated antiviral signal transduction. Proc. Natl. Acad. Sci. U. S. A. 105:16743–16748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gack M. U., et al. 2007. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature 446:916–920 [DOI] [PubMed] [Google Scholar]
- 10. Higgs R., et al. 2008. The E3 ubiquitin ligase Ro52 negatively regulates IFN-beta production post-pathogen recognition by polyubiquitin-mediated degradation of IRF3. J. Immunol. 181:1780–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Hilton L., et al. 2006. The NPro product of bovine viral diarrhea virus inhibits DNA binding by interferon regulatory factor 3 and targets it for proteasomal degradation. J. Virol. 80:11723–11732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ishikawa H., Ma Z., Barber G. N. 2009. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature 461:788–792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 14. Kong H. J., et al. 2007. Autoantigen Ro52 is an interferon inducible E3 ligase that ubiquitinates IRF-8 and enhances cytokine expression in macrophages. J. Immunol. 179:26–30 [DOI] [PubMed] [Google Scholar]
- 15. Lee Y. M., Tscherne D. M., Yun S. I., Frolov I., Rice C. M. 2005. Dual mechanisms of pestiviral superinfection exclusion at entry and RNA replication. J. Virol. 79:3231–3242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Li K., Chen Z., Kato N., Gale M., Jr., Lemon S. M. 2005. Distinct poly(I-C) and virus-activated signaling pathways leading to interferon-beta production in hepatocytes. J. Biol. Chem. 280:16739–16747 [DOI] [PubMed] [Google Scholar]
- 17. Lindenbach B. D., et al. 2005. Complete replication of hepatitis C virus in cell culture. Science 309:623–626 [DOI] [PubMed] [Google Scholar]
- 18. Meroni G., Diez-Roux G. 2005. TRIM/RBCC, a novel class of ‘single protein RING finger’ E3 ubiquitin ligases. Bioessays 27:1147–1157 [DOI] [PubMed] [Google Scholar]
- 19. Munir M. 2010. TRIM proteins: another class of viral victims. Sci. Signal. 3:jc2. [DOI] [PubMed] [Google Scholar]
- 20. Nisole S., Stoye J. P., Saib A. 2005. TRIM family proteins: retroviral restriction and antiviral defence. Nat. Rev. Microbiol. 3:799–808 [DOI] [PubMed] [Google Scholar]
- 21. Ozato K., Shin D. M., Chang T. H., Morse H. C., III 2008. TRIM family proteins and their emerging roles in innate immunity. Nat. Rev. Immunol. 8:849–860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Ponting C., Schultz J., Bork P. 1997. SPRY domains in ryanodine receptors (Ca(2+)-release channels). Trends Biochem. Sci. 22:193–194 [DOI] [PubMed] [Google Scholar]
- 23. Rajsbaum R., Stoye J. P., O'Garra A. 2008. Type I interferon-dependent and -independent expression of tripartite motif proteins in immune cells. Eur. J. Immunol. 38:619–630 [DOI] [PubMed] [Google Scholar]
- 24. Reymond A., et al. 2001. The tripartite motif family identifies cell compartments. EMBO J. 20:2140–2151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Rice C. M. 1996. Flaviviridae: the viruses and their replication, p. 931–959In Fields B. N., Knipe D. M., Howley P. M. (ed.), Fields virology, 3rd ed. Lippincott-Raven Publishers, Philadelphia, PA [Google Scholar]
- 26. Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Seago J., Goodbourn S., Charleston B. 2010. The classical swine fever virus Npro product is degraded by cellular proteasomes in a manner that does not require interaction with interferon regulatory factor 3. J. Gen. Virol. 91:721–726 [DOI] [PubMed] [Google Scholar]
- 28. Shi M., et al. 2008. TRIM30 alpha negatively regulates TLR-mediated NF-kappa B activation by targeting TAB2 and TAB3 for degradation. Nat. Immunol. 9:369–377 [DOI] [PubMed] [Google Scholar]
- 29. Short K. M., Cox T. C. 2006. Subclassification of the RBCC/TRIM superfamily reveals a novel motif necessary for microtubule binding. J. Biol. Chem. 281:8970–8980 [DOI] [PubMed] [Google Scholar]
- 30. Stremlau M., et al. 2004. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature 427:848–853 [DOI] [PubMed] [Google Scholar]
- 31. Sun W., et al. 2009. ERIS, an endoplasmic reticulum IFN stimulator, activates innate immune signaling through dimerization. Proc. Natl. Acad. Sci. U. S. A. 106:8653–8658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Tscherne D. M., et al. 2006. Time- and temperature-dependent activation of hepatitis C virus for low-pH-triggered entry. J. Virol. 80:1734–1741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsuchida T., et al. 2010. The ubiquitin ligase TRIM56 regulates innate immune responses to intracellular double-stranded DNA. Immunity 33:765–776 [DOI] [PubMed] [Google Scholar]
- 34. Uchil P. D., Quinlan B. D., Chan W. T., Luna J. M., Mothes W. 2008. TRIM E3 ligases interfere with early and late stages of the retroviral life cycle. PLoS Pathog. 4:e16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Wakita T., et al. 2005. Production of infectious hepatitis C virus in tissue culture from a cloned viral genome. Nat. Med. 11:791–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang N., et al. 2010. Viral induction of the zinc finger antiviral protein is IRF3-dependent but NF-kappaB-independent. J. Biol. Chem. 285:6080–6090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Wang N., et al. 2009. Toll-like receptor 3 mediates establishment of an antiviral state against hepatitis C virus in hepatoma cells. J. Virol. 83:9824–9834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yamane D., et al. 2009. Inhibition of sphingosine kinase by bovine viral diarrhea virus NS3 is crucial for efficient viral replication and cytopathogenesis. J. Biol. Chem. 284:13648–13659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Yoneyama M., et al. 2004. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat. Immunol. 5:730–737 [DOI] [PubMed] [Google Scholar]
- 40. Zhong B., et al. 2008. The adaptor protein MITA links virus-sensing receptors to IRF3 transcription factor activation. Immunity 29:538–550 [DOI] [PubMed] [Google Scholar]
- 41. Zhou Z., et al. 2011. Antiviral activities of ISG20 in positive-strand RNA virus infections. Virology 409:175–188 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.