Abstract
Human parainfluenza virus type 2 (HPIV-2), an important pediatric respiratory pathogen, encodes a V protein that inhibits type I interferon (IFN) induction and signaling. Using reverse genetics, we attempted the recovery of a panel of V mutant viruses that individually contained one of six cysteine-to-serine (residues 193, 197, 209, 211, 214, and 218) substitutions, one of two paired charge-to-alanine (R175A/R176A and R205A/K206A) substitutions, or a histidine-to-phenylalanine (H174F) substitution. This mutagenesis was performed using a cDNA-derived HPIV-2 virus that expressed the V and P coding sequences from separate mRNAs. Of the cysteine substitutions, only C193S, C214S, and C218S yielded viable virus, and only the C214S mutant replicated well enough for further analysis. The H174F, R175A/R176A, and R205A/K206A mutants were viable and replicated well. The H174F and R205A/K206A mutants did not differ from the wild-type (WT) V in their ability to physically interact with MDA5, a cytoplasmic sensor of nonself RNA that induces type I IFN. Like WT HPIV-2, these mutants inhibited IFN-β induction and replicated efficiently in African green monkeys (AGMs). In contrast, the C214S and R175A/R176A mutants did not bind MDA5 efficiently, did not inhibit interferon regulatory factor 3 (IRF3) dimerization or IFN-β induction, and were attenuated in AGMs. These findings indicate that V binding to MDA5 is important for HPIV-2 virulence in nonhuman primates and that some V protein residues involved in MDA5 binding are not essential for efficient HPIV-2 growth in vitro. Using a transient expression system, 20 additional mutant V proteins were screened for MDA5 binding, and the region spanning residues 175 to 180 was found to be essential for this activity.
INTRODUCTION
Human parainfluenza viruses (HPIVs) are enveloped, nonsegmented, negative-stranded RNA viruses. The HPIVs belong to the Paramyxoviridae family, and HPIV type 1 (HPIV-1) and HPIV-3 are members of the genus Respirovirus, whereas HPIV-2 is a member of the genus Rubulavirus. These three HPIVs cause disease in humans, ranging from mild upper respiratory tract illness (URTI) to severe lower respiratory tract illness (LRTI), including croup, bronchiolitis, and pneumonia (35). HPIV-2 is an important cause of URTI, LRTI, and undifferentiated febrile illness in children, as well as of severe LRTI, with high mortality rates in transplant and other immunocompromised patients of all ages (16, 23, 45). Currently, no effective vaccines or antiviral therapies are licensed to prevent HPIV disease.
The HPIV-2 genome is 15,654 nucleotides in length and encodes 7 polypeptides from 6 genes arranged in the order 3′-N-P/V-M-F-HN-L-5′. The viral nucleocapsid protein (N), the phosphoprotein (P), and the large polymerase (L) protein direct transcription and replication (24). The fusion (F) and hemagglutinin-neuraminidase (HN) transmembrane glycoproteins are the major protective antigens that induce neutralizing antibodies, and the matrix protein (M) supports virion morphogenesis (35). The V protein is expressed from unedited P/V mRNA (illustrated in Fig. 1A), and two of its reported functions are to inhibit production of type I interferon (IFN) (48) and to block IFN signaling by targeting STAT2 for proteasomal degradation (1a, 2, 20, 21, 38, 41, 48, 67).
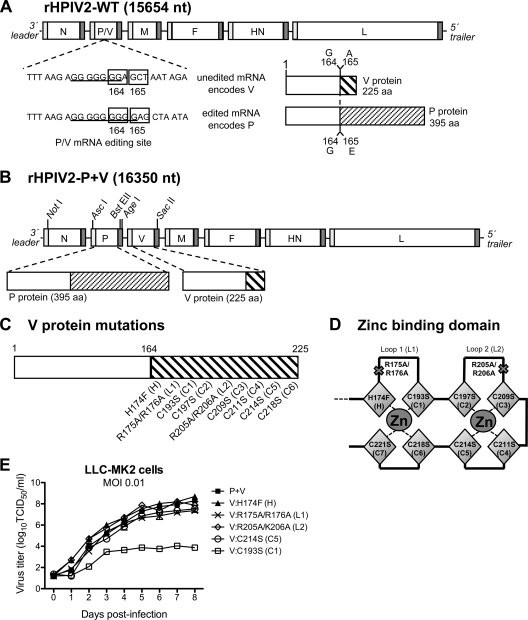
Fig. 1.
