Abstract
Cryptococcus neoformans is a leading cause of life-threatening fungal infection in immunocompromised patients. Inositol-phosphoryl ceramide synthase 1 (Ipc1) is a fungus-specific enzyme, encoded by the essential IPC1 gene, that catalyzes the formation of complex sphingolipids and may also regulate the levels of phytoceramide and diacylglycerol. Here, we investigated the functions of this essential gene by modulating its expression in C. neoformans using a galactose-inducible promoter. Down-regulation of IPC1 significantly lowers the expression of certain virulence traits such as melanin pigmentation and, remarkably, impairs pathogenicity of C. neoformans in an established rabbit model. Interestingly, we found that IPC1 down-regulation significantly decreases the intracellular growth of C. neoformans in the J774.16 murine macrophage-like cells. Finally, we studied the effect of IPC1 expression under different stress conditions and found that down-regulation of IPC1 confers a defect on in vitro growth at low pH. Because this environment is similar to that in the phagolysosome of J774.16 macrophage-like cells, our findings indicate that down-regulation of IPC1 confers a growth defect in vivo through a pH-dependent mechanism. In conclusion, our study is the first to define a novel and crucial function of Ipc1 in fungal pathogenesis.
Keywords: sphingolipids, Cryptococcus neoformans, virulence
Cryptococcus neoformans is a common invasive opportunistic pathogen in AIDS patients, and it is the most common cause of fungal meningitis worldwide (Casadevall and Perfect 1998). Existing treatments are often inadequate for eradicating infection and have toxic side effects, and resistant strains are emerging. Novel approaches for the discovery of new antifungal targets and their inhibitors have been reported, but as of yet, none has led to therapeutic interventions (Fostel and Lartey 2000). Recently, several studies have suggested the sphingolipid pathway as a potential novel target for antifungal therapy (Mandala et al. 1998; Wills et al. 2000).
Sphingolipids are essential components of all eukaryotic membranes, and their presence or absence can have a major impact on cell viability and growth in both mammalian and yeast cells (Dickson and Lester 1999; Hannun and Luberto 2000). Although eukaryotic organisms are characterized by the presence of sphingolipids, differences between yeast and mammals throughout the biosynthetic pathways have been described (Mandala et al. 1998; Wills et al. 2000). One of the fungus-specific steps is the synthesis of IPC from phytoceramide, regulated by the inositol-phosphoryl ceramide synthase 1 (Ipc1), encoded by the IPC1 gene (also called AUR1) (Heidler and Radding 1995). Concomitant to IPC formation, Ipc1 also produces diacylglycerol (DAG) and consumes phytoceramide. The importance of Ipc1 therefore may be due not only to the formation of IPC itself, one of the more abundant sphingolipids in the membranes, but also to the regulation of phytoceramide, implicated in growth arrest and yeast stress responses (Jenkins et al. 1997; Chung et al. 2000), and DAG, a well-established mitogen and activator of protein kinase C (PKC) (Fig. 6).
Figure 6.
A model for the role of Inositol-phosphoryl ceramide synthase 1 (Ipc1) in regulation of virulence traits and pathogenesis of C. neoformans. Ipc1 transfers the phosphoryl-inositol moiety from phosphatidylinositol (PI) to phytoceramide, forming inositol-phosphoryl ceramide (IPC) and diacylglycerol (DAG).
Because IPC1 is both an essential gene and specific to different fungi (Heidler and Radding 1995; Kuroda et al. 1999; Heidler and Radding 2000), modulation of its expression may assume a critical role in vital biological functions. Importantly, in the specific case of an opportunistic pathogen (such as C. neoformans), alteration of these crucial functions could lead to an impaired pathogenicity by affecting host–fungus interaction, modifying the production of virulence traits (fungus factor), and/or altering the immune responses (host factor). A key factor for understanding the mechanisms by which Ipc1 exerts its cellular effects is the modulation of its expression by a promoter that allows the study of both up- and down-regulation of IPC1. The C. neoformans GAL7 promoter allows these studies because even under repressing conditions (glucose medium) transcription can still be detected (Del Poeta et al. 1999a). Moreover, C. neoformans is ideally suited for studies of fungal pathogenesis because it has a defined sexual life cycle (Kwon-Chung and Popkin 1976), stable isogenic haploid strains of opposite mating type are available (Moore and Edman 1993), it is amenable to genetic analysis, transformation, gene disruption and regulation by homologous recombination (Toffaletti et al. 1993; Chang and Kwon-Chung 1994; Lodge et al. 1994), and virulence of strains can be readily assessed both in animal models and in in vitro assays (Perfect et al. 1993; Chaturvedi et al. 1996; Alspaugh et al. 1997; Odom et al. 1997; Franzot et al. 1998; Cruz et al. 1999; Del Poeta et al. 1999b; Fries et al. 1999; Liu et al. 1999).
Here, we regulated the expression of the C. neoformans IPC1 gene under the GAL7 promoter, and we report for the first time that a gene of the sphingolipid pathway has a role in pathogenesis. In particular, we find that modulation of Ipc1 levels in vitro has important effects on melanin pigmentation. Down-regulation of IPC1 generated a strain no longer pathogenic in a rabbit model of cryptococcal meningitis. Importantly, we found that a decreased Ipc1 level impaired the C. neoformans growth into J774.16 murine macrophage-like cell line. Finally, we found that an acidic environment significantly altered growth of C. neoformans with low Ipc1 activity. Our studies clearly indicate an important role for Ipc1 in pathogenesis by modulating different virulence traits of C. neoformans.
Results
Cloning the IPC1 gene under GAL7 regulation
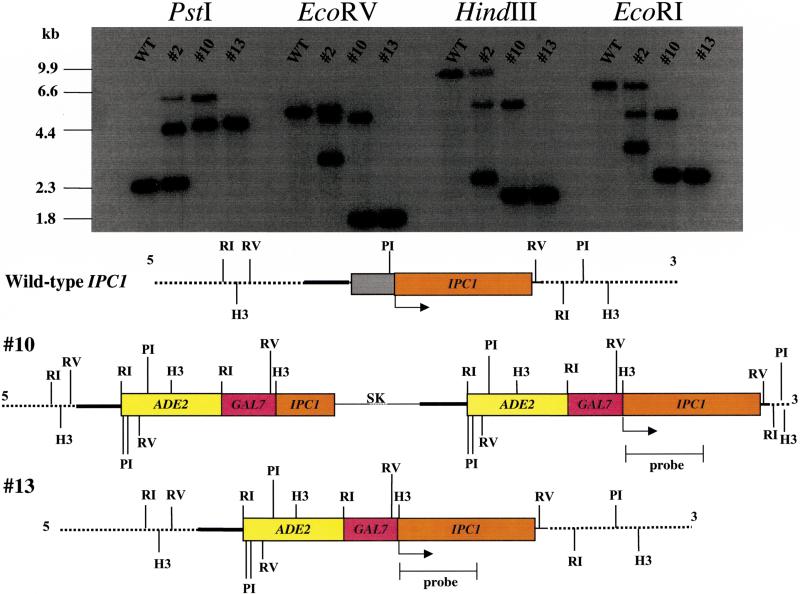
Because the IPC1 gene is essential, a key factor for understanding the mechanisms by which Ipc1 mediates its cellular effects is the need to modulate its expression by a promoter that allows both up- and down-regulation of IPC1. The C. neoformans GAL7 promoter allows these studies because even under repressing conditions (glucose medium) the transcription of the gene still can be detected (Del Poeta et al. 1999a). Therefore, we placed IPC1 under the control of the GAL7 promoter by homologous recombination in the ade2 strain M001 using the plasmid pGAL7::IPC1/ADE2. Out of 30 ADE+ transformants, two showed that the endogenous IPC1 gene had been replaced by the pGAL7::IPC1/ADE2 construct (Fig. 1). The two transformants (#10 and #13) in which the GAL7::IPC1/ADE2 fusion gene integrated at the IPC1 locus, and at no other ectopic sites, were chosen and designated the GAL7::IPC1 strains #10 and #13, respectively. Transformant #10 showed a double crossover with insert of plasmid loop (or single crossover followed by a double crossover event). Note that transformant #2 showed a single crossover event at the upstream region of the IPC1 locus and an ectopic integration. We have used the GAL7::IPC1 strains in which IPC1 expression can be regulated to study the effects of different levels of Ipc1 activity on biology and pathogenesis of C. neoformans.
Figure 1.
Replacement of the endogenous wild-type IPC1 with pGAL7::IPC1/ADE2. Southern analysis of wild-type strain H99 and transformants with genomic DNA digested with PstI (PI), EcoRV (RV), HindIII (H3), and EcoRI (RI), as indicated. Transformant #2 showed a single crossover event at the upstream region of the IPC1 locus. Transformant #10 showed a double crossover event with insert of plasmid loop (or single crossover followed by a double crossover event). Transformant #13 showed a double crossover event. Confirmation of the GAL7::IPC1 gene replacement was also confirmed by PCR analysis using different primer combinations (data not shown). (SK) pBluescript SK plasmid.
Expression levels of IPC1 under inducing and repressing conditions
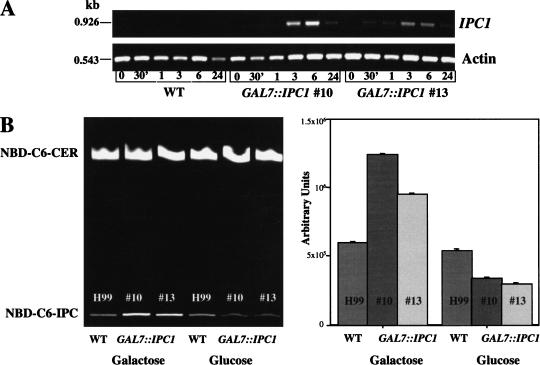
The expression levels of the IPC1 gene under the GAL7 promoter were determined at its transcription and translation levels by determining both mRNA and Ipc1 activity. The IPC1 mRNA levels were determined by semiquantitative RT-PCR in both GAL7::IPC1 strains. Figure 2A shows that the IPC1 mRNA expression level after 6 h on galactose medium was increased ∼50- and ∼30-fold in the GAL7::IPC1 strains #10 and #13, respectively, and the expression was decreased after 24 h of induction. We were unable to determine the expression level under repressing glucose conditions because the basal level of IPC1 mRNA in wild-type cells is extremely low, although overexposure to UV light allowed the detection of IPC1 mRNA in the wild-type cells but not the repressed ones (data not shown). No significant differences were found in actin mRNA expression levels. Because these portions of the 3′ regions of these two genes contain two small introns (52 bp for the IPC1 gene and 51 bp for the actin gene), we could establish that the RT-PCR products derived from cDNA and not contaminating genomic DNA, based on the sizes of the RT-PCR products.
Figure 2.
(A) In vitro quantitative analysis of C. neoformans IPC1 mRNA in H99 and GAL7::IPC1 strains. The isogenic wild-type strain H99 (WT) and the GAL7::IPC1 strains were grown on galactose (inducing conditions). RNA was extracted at different time points (30 min; 1, 3, 6, and 24 h), converted to cDNA, and amplified with primers directed to IPC1 or actin genes. RT-PCR products were separated by electrophoresis in a 1% agarose gel and stained with ethidium bromide. The IPC1 and actin RT-PCR products are 926 and 543 bp, as indicated. (B) In vitro quantitative analysis of Ipc1 activity in C. neoformans wild-type (H99) and GAL7::IPC1 strains grown on glucose (repressing conditions) and galactose (inducing conditions). (Left) The Ipc1 synthase in vitro activity was determined as conversion of NBD-C6-ceramide (NBD-C6-CER) into NBC-C6-IPC by thin layer chromatography (TLC) analysis. (Right) Numerical analysis of Ipc1 activity from the TLC is reported.
In vitro Ipc1 activity levels were determined in the wild-type H99 and GAL7::IPC1 strains using NBD-C6-ceramide as a substrate. Ipc1 activity was significantly increased under galactose-inducing medium after 24 h (Fig. 2B), whereas no significant differences were found after 30 min, 1 h, 3 h, and 6 h of incubation (data not shown). Decreased Ipc1 activity was observed under glucose-repressing conditions, although residual Ipc1 activity is still available to support cell viability. In addition, the in vitro Ipc1 activity levels were determined in a prototrophic transformant (M001 + genomic ADE2) under glucose and galactose growth conditions. No differences were found between this prototrophic transformant and the parental H99 strain (data not shown).
Modulation of IPC1 gene expression did not exert any effect on cell growth at both 30°C and 37°C. In fact, the growth curves relative to each strains and conditions (temperature and/or Ipc1 levels) were superimposable (data not shown).
IPC1 regulates melanin production in C. neoformans
Melanin production is considered one of the virulence factors in pathogenic fungi. In fact, melanin is thought to provide cellular protection from oxygen- and nitrogen-reactive species produced by host immune cells (Wang et al. 1995). Indeed, melanin-deficient mutant strains show reduction of virulence in animal models of cryptococcosis (Kwon-Chung et al. 1982; Rhodes et al. 1982; Alspaugh et al. 1997; Alspaugh et al. 1998; Franzot et al. 1998; Perfect et al. 1998; Williamson et al. 1998; Doering et al. 1999; Nosanchuk et al. 1999). Recent studies have implicated DAG-induced activation of PKC in the stimulation of this process in mammalian cells (Agin et al. 1991; De Oliviera et al. 1996; Gilchrest et al. 1996). Because DAG is one of the products of the Ipc1 reaction, we wondered whether the modulation of IPC1 expression would exert an effect on melanin production in C. neoformans.
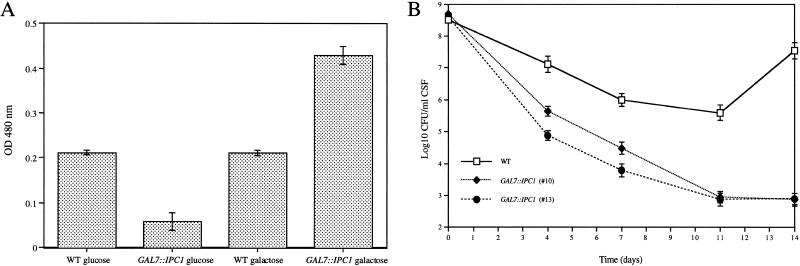
By using a quantitation method previously described (Alspaugh et al. 1997), we addressed whether the activity of phenoloxidase (the key cryptococcal enzyme involved in melanin production) was affected in the GAL7::IPC1 strain (#10) under inducing and repressing conditions. We found that melanin production was reduced by 60% in the GAL7::IPC1 strain under glucose-repressing conditions, and it was increased by ∼80% under galactose-inducing conditions (Fig. 3A). These data show that altered Ipc1 activity regulates melanin production in C. neoformans. These results were confirmed using a second independent GAL7::IPC1 transformant (#13) (data not shown).
Figure 3.
(A) IPC1 regulates melanin pigmentation in C. neoformans. The wild-type (WT) and GAL7::IPC1 strains were grown for 16 h at 30°C under inducing and repressing conditions. Cells were permeabilized with toluene:ethanol, incubated for 16 h in presence of L-dopamine, and the cellular phenoloxidase activity was assayed spectrophotometrically by measuring the appearance of melanin in the supernatant by the change in absorbance at 480 nm. The A480 of samples to which L-dopamine had been added was compared with the blank sample in which no L-dopamine had been added. Data represent geometric means ± standard deviations of three separate experiments. (B) IPC1 regulates virulence of C. neoformans in a rabbit animal model of cryptococcal meningitis. Rabbits (three for each strain) were immunosuppressed with steroids and inoculated with 3 × 108 cells. CSF was removed on days 4, 7, 11, and 14 following inoculation, and the number of surviving organisms was determined by serial dilutions and plating on YPD medium. Each data point represents the geometric means of all cultures for each strain, and the standard deviations of the mean are indicated.
Down-regulation of Ipc1 renders C. neoformans nonpathogenic
Because we found that Ipc1 could be involved in the modulation of melanin production, we next tested whether Ipc1 down-regulation would affect the pathogenicity in the rabbit animal model of cryptococcal meningitis, in which glucose is available as a carbon source instead of galactose.