Generation and replication of cDNA-derived HPIV-2 with mutations in V only. (A) Schematic representation of the rHPIV-2-WT genome. The nucleotide sequence at the P/V mRNA editing site is shown for the unedited mRNA encoding V and the edited mRNA encoding P, with underlining denoting the seven (V mRNA) or nine (P mRNA) G residues. Codons 164 and 165 in each mRNA, marking the junction at which editing changes the reading frame, are boxed. Diagrams of the complete V and P proteins are shown to the right; as indicated, P and V share identical N-terminal sequences (164 amino acids in length) but have unique C-terminal sequences after the editing site. (B) Diagram of the rHPIV-2-P+V genome with separated P and V genes, which was used as the backbone to create the mutants described in the present study. The unique restriction enzyme sites used to insert mutated V open reading frames (ORFs) are indicated. (C) Using the HPIV-2-P+V cDNA, mutations were introduced separately into the C-terminal domain (CTD) of the HPIV-2 V protein encoded by the separated V-only gene unit, without affecting P. (D) Representation of the CTD zinc-finger fold predicted from the crystal structure of the PIV5 V protein (30). Two loops coordinate two zinc ions via amino acids H174 (H), C193 (C1), C197 (C2), C209 (C3), C211 (C4), C214 (C5), C218 (C6), and C221 (C7). (E) Multicycle replication in LLC-MK2 cells infected at an MOI of 0.01 TCID50/cell. Replication studies were performed in triplicate, and mean titers for each time point are indicated.
The deletion or mutation of virus-encoded IFN antagonists has been proposed as one strategy to develop live attenuated virus vaccines. This would be expected to restore an effective type I IFN response that would restrict vaccine virus replication and induce a protective immune response (10, 15, 57). In addition, increased expression of type I IFN has the potential to result in increased immunogenicity. The deletion of IFN antagonists, such as the NS1 protein of influenza virus or the NS1 and NS2 proteins of respiratory syncytial virus (RSV), has been shown to attenuate growth in vivo and to have potential as mutations in live virus vaccines (11, 18, 57, 58, 64, 65). We previously attempted to similarly attenuate HPIV-2 through the deletion of the IFN antagonist V protein. However, this recombinant HPIV-2 mutant, the rHPIV-2-Vko mutant, was overattenuated in vivo due to the strong induction of antiviral host cell responses, increased cytopathology, and highly restricted replication (53).
The overattenuation of rHPIV-2-Vko suggested that complete deletion of the HPIV-2 V protein would not be an effective strategy for developing a live attenuated vaccine candidate. Studies with model paramyxoviruses, including Sendai virus (SeV; also referred to as murine PIV1) and parainfluenza virus 5 (PIV5; previously known as simian virus 5 [SV5]) showed that, in addition to inhibiting the innate antiviral response, the V protein contributes to virus replication and pathogenesis by preventing apoptosis, regulating viral RNA synthesis, and supporting virion morphogenesis (7, 8, 20, 30, 46, 48, 62). Studies with HPIV-2, including the results with the rHPIV-2-Vko mutant described above, suggest that the V protein of HPIV-2 has similar activities (26, 53). Therefore, as an alternative strategy to the complete deletion of V, we sought to use site-directed mutagenesis to identify the functional domains of V, to dissociate its various activities individually, and to assess their contribution to the attenuation of HPIV-2 in vivo. We previously generated two recombinant strains of HPIV-2, the rHPIV-2-L101E/L102E and rHPIV-2-Δ122-127 mutants, with the indicated mutations in the V protein. We found that these mutations prevented HPIV-2 from inhibiting IFN signaling (52). The wild-type (WT) V protein (V-WT) of HPIV-2, like that of PIV5, engages STAT1 and STAT2 proteins in a cytoplasmic complex with the cellular E3 ubiquitin ligase DDB1, thereby promoting ubiquitination and proteasomal degradation of STAT2 (1a, 2, 20, 21, 38, 41, 48, 67). Our V mutants were unable to interact with DDB1 and therefore could not cause STAT2 degradation or inhibit IFN signaling (52). However, although both of these HPIV-2 V mutants permitted IFN signaling, they were not attenuated in vivo. This indicated that the loss of the ability of HPIV-2 to inhibit IFN signaling was insufficient to attenuate virus replication in vivo as long as IFN induction was still inhibited (52). Thus, it was necessary to identify mutations that target IFN production or possibly other activities of V to derive appropriately attenuated V mutant HPIV-2 viruses that could be used as live attenuated vaccines.