To test this hypothesis, we inoculated intrathecally immunosuppressed rabbits with the C. neoformans wild-type or GAL7::IPC1 strains. Survival of C. neoformans in the central nervous system (CNS) was determined by removal of cerebral spinal fluid (CSF) and quantitation of colony-forming units (CFU) by serial dilution and plating. Several studies have established the value of this experimental model in studying C. neoformans virulence (Perfect et al. 1980; Lodge et al. 1994; Alspaugh et al. 1997; Odom et al. 1997; Del Poeta et al. 1999a; Del Poeta et al. 1999b; Wang and Heitman 1999; Yue et al. 1999).
As shown in Figure 3B, whereas the wild-type C. neoformans strain H99 persists for 14 d in the CSF of immunocompromised animals, the number of GAL7::IPC1 cells was significantly reduced (P < 0.01) in the immunosuppressed hosts. By days 7 and 14 following inoculation, survival of the GAL7::IPC1 strain was reduced by ∼350- and ∼10,000-fold, respectively, compared with the wild-type strain, as measured by quantitative colony counts. This experiment was performed with the two independent GAL7::IPC1 transformants (#10 and #13). These findings indicate that full expression of IPC1 is required for growth of C. neoformans in the host.
The intracellular growth of the GAL7::IPC1 strain is impaired in J774.16 murine macrophage-like cells under glucose-repressing conditions
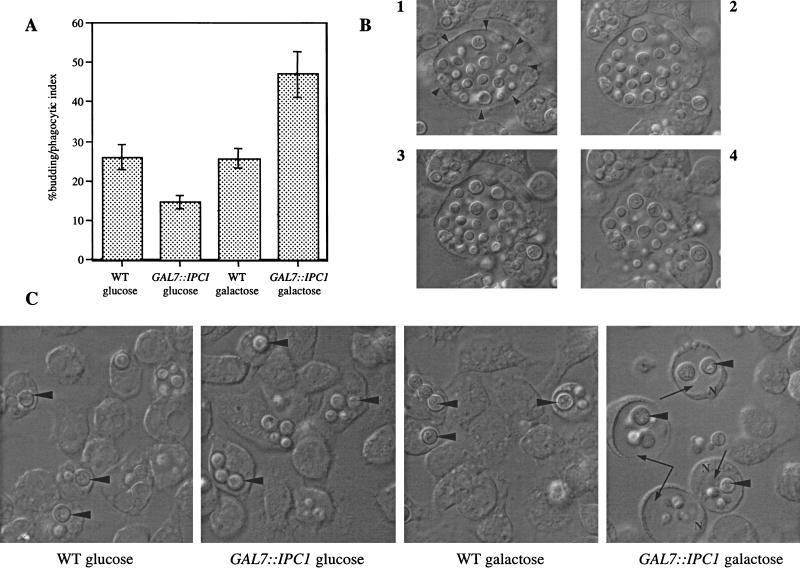
C. neoformans is a facultative intracellular pathogen in vivo (Feldmesser et al. 2000). To investigate the mechanism by which Ipc1 contributes to virulence, we explored the effect of reduced Ipc1 on macrophage intracellular growth. We infected the J774.16 murine macrophage-like cell line with the C. neoformans wild-type H99 and GAL7::IPC1 strains and monitored the fungal growth inside the host cells under glucose-repressing and galactose-inducing conditions. We found that under glucose conditions, the GAL7::IPC1 strain replicated slower than the wild type, whereas under galactose conditions, it replicated faster than the wild type (Fig. 4A). In addition, we found that under glucose-repressing conditions the GAL7::IPC1 cells remained in a tight cluster after lysis of the macrophage-like cells (data not shown), whereas they appeared to disperse under galactose conditions (Fig. 4B). These findings indicated to us that expression of IPC1 is required for normal in vivo growth, by modulating both replication and diffusion of the infection. Moreover, we observed that the GAL7::IPC1 strain produced a larger phagolysosome in galactose-inducing conditions (Fig. 4C). Taken together, these observations indicate that IPC1 regulates the growth of C. neoformans in J774.16 murine macrophage-like cell line.
Figure 4.
IPC1 regulates intracellular growth in the J774.16 macrophage-like cell line. (A) Five hours postinoculation, buds were counted only in yeast cells inside the macrophages (% budding/phagocytic index). Counts are geometric means ± standard deviations of four different fields. (B) Lysis of macrophage infected by the GAL7::IPC1 strain on galactose medium at 18 h postinoculation. (Frame 1) Intact macrophage. Arrowheads indicate the phagolysosome. (Frames 2–4) Lysis of macrophage. (C) Phagolysosomes size. Arrowheads indicate yeast cells inside the phagolysosome, whereas long arrows in the last right panel indicate the larger phagolysosome. (N) nucleus. Cells were photographed at ∼18 h postinoculation.
Down-regulation of IPC1 gene increases the sensitivity to low pH
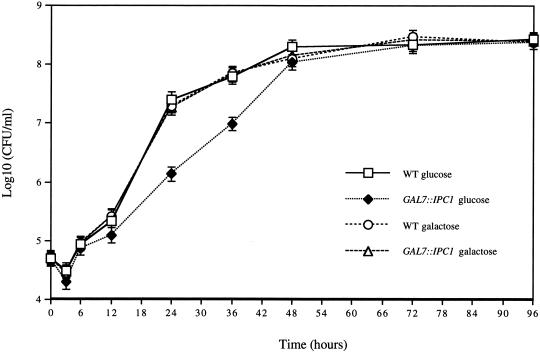
As shown in Figure 4C, the intracellular growth of C. neoformans into macrophages occurs in the phagolysosome, an acidic environment. Therefore, we wondered whether the lack of replication of the Ipc1 down-regulated cells in the phagolysosome was due to a lower tolerance of acidic pH. Thus, we monitored the in vitro growth of the GAL7::IPC1 and wild-type H99 strains at pH 4.0, under glucose-repressing and galactose-inducing conditions. As shown in Figure 5, we found that during the exponential proliferation phase, the growth of the C. neoformans GAL7::IPC1 strain (#10) in glucose was reduced by ∼20- and 15-fold compared with the wild type at 24 h and 36 h of incubation, respectively. However, it is interesting to note that the cell concentrations at the stationary phase were the same for both strains.
Figure 5.
IPC1 expression modulates in vitro growth under low-pH stress condition. C. neoformans wild-type (WT) and GAL7::IPC1 strains grown on YP medium (pH 4.0), supplemented with 2% glucose or 2% galactose. Colony forming units/mL (CFU/mL) are geometric means ± SD of three separate experiments.
Discussion
The study of the role and function of essential genes in yeast is particularly difficult because often, under repressing conditions, strains are not viable. In our previous studies we showed that the GAL7 promoter, when used for the regulation of essential genes, maintains viable cells (Del Poeta et al. 1999a) and can be successfully used in C. neoformans to study its virulence traits and pathogenesis. By regulating the IPC1 gene under the GAL7 promoter, we showed that down-regulation of IPC1 impairs pathogenicity of C. neoformans. Indeed, we found that down-regulation of IPC1 significantly lowers melanin production, in vivo growth in macrophages, and diffusion ability of cell progenies. Our results indicate that Ipc1 exerts a major role in the pathogenesis of C. neoformans through crucial and perhaps diverse mechanisms.
First, we addressed the question whether IPC1 expression has a role in virulence traits of C. neoformans, such as melanin pigmentation. We found that overexpression of the IPC1 gene induces more melanin production, whereas down-regulation decreases melanin pigmentation. Melanin synthesis in C. neoformans is catalyzed by a single laccase gene (Williamson et al. 1998). Its product, a laccase, converts exogenous diphenolic substrates through a series of autooxidation reactions to the final pigmented product (Polacheck and Kwon-Chung 1988). Melanin has been proposed as a virulence factor for C. neoformans because melanin-deficient mutants are not pathogenic in the animal models of cryptococcosis (Kwon-Chung et al. 1982; Wang et al. 1995; Salas et al. 1996; Williamson et al. 1998; Nosanchuk et al. 1999).
C. neoformans melanizes during infection, and it has been suggested that the neurotrophism of this pathogenic fungus is associated with its ability to produce melanin from cathecolamines, which are found in concentrated amounts in the CNS (Polacheck et al. 1990). Recent evidence indicates that this pigment protects the fungus against immune defense mechanisms, possibly because of its autooxidants properties, and/or because of its ability to mask potential antigens (Wang et al. 1995; Nosanchuk et al. 1998; Doering et al. 1999; Liu et al. 1999; Schnitzler et al. 1999; Casadevall et al. 2000). Interestingly, it has been proposed that melanin production by C. neoformans could affect cytokines release from the host immune defense (Huffnagle and McNeil 1999; Barluzzi et al. 2000). Several studies have proposed that DAG stimulates melanogenesis in human, poikothermic vertebrates, and in Skh-2 pigmented hairless mice through activation of PKC (Agin et al. 1991; De Oliviera et al. 1996; Gilchrest et al. 1996). Whether this mechanism is also involved in C. neoformans is not known. Because Ipc1 regulation directly modulates DAG formation, the hypothesis that the DAG–PKC–melanin connection occurs also in C. neoformans is attractive, and we will investigate this possibility in the future.
Because Ipc1 was involved in the modulation of melanin production, we studied whether Ipc1 down-regulation affected the pathogenicity in the rabbit animal model of cryptococcal meningitis, in which glucose is available as a carbon source instead of galactose. Glucose level in the CSF is ∼50%–60% of glucose level in the blood, which is 80 mg/100 mL or 800 μg/mL. At glucose concentrations equal, below, and above blood level, the in vitro Ipc1 activity of the GAL7::IPC1 strains (#10 and #13) is similarly reduced by approximately fourfold compared with the wild-type H99 (data not shown). These observations lead us to hypothesize that if full expression of IPC1 is required in vivo, then the survival rate of the GAL7::IPC1 strains will be impaired. To test this hypothesis, we inoculated immunosuppressed rabbits intrathecally with the C. neoformans wild-type or GAL7::IPC1 strains. By testing two independent C. neoformans GAL7::IPC1 transformants, we observed that these strains are no longer pathogenic in the rabbit animal model of cryptococcal meningitis. Because the in vitro growth of the GAL7::IPC1 strains is not impaired under normal conditions at 30°C and 37°C, and at low and high glucose concentrations (data not shown), the data obtained from the animal correlate the pathogenic trait of C. neoformans to its growth in vivo. Our results for the first time implicate IPC1 in the virulence of a human pathogen.
One major factor for C. neoformans to produce infection is its ability to grow in the host cells (Feldmesser et al. 2000). Therefore, to investigate this mechanism, we studied the effects of the up- and down-regulation of the IPC1 gene on intracellular growth in the J774.16 murine macrophage-like cell line. As shown in Figure 4A, the replication time of the GAL7::IPC1 strain in glucose medium was reduced compared with the wild type. In galactose medium, the replication time of the GAL7::IPC1 strain was increased. We found by electron microscopy that the phagolysosome membrane is not intact (Feldmesser et al. 2000). Therefore, we hypothesize that small molecules (e.g., glucose or galactose) will also enter the phagolysosome. Recently, it has been shown that survival and growth of C. neoformans in macrophages occur in the acidic phagolysosome (Levitz et al. 1999). Unlike other pathogens, such as Mycobacterium tuberculosis and Histoplasma capsulatum, C. neoformans does not regulate the pH of the phagolysosome, and it is resistant to the action of macrophage lysosomal enzymes (Howard 1961; Diamond and Bennett 1973; Levitz and Tabuni 1991; Levitz et al. 1997).
Therefore, we next investigated the effect of IPC1 regulation on the ability of C. neoformans to grow in acidic conditions. In our case, down-regulation of IPC1 conferred a defect on in vitro growth of GAL7::IPC1 strain at acidic pH, whereas it does not affect growth under neutral pH (7.0) (data not shown). Because these conditions are similar to those in the phagolysosome of the J774.16 macrophage-like cell line, our findings indicate that the down-regulation of IPC1 confers a growth defect in vivo through a pH-dependent mechanism. This hypothesis is further supported by the observation that, after 5 h post-infection, the GAL7::IPC1 strain into J774.16 cells is able to replicate faster than the wild-type cells under galactose-inducing conditions (Fig. 4A).
The involvement of the IPC1 gene in the adjustment to pH-induced stress may indicate a more general role of Ipc1 in regulation of other stress responses. Indeed, we found that Ipc1 levels also regulate the response to osmotic stress (data not shown). One possible mechanism by which modulation of Ipc1, and therefore sphingolipid levels, can regulate pH and osmotic stress is through the alteration of the physical properties of the plasma membrane (Levine et al. 2000). The acyl chains, the association with sterols, and the capacity for hydrogen bonding lead to sphingolipids promoting a more compact, thicker, and less permeant bilayer.
Such a role for sphingolipids is supported by the determination that yeast cells lacking complex sphingolipids are not viable (Lester et al. 1993; Nagiec et al. 1993; Hashida-Okado et al. 1996). In our studies, we found that the GAL7::IPC1 strain produces less complex sphingolipids under glucose-repressing conditions (data not shown), indicating that the resulting plasma membrane may be more permeant to solutes and cations and, thus, be in an altered intracellular homeostasis of Na+ and H+.
To produce chronic infection, microbial pathogens must escape host immune responses, and infection by C. neoformans is typically chronic. In early infection, C. neoformans cells were found predominantly in the intracellular compartment. A shift toward extracellular predominance occurred by ∼24 h of infection and was accompanied by macrophage cytotoxicity and disruption (Feldmesser et al. 2000). Interestingly, after macrophage disruption we observed that the release of the GAL7::IPC1 strain in glucose differed from that in galactose and the wild-type H99. Cells were compacted, forming a cluster of a “grape-like” shape (data not shown), whereas the GAL7::IPC1 on galactose and the wild type were dispersed as single cells after the macrophage disruption (Fig. 4B). Perhaps this decreases the ability of the organism to cause tissue damage because the cells are clumped together and possibly less likely to infect other macrophages.
As a direct consequence, it is reasonable to hypothesize that such yeast cells would not be able to readily infect other host cells, and, thus, they would be easily attacked by the immune responses, resulting in a decreased dissemination of the infection.
Although the IPC1 gene has been isolated in several microorganisms, the biological roles of this “fungus-specific step” have not been described. Our studies define novel functions for Ipc1 in pathogenesis of the opportunistic pathogen C. neoformans. In particular, we find that modulation of IPC1 expression regulates virulence traits, such as melanin pigmentation. Also, IPC1 down-regulation impairs pathogenicity of C. neoformans in the animal model and significantly decreases its intracellular growth in the phagolysosome of J774.16 macrophage-like cells. Finally, we find that down-regulation of the IPC1 gene confers a defect on in vitro growth at low pH, and, because this environment is similar to that in the phagolysosome of J774 cells, our results indicate that down-regulation of IPC1 confers a growth defect in vivo through a pH-dependent mechanism. These findings are summarized in the model presented in Figure 6. Our ongoing epistasis analysis begins to determine the mechanisms of how the sphingolipid pathway regulates virulence in pathogenic fungi and, for the first time, implicates Ipc1 in the virulence of a human pathogen. Finally, these results should prompt the development of novel antifungal drugs against this fungus-specific target.
Materials and methods
Strains and growing media
C. neoformans var. grubii serotype A strain H99 (wild-type) and M001 strain, an ade2 isogenic derivative of H99, were used in this study. A GAL7::IPC1 strain was created from M001. A synthetic medium, containing 6.7 g/L of Yeast Nitrogen Base (YNB) without amino acids, 1.3 g/L amino acid mix lacking adenine, 180 g/L sorbitol, 20 g/L galactose, and 20 g/L agar, was used for selecting the GAL7::IPC1 strain obtained from biolistic transformation. C. neoformans H99, M001, and GAL7::IPC1strains were routinely grown on yeast extract peptone dextrose (YPD) medium. Yeast extract peptone (YP) supplemented with 20 g/L glucose or 20 g/L galactose was used for down- and up-regulated IPC1 expression, respectively.