The paramyxovirus V protein has been proposed to prevent the induction of IFN by viral RNA through an interaction with the constitutively expressed, cytoplasmic RNA helicase MDA5 (1a, 5). Viral sensing by MDA5 or RIG-I initiates a common signaling cascade through the mitochondrial antiviral signaling protein MAVS (also known as IPS-1, Cardif, or VISA), leading to activation of transcription factors, such as interferon regulatory factor 3 (IRF3) and IRF7, as well as NF-κB and ATF-2/c-JUN. The V proteins of several paramyxoviruses, including HPIV-2, have been shown to inhibit IRF3 dimerization and NF-κB activation and to limit IFN-β promoter activation (20, 48, 66). In the context of virus infection, PIV5 and SeV mutants that lack either the entire V protein or its C terminus were unable to block IRF3 activation and subsequent IFN-β induction (20, 28). Furthermore, inhibition of IFN-β promoter activation was linked to an interaction between the C terminus of the V protein and MDA5—an interaction that is highly conserved among V proteins of paramyxoviruses, including HPIV-2, PIV5, SeV, bovine PIV3, mumps virus (MuV), measles virus (MeV), and Hendra virus (HeV) (1a, 3, 5, 27, 66). Additionally, recent studies have shown that the V protein appears to block double-stranded RNA (dsRNA) activation of MDA5 by binding the helicase domain of MDA5 and preventing its oligomerization (6, 44).
To test the hypothesis that cDNA-derived HPIV-2 mutants that cannot block IFN-β induction may be attenuated in vivo and to identify residues that could be deleted without being lethal to the virus, we targeted conserved residues in the C terminus of the HPIV-2 V protein for mutagenesis. These included six individual cysteine-to-serine substitutions (at residues 193, 197, 209, 211, 214, and 218), one histidine-to-phenylalanine substitution at position 174 (H174F), and two paired charge-to-alanine mutations (R175A/R176A and R205A/K206A). Two of these virus mutants, with substitutions at residues C214 or R175 and R176, were viable, were unable to bind to MDA5, and could not inhibit IRF3 dimerization or IFN-β induction as efficiently as the WT virus (HPIV-2-WT). Both mutants were attenuated in the respiratory tract of African green monkeys (AGMs). This suggested that the binding of the V protein to MDA5 and subsequent inhibition of downstream signaling contributes to HPIV-2 virulence in primates and identified mutations that could be used to make viable attenuated derivatives for vaccine purposes.
MATERIALS AND METHODS
Cell lines and viruses.
Human HEp-2 (ATCC CCL 23) and rhesus monkey LLC-MK2 (ATCC CCL 7.1) cells were maintained in Opti-MEM I (Invitrogen, Grand Island, NY) supplemented with 5% fetal bovine serum (FBS) and 50 μg/ml gentamicin sulfate. BHK-T7 baby hamster kidney cells, which constitutively express T7 RNA polymerase (4), were kindly supplied by Ursula Buchholz (NIAID) and were maintained in Glasgow minimal essential medium (GMEM) supplemented with 2 mM l-glutamine, 1× MEM amino acid solution (Invitrogen), 10% FBS, and 1 mg/ml Geneticin (Invitrogen). African green monkey Vero cells (ATCC CCL-81) were maintained in MEM (Invitrogen) supplemented with 10% FBS, 50 μg/ml gentamicin sulfate, and 4 mM l-glutamine. Human type II alveolar A549 cells (ATCC CCL-185) were maintained in F-12 nutrient mixture (Invitrogen) supplemented with 10% FBS, 50 μg/ml gentamicin sulfate, and 2 mM l-glutamine. Human embryonic kidney 293T cells were maintained in Dulbecco modified Eagle medium (DMEM) supplemented with 10% FBS and 50 μg/ml gentamicin sulfate.
The cDNA-derived recombinant HPIV-2 (rHPIV-2) mutants described here are based on the biologically derived HPIV-2 strain V9412-6 (V94), which was isolated from a nasal wash specimen of an infected infant and was kindly provided by Peter Wright of Vanderbilt University (55). The consensus sequence for the V94 strain was determined previously (GenBank accession number AF533010) (42). The recombinant wild-type virus, rHPIV-2-WT, was previously referred to as rV94(15T) and was derived from an antigenomic cDNA copy of the HPIV-2 V94 genome (42, 43, 55). An rHPIV-2 virus with separated P and V genes (rHPIV-2-P+V) was previously described (52). Recombinant