Preparation of genomic DNA
DNA was isolated from C. neoformans strains as follows: 10 mL of mid- to late-log phase C. neoformans grown in appropriate media was pelleted and washed three times in sterile deionized water (SDW). Cells were suspended in 0.5 mL TES buffer (10 mM Tris at pH 7.5, 1 mM EDTA at pH 8.0, 1% sodium dodecyl sulfate [SDS]). Then, 0.2 mL of 0.5-mm acid-washed glass beads was added and samples were incubated for 1 min on ice, followed by homogenization for 1 min by mini Bead-Beader-8 (BioSpec). Cells were incubated for 10 min at 70°C, briefly vortexed, and, after adding 200 μL of 3M potassium acetate and 150 μL of 5M NaCl, cells were incubated for 20 min on ice. After centrifugation for 20 min in a microfuge at 13,000 rpm, the aqueous phase (∼600 μL) was transferred to a new tube, 300 μL of 30% PEG 6000 was added, and samples were incubated for 10 min on ice. Precipitated DNA was collected by centrifugation at 13,000 rpm, resuspended in 40 μL TE (pH 8.0), and stored at −20°C.
Cloning the C. neoformans IPC1 gene under the GAL7 promoter
The C. neoformans IPC1 gene has been isolated. However, the sequence submitted to the GenBank (accession #AAD28749) is truncated at the promoter region. To isolate the promoter and the untranslated upstream regions, the following strategy was used. Amplification of a part of C. neoformans IPC1 from genomic DNA was performed by PCR using the following primers: IPC1-5: 5′-ATG TCC GCC ATC CGC GCA CTC-3′ and IPC1-3: 5′-GAG CTA TTC TCT GAA GCC ACC-3′. PCR conditions were 5 min at 95°C (1 cycle); 50 sec at 93°C, 50 sec at 50°C, 80 sec at 72°C (30 cycles); 10 min at 72°C (1 cycle). This amplification strategy produced a 1450-bp fragment that was cloned into pCRII-TOPO vector (Invitrogen), sequenced, and named pCR-CnIPC1. Then, a genomic library of H99 in EMBL3 was screened with the PstI- and EcoRV-digested 1.4-kb probe fragment from the pCR-CnIPC1 plasmid. Positive plaques were then purified through three rounds of repeated screening. One strongly positive λ clone (#2.1) was purified using the plate lysate method, as described by Fritsch (Sambrook et al. 1989). The 2.1 genomic clone was further digested with NcoI and EcoRV, generating 2.1-kb and 0.7-kb hybridizing fragments. The 0.7-kb fragment corresponded to the 3′ end of the IPC1 gene, whereas the 2.1-kb fragment contained the 5′, promoter, and upstream untranslated regions of the IPC1 gene. This 2.1-kb fragment was extracted from agarose gel using the Qiagen Gel Extraction Kit (Qiagen), partially sequenced in both directions, and shown to be the 5′, promoter, and upstream regions of the C. neoformans IPC1 gene. Presumptive TATA and CAAT boxes were present at −199 bp and −157 bp from the ATG start site, respectively (GenBank accession no. AY007247).
The pGAL7::IPC1/ADE2 construct was generated by using a series of molecular manipulations involving the IPC1 gene, the 585-bp C. neoformans GAL7 promoter from serotype D, strain 3501, and the 3000-bp ADE2 marker genomic DNA fragment obtained from serotype A, strain H99. (1) Fragment A (526 bp) was generated by PCR using IPC1 λ DNA (clone 2.1) as a template and the primers #IPC-10, 5′-CTCA GGTACC TTA TCC CCA CTT GAA AGA GCT CG-3′, and #IPC-11, 5′-GCGA GAATTC AAG ACT ATC TTC ACG GCC AGC-3′, containing a KpnI and EcoRI site, respectively (bolded and underlined). The 526-bp fragment contains a sequence corresponding to the upstream untranslated region of the IPC1 promoter.
This fragment was then digested with KpnI and EcoRI, producing fragment B (518 bp). (2) Fragment C (1104 bp) was generated using IPC1 λ DNA (clone 2.1) as a template and the primers #IPC-12, 5′-GTAT AAGCTT ATG TCC GCC ATC CGC GCA CTCAC-3′, and #IPC-13, 5′-GACA TCTAGA CCT CCC ACT GTA TCT GAT CAA CG-3′, which contain sequence homologous to the 5′ and internal regions of IPC1, respectively. The primers #IPC-12 and #IPC-13 contain a HindIII and XbaI site, respectively (bolded and underlined). This fragment was then digested with HindIII and XbaI, producing fragment D (1092 bp). (3) The GAL7 promoter was amplified from the plasmid pAUG-MF, made by Wickes and Edman (1995), using primers 6C, 5′-CAGG GAATTC GTG GAA AGA AGC AGG TCT TGT CGA-3′, and 6H, 5′-ATTA AAGCTT TCT CAA GAG GGG ATT GAG CGC TGA-3′, containing an EcoRI and HindIII site, respectively. This fragment was then cleaved with EcoRI and HindIII, generating fragment E (585 bp). (4) Fragment B (upstream untranslated region) was cloned into the KpnI and EcoRI sites of an SK+ pBluescript vector, generating plasmid #1. (5) Fragment D (5′-region of IPC1 gene) was combined with fragment E (GAL7 promoter), ligated, and cloned into the EcoRI and XbaI sites of plasmid #1, generating plasmid #2. The resulting plasmid contains the upstream untranslated region of the IPC1 locus and the GAL7 promoter in frame with the ATG start site of the 5′-truncated IPC1 gene. (6) Finally, the EcoRI-restricted ADE2 fragment was then inserted into the EcoRI site located at the 5′-end of the GAL7 promoter to form pGAL7::IPC1/ADE2. This GAL7::IPC1 fusion construct was sequenced to make sure that no mutations were introduced by PCR manipulations.
Biolistic transformation
The pGAL7::IPC1-ADE2 plasmid was transformed into C. neoformans strain M001 using biolistic delivery of DNA, following the protocol described by Toffaletti et al. (1993). Transformants were grown on YNB-galactose without adenine (see above). Colonies were chosen randomly and purified. Thirty stable transformants were chosen and transferred in YPD plates. Then, 10 mL YPD broth cultures were inoculated with a single colony and incubated for 48–72 h at 30°. Genomic DNA preparations for PCR and/or Southern blot analysis of transformants were performed as described by Sambrook et al. (1989). The transformants showing integration of the GAL7::IPC1-ADE2 fusion construct at the IPC1 locus were chosen and designated GAL7::IPC1 strains (Fig. 1).
In vitro growth studies
From overnight YPD broth cultures of C. neoformans H99 and GAL7::IPC1 strains, cells were washed twice in SDW, resuspended, and diluted into 10 mL fresh YP broth with either glucose or galactose to a final density of 104 cells/mL and incubated at 250 rpm/min in a shaker incubator at 30° and 37°C. For in vitro growth kinetics under pH stress condition, YP broth (pH 4.0) was used.
Samples were taken from the tubes at various time points (0, 3, 6, 12, 24, 36, 48, 72, and 96 h), diluted with SDW, and plated onto YPD plates for assessment of colony-forming units. All cultures were performed in triplicate.
RNA extraction and RT-PCR
From overnight YPD broth cultures of C. neoformans H99 and GAL7::IPC1 strains, cells were washed twice in SDW, resuspended, and diluted into 10 mL fresh YP broth with either glucose or galactose to a final density of 106 cells/mL and incubated in a shaker incubator at 250 rpm/min at 30°. Samples were taken from the tubes at various time points (0 and 30 min; 1, 3, 6, and 24 h). Then, total RNA was prepared as described by RNeasy Mini-Kit (Qiagen, Cat. #74104), following the mechanical disruption protocol for yeasts. First-strand synthesis of cDNA was performed using 1μg of total RNA for each group following the protocol described by SuperScript Preamplication System (GIBCO BRL). First-strand synthesis was made with oligo(dT) primer. Second-strand synthesis of the IPC1 gene was with specific primers IPC-15 (5′-CTA AGA GCT ATT CTC TGA AGC) and IPC-18 (5′-GAT ATA CCA ACG CTT TCC TTG), yielding a 926-bp fragment.
Because of the presence of a 52-bp intron in this region, this pair of primers yielded a 978-bp fragment when genomic DNA was used as template for PCR. Actin-specific primers AC-2 (5′-CAG CTG GAA GGT AGA CAA AGA GGC) and AC-1 (5′-CGC TAT CCT CCG TAT CGA TCT TGC) were used for the second-strand synthesis of the actin gene as a control, yielding a 543-bp fragment. Because of the presence of a 51-bp intron in this region, this pair of primers yielded a 594-bp fragment when genomic DNA was used as template for PCR. The analysis of mRNA levels of the IPC1 gene was performed with a second independent GAL7::IPC1 transformant. RT-PCR of control RNA was performed in each run of experiments.
In vitro activity of Ipc1
Ipc1 activity was measured by using the fluorescent ceramide analog NBD-C6-ceramide (Avanti Polar Lipids) as substrate and monitoring the formation of NBC-C6-IPC, as described by Fischl et al. (2000) with some modifications. Briefly, wild-type and GAL7::IPC1 C. neoformans strains were grown on YP-glucose and YP-galactose media in a shaker incubator for 24 h at 30°C. Cells were harvested by centrifugation, washed with SDW, and the pellets were stored at−80°C. Cell pellets were resuspended in lysis buffer (25 mM Tris at pH 7.4, 5 mM EDTA, 1 mM PMSF [Sigma # P-7626], and 10 μg/mL each Chymostatin [Sigma #C-7268], Leupeptin [Sigma #L-2884], Antipain [Sigma #A-6271], and Pepstatin A [Sigma #P-4265]). Then, acid-washed glass beads (for a volume equal to 3/4 of the cell suspension) were added, and cells were homogenized three times for 45 sec using the Bead-Beader-8. After centrifugation at 2500g for 10 min at 4°C, supernatant (∼100 μL) was transferred to a sterile 1.5-mL microcentrifuge tube for proteins quantitation, which was determined by the method of Bradford (1976). Proteins from the cell lysates (60–100 μg) were incubated for 30 min at 30°C in 50 mM bis-Tris-HCl buffer (pH 6.5) containing 1 mM PI, 5 mM Triton X-100, 1 mM MnCl2, 5 mM MaCl2, 20 μM NBD-C6-ceramide in a final reaction volume of 100 μL. The reaction was terminated by the addition of 0.5 mL of 0.1 N HCl in methanol. Chloroform (1 mL) and 1 M MgCl2 (1.5 mL) were added, the solution was mixed, and the phases were separated by a 10-min centrifugation at 1000g. The chloroform-soluble product, NBD-IPC, was analyzed by analytical thin-layer chromatography (TLC) on silica gel 60 plates (EM Science) using the solvent system chloroform/methanol/water (65:25:4). NBD-IPC was identified and quantified by direct fluorescence using a Molecular Dynamics 840 STORM unit.
Melanin production
Phenoloxidase activity was assayed as described previously (Alspaugh et al. 1997) with minor modifications. Cells were inoculated from fresh cultures in YPD into YP broth with 2% glucose or 2% galactose and incubated for 16 h at 30°C in a shaking incubator. Cells were pelleted, washed once with liquid YNB (6.7 g/L) without sugar, and resuspended in 10 mL of the same medium. Cells were incubated for an additional 7 h at 30°C, pelleted, washed once with SDW, and resuspended in 50 mM sodium phosphate (pH 7.0) (1 mL sodium phosphate solution for each 100 mg of cells, wet weight).
Toluene:ethanol (1:4, v/v), 100 μL per 1 mL of cell suspension, was added to permeabilize the cells, and the mixture was vortexed for 90 sec. The cell suspensions were incubated for 16 h in a 30°C water bath either with or without 1 mM L-dopamine. The suspensions were pelleted in a microcentrifuge at 14,000 rpm for 30 sec, and the supernatants were analyzed in a Beckman DU 640 spectrophotometer at 480 nm. The A480 of samples to which L-dopamine had been added was compared with the blank sample in which no L-dopamine had been added. Data represent geometric means ± standard deviations of three separate experiments.
Animal model of cryptococcal meningitis
New Zealand White rabbits weighing 2–3 kg were housed in separate cages and provided with water ad libitum and Purina rabbit chow. Wild-type H99 and the isogenic GAL7::IPC1 strains were prepared by growth for 48 h at 30° in YPD medium. The cells were pelleted, washed twice, and suspended in PBS at a concentration of 5 × 108 cells/mL. After sedation with ketamine (Fort Dodge) and xylazine (Vedco), ∼3 × 108 viable cells of each strain were inoculated intracisternally into three rabbits that had received an intramuscular injection of cortisone acetate at 2.5 mg/kg (Merck) 1 d earlier and then daily for 14 d. Three rabbits received the wild-type H99 strain, three rabbits received the GAL7::IPC1 strain (#10), and three rabbits received a second independent GAL7::IPC1 transformant (#13). Rabbits were sedated with ketamine and xylazine on days 4, 7, 11, and 14 after inoculation, and CSF was withdrawn. Quantitative yeast cultures were performed by diluting the CSF in PBS, plating on YPD medium, and incubating for 72 h at 30°C.
Intracellular growth in J774.16 macrophage-like cell line
J774.16 is a murine reticulum sarcoma macrophage-like cell line that has been extensively characterized (Chang et al. 1992). The intracellular growth of C. neoformans into J774.16 macrophage-like cells was studied as previously described (Mukherjee et al. 1995; Mukherjee et al. 1996; Cox et al., in prep.). In brief, J774.16 cells were used up to passage #10, with each passage dilution equal to 1:15. Cells were plated on 35 × 10-mm plates (Constar Corp.) and maintained in DMEM supplemented with 10% fetal calf serum and 1× penicillin/streptomycin at 37°C with 10% CO2. Just before the infection, 2 mL of the fresh medium containing 2% glucose or 2% galactose was added to the cells. Then, 50 units/mL of IFN-γ (Genzyme Corp.), 0.3 μg/mL of LPS (Sigma Chemical Co.), and 10 μg/mL of 18B7 (IgG-specific anti-GXM monoclonal antibody) were added to the cells. Finally, C. neoformans wild-type H99 and GAL7::IPC1 (#10) strains were grown on YPD for 48 h at 30°C, washed three times in PBS, and added to the J774.16 cells at a multiplicity of infection (effector to target ratio of 1:1). After a short incubation (30 min to 1 h), extracellular C. neoformans cells were washed away with three changes in cell culture media. The imaging of cells was performed with a Photometrics Sensys cooled CCD camera on an Olympus IX 70 microscope. The temperature was kept constant at 37°C by both a heated stage and heated air unit supplied by Olympus. The Plexiglas environment surrounding the microscope was kept under positive pressure with 5% CO2/95% air. Imaging software was IP Lab Spectrum running on a Macintosh G3. At the 5 h time point buds were counted, and the percentage of budding cells per the phagocytic index was recorded. The phagocytic index is the number of internalized yeast per number of macrophages per field. Data are geometric means ± standard deviations of four different fields.
Acknowledgments
We thank Meiquing Shen and Allyson Plowden for technical assistance, Ogretmen Besim for helping in RNA extraction, Lina Obeid, Cungui Mao, Gary M. Jenkins, Yasuo Okamoto, Cletus A. D'Souza, Andrew Alspaugh, and Joseph Heitman for discussion, and LuAnne Harley for helping on the preparation of this manuscript. This work was supported in part by MUCU Institutional Project 21363, in part by grant GM 43825, in part by PO1 grant AI44975 from NIAID to the Duke University Mycology Research Units, in part by NIAID grant AI28388, and in part by grant HL59842.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL delpoeta@musc.edu; FAX (843) 792–1627.
Article and publication are at www.genesdev.org/cgi/doi/10.1101/gad.856001.
References
- Agin PP, Dowdy JC, Costlow ME. Diacylglycerol-induced melanogenesis in Skh-2 pigmented hair-less mice. Photodermatol Photoimmunol Photomed. 1991;8:51–56. [PubMed] [Google Scholar]
- Alspaugh JA, Perfect JR, Heitman J. Cryptococcus neoformansmating and virulence are regulated by the G-protein α subunit GPA1 and cAMP. Genes & Dev. 1997;11:3206–3217. doi: 10.1101/gad.11.23.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alspaugh JA, Perfect JR, Heitman J. Signal transduction pathways regulating differentiation and pathogenicity of Cryptococcus neoformans. Fungal Genet Biol. 1998;25:1–14. doi: 10.1006/fgbi.1998.1079. [DOI] [PubMed] [Google Scholar]
- Barluzzi R, Brozzetti A, Mariucci G, Tantucci M, Neglia RG, Bistoni F, Blasi E. Establishment of protective immunity against cerebral cryptococcosis by means of an avirulent, non melanogenic Cryptococcus neoformansstrain. J Neuroimmunol. 2000;109:75–86. doi: 10.1016/s0165-5728(00)00319-2. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Casadevall A, Perfect JR. Cryptococcus neoformans. Washington, DC.: American Society for Microbiology; 1998. [Google Scholar]
- Casadevall A, Rosas AL, Nosanchuk JD. Melanin and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 2000;3:354–358. doi: 10.1016/s1369-5274(00)00103-x. [DOI] [PubMed] [Google Scholar]
- Chang J, Xing Y, Magliozzo RS, Bloom BR. Killing of virulent Mycobacterium tuberculosisby reactive nitrogen intermediates produced by activated murine macrophages. J Exp Med. 1992;175:1111–1122. doi: 10.1084/jem.175.4.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang YC, Kwon-Chung KJ. Complementation of a capsule-deficient mutation of Cryptococcus neoformansrestores its virulence. Mol Cell Biol. 1994;14:4912–4919. doi: 10.1128/mcb.14.7.4912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaturvedi V, Flynn T, Niehaus WG, Wong B. Stress tolerance and pathogenic potential of a mannitol mutant of Cryptococcus neoformans. Microbiology. 1996;142:937–943. doi: 10.1099/00221287-142-4-937. [DOI] [PubMed] [Google Scholar]
- Chung N, Jenkins G, Hannun YA, Heitman J, Obeid LM. Sphingolipids signal heat stress-induced ubiquitin-dependent proteolysis. J Biol Chem. 2000;275:17229–17232. doi: 10.1074/jbc.C000229200. [DOI] [PubMed] [Google Scholar]
- Cruz MC, Cavallo LM, Gorlach JM, Cox G, Perfect JR, Cardenas ME, Heitman J. Rapamycin antifungal action is mediated via conserved complexes with FKBP12 and TOR kinase homologs in Cryptococcus neoformans. Mol Cell Biol. 1999;19:4101–4112. doi: 10.1128/mcb.19.6.4101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Oliviera AR, Castrucci AM, Visconti MA. Cellular signalling in vertebrate pigment cells. Braz J Med Biol Res. 1996;29:1743–1749. [PubMed] [Google Scholar]
- Del Poeta M, Toffaletti DL, Rude TH, Dykstra CC, Heitman J, Perfect JR. Topoisomerase I is essential in Cryptococcus neoformans: Role in pathobiology and as an antifungal target. Genetics. 1999a;152:167–178. doi: 10.1093/genetics/152.1.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Del Poeta M, Toffaletti DL, Rude TH, Sparks SD, Heitman J, Perfect JR. Cryptococcus neoformansdifferential gene expression detected in vitro and in vivo with green fluorescent protein. Infect Immun. 1999b;67:1812–1820. doi: 10.1128/iai.67.4.1812-1820.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diamond RD, Bennett JE. Disseminated cryptococcosis in man: Decreased lymphocyte transformation in response to Cryptococcus neoformans. J Infect Dis. 1973;127:694–697. doi: 10.1093/infdis/127.6.694. [DOI] [PubMed] [Google Scholar]
- Dickson RC, Lester RL. Yeast sphingolipids. Biochim Biophys Acta. 1999;1426:347–357. doi: 10.1016/s0304-4165(98)00135-4. [DOI] [PubMed] [Google Scholar]
- Doering TL, Nosanchuk JD, Roberts WK, Casadevall A. Melanin as a potential cryptococcal defence against microbicidal proteins. Med Mycol. 1999;37:175–181. [PubMed] [Google Scholar]
- Feldmesser M, Kress Y, Novikoff P, Casadevall A. Cryptococcus neoformansis a facultative intracellular pathogen in murine pulmonary infection. Infect Immunol. 2000;68:4225–4237. doi: 10.1128/iai.68.7.4225-4237.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fischl AS, Liu Y, Browdy A, Cremesti AE. Inositolphosphoryl ceramide synthase from yeast. Meth Enzymol. 2000;311:123–130. doi: 10.1016/s0076-6879(00)11073-0. [DOI] [PubMed] [Google Scholar]
- Fostel JM, Lartey PA. Emerging novel antifungal agents. Drug Discov Today. 2000;5:25–32. doi: 10.1016/s1359-6446(99)01430-0. [DOI] [PubMed] [Google Scholar]
- Franzot SP, Mukherjee J, Cherniak R, Chen LC, Hamdan JS, Casadevall A. Microevolution of a standard strain of Cryptococcus neoformansresulting in differences in virulence and other phenotypes. Infect Immunol. 1998;66:89–97. doi: 10.1128/iai.66.1.89-97.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fries BC, Goldman DL, Cherniak R, Ju R, Casadevall A. Phenotypic switching in Cryptococcus neoformansresults in changes in cellular morphology and glucuronoxylomannan structure. Infect Immunol. 1999;67:6076–6083. doi: 10.1128/iai.67.11.6076-6083.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilchrest BA, Park HY, Eller MS, Yaar M. Mechanisms of ultraviolet light-induced pigmentation. Photochem Photobiol. 1996;63:1–10. doi: 10.1111/j.1751-1097.1996.tb02988.x. [DOI] [PubMed] [Google Scholar]
- Hannun YA, Luberto C. Ceramide in the eukaryotic stress response. Trends Cell Biol. 2000;10:73–80. doi: 10.1016/s0962-8924(99)01694-3. [DOI] [PubMed] [Google Scholar]
- Hashida-Okado T, Ogawa A, Endo M, Yasumoto R, Takesako K, Kato I. AUR1, a novel gene conferring aureobasidin resistance on Saccharomyces cerevisiae: A study of defective morphologies in Aur1p-depleted cells. Mol Gen Genet. 1996;251:236–244. doi: 10.1007/BF02172923. [DOI] [PubMed] [Google Scholar]
- Heidler SA, Radding JA. The AUR1 gene in Saccharomyces cerevisiaeencodes dominant resistance to the antifungal agent aureobasidin A (LY295337) Antimicrob Agents Chemother. 1995;39:2765–2769. doi: 10.1128/aac.39.12.2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Inositol phosphoryl transferases from human pathogenic fungi. Biochim Biophys Acta. 2000;1500:147–152. doi: 10.1016/s0925-4439(99)00097-6. [DOI] [PubMed] [Google Scholar]
- Howard DH. Some factors which affect the initiation of growth of Cryptococcus neoformans. J Bacteriol. 1961;82:430–435. doi: 10.1128/jb.82.3.430-435.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huffnagle GB, McNeil LK. Dissemination of C. neoformansto the central nervous system: Role of chemokines, Th1 immunity and leukocyte recruitment. J Neurovirol. 1999;5:76–81. doi: 10.3109/13550289909029748. [DOI] [PubMed] [Google Scholar]
- Jenkins GM, Richards A, Wahl T, Mao C, Obeid L, Hannun Y. Involvement of yeast sphingolipids in the heat stress response of Saccharomyces cerevisiae. J Biol Chem. 1997;272:32566–32572. doi: 10.1074/jbc.272.51.32566. [DOI] [PubMed] [Google Scholar]
- Kuroda M, Hashida-Okado T, Yasumoto R, Gomi K, Kato I, Takesako K. An aureobasidin A resistance gene isolated from Aspergillus is a homolog of yeast AUR1, a gene responsible for inositol phosphorylceramide (IPC) synthase activity. Mol Gen Genet. 1999;261:290–296. doi: 10.1007/s004380050969. [DOI] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Popkin TJ. Ultrastructure of septal complex in Filobasidiella neoformans (Cryptococcus neoformans) J Bacteriol. 1976;126:524–528. doi: 10.1128/jb.126.1.524-528.1976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon-Chung KJ, Polacheck I, Popkin TJ. Melanin-lacking mutants of Cryptococcus neoformansand their virulence for mice. J Bacteriol. 1982;150:1414–1421. doi: 10.1128/jb.150.3.1414-1421.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lester RL, Wells GB, Oxford G, Dickson RC. Mutant strains of Saccharomyces cerevisiaelacking sphingolipids synthesize novel inositol glycerophospholipids that mimic sphingolipid structures. J Biol Chem. 1993;268:845–856. [PubMed] [Google Scholar]
- Levine TP, Wiggins CAR, Munro S. Inositol phosphorylceramide synthase is located in the Golgi apparatus of Saccharomyces cerevisiae. Mol Biol Cell. 2000;11:2267–2281. doi: 10.1091/mbc.11.7.2267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, Tabuni A. Binding of Cryptococcus neoformansby human cultured macrophages. Requirements for multiple complement receptors and actin. J Clin Invest. 1991;87:528–535. doi: 10.1172/JCI115027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, Harrison TS, Tabuni A, Liu X. Chloroquine induces human mononuclear phagocytes to inhibit and kill Cryptococcus neoformansby a mechanism independent of iron deprivation. J Clin Invest. 1997;100:1640–1646. doi: 10.1172/JCI119688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitz SM, Nong S-H, Seeto KF, Harrison TS, Speizer RA, Simons ER. Cryptococcus neoformansresides in an acidic phagolysosome of human macrophages. Infect Immunol. 1999;67:885–890. doi: 10.1128/iai.67.2.885-890.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Tewari RP, Williamson PR. Laccase protects Cryptococcus neoformansfrom antifungal activity of alveolar macrophages. Infect Immunol. 1999;67:6034–6039. doi: 10.1128/iai.67.11.6034-6039.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge JK, Jackson-Machelski E, Toffaletti DL, Perfect JR, Gordon JI. Targeted gene replacement demonstrates that myristoyl-CoA: Protein N-myristoyltransferase is essential for viability of Cryptococcus neoformans. Proc Natl Acad Sci. 1994;91:12008–12012. doi: 10.1073/pnas.91.25.12008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandala SM, Thornton RA, Milligan J, Rosenbach M, Garcia-Calvo M, Bull HG, Harris G, Abruzzo GK, Flattery AM, Gill CJ, et al. Rustmicin, a potent antifungal agent, inhibits sphingolipid synthesis at inositol phosphoceramide synthase. J Biol Chem. 1998;273:14942–14949. doi: 10.1074/jbc.273.24.14942. [DOI] [PubMed] [Google Scholar]
- Moore TD, Edman JC. The alpha-mating type locus of Cryptococcus neoformanscontains a peptide pheromone gene. Mol Cell Biol. 1993;13:1962–1970. doi: 10.1128/mcb.13.3.1962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Lee SC, Casadevall A. Antibodies to Cryptococcus neoformansglucuronoxylomannan enhance antifungal activity of murine macrophages. Infect Immunol. 1995;63:573–579. doi: 10.1128/iai.63.2.573-579.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee S, Feldmesser M, Casadevall A. J774 murine macrophage-like cell interactions with Cryptococcus neoformansin the presence and absence of opsonins. J Infect Dis. 1996;173:1222–1231. doi: 10.1093/infdis/173.5.1222. [DOI] [PubMed] [Google Scholar]
- Nagiec RM, Wells GB, Lester RL, Dickson RC. A suppressor gene enables Saccharomyces cerevisiae to grow without making sphingolipids encodes a protein that resembles an Escherichia colifatty acyltransferase. J Biol Chem. 1993;268:22156–22163. [PubMed] [Google Scholar]
- Nosanchuk JD, Rosas AL, Casadevall A. The antibody response to fungal melanin in mice. J Immunol. 1998;160:6026–6031. [PubMed] [Google Scholar]
- Nosanchuk JD, Valadon P, Feldmesser M, Casadevall A. Melanization of Cryptococcus neoformansin murine infection. Mol Cell Biol. 1999;19:745–750. doi: 10.1128/mcb.19.1.745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Odom A, Muir S, Lim E, Toffaletti DL, Perfect J, Heitman J. Calcineurin is required for virulence of Cryptococcus neoformans. EMBO J. 1997;16:2576–2589. doi: 10.1093/emboj/16.10.2576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Lang SD, Durack DT. Chronic cryptococcal meningitis: A new experimental model in rabbits. Am J Pathol. 1980;101:177–194. [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Toffaletti DL, Rude TH. The gene encoding phosphoribosylaminoimidazole carboxylase (ADE2) is essential for growth of Cryptococcus neoformansin cerebrospinal fluid. Infect Immunol. 1993;61:4446–4451. doi: 10.1128/iai.61.10.4446-4451.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perfect JR, Wong B, Chang YC, Kwon-Chung KJ, Williamson PR. Cryptococcus neoformans: Virulence and host defences. Med Mycol. 1998;36:79–86. [PubMed] [Google Scholar]
- Polacheck I, Kwon-Chung KJ. Melanogenesis in Cryptococcus neoformans. J Gen Microbiol. 1988;134:1037–1041. doi: 10.1099/00221287-134-4-1037. [DOI] [PubMed] [Google Scholar]
- Polacheck I, Platt Y, Aronovitch J. Catecholamines and virulence of Cryptococcus neoformans. Infect Immunol. 1990;58:2919–2922. doi: 10.1128/iai.58.9.2919-2922.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhodes JC, Polacheck I, Kwon-Chung KJ. Phenoloxidase activity and virulence in isogenic strains of Cryptococcus neoformans. Infect Immunol. 1982;36:1175–1184. doi: 10.1128/iai.36.3.1175-1184.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salas SD, Bennett JE, Kwon-Chung KJ, Perfect JR, Williamson PR. Effect of the laccase gene CNLAC1, on virulence of Cryptococcus neoformans. J Exp Med. 1996;184:377–386. doi: 10.1084/jem.184.2.377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Schnitzler N, Peltroche-Llacsahuanga H, Bestier N, Zundorf J, Lutticken R, Haase G. Effect of melanin and carotenoids of Exophiala (Wangiella) dermatitidison phagocytosis, oxidative burst, and killing by human neutrophilis. Infect Immunol. 1999;67:94–101. doi: 10.1128/iai.67.1.94-101.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toffaletti DL, Rude TH, Johnston SA, Durack DT, Perfect JR. Gene transfer in Cryptococcus neoformansby use of biolistic delivery of DNA. J Bacteriol. 1993;175:1405–1411. doi: 10.1128/jb.175.5.1405-1411.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P, Heitman J. Signal transduction cascades regulating mating, filamentation, and virulence in Cryptococcus neoformans. Curr Opin Microbiol. 1999;2:358–362. doi: 10.1016/S1369-5274(99)80063-0. [DOI] [PubMed] [Google Scholar]
- Wang Y, Aisen P, Casadevall A. Cryptococcus neoformansmelanin and virulence: Mechanism of action. Infect Immunol. 1995;63:3131–3136. doi: 10.1128/iai.63.8.3131-3136.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickes BL, Edman JC. The Cryptococcus neoformans GAL7gene and its use as an inducible promoter. Mol Microbiol. 1995;16:1099–1109. doi: 10.1111/j.1365-2958.1995.tb02335.x. [DOI] [PubMed] [Google Scholar]
- Williamson PR, Wakamatsu K, Ito S. Melanin biosynthesis in Cryptococcus neoformans. J Bacteriol. 1998;180:1570–1572. doi: 10.1128/jb.180.6.1570-1572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wills EA, Rebindo MR, Perfect JR, Del Poeta M. New potential targets for antifungal development. Emerg Ther Targets. 2000;4:265–296. [Google Scholar]
- Yue C, Cavallo LM, Alspaugh JA, Wang P, Cox GM, Perfect JR, Heitman J. The STE12 α homolog is required for aploid filamentation but largely dispensable for mating and virulence in Cryptococcus neoformans. Genetics. 1999;153:1601–1615. doi: 10.1093/genetics/153.4.1601. [DOI] [PMC free article] [PubMed] [Google Scholar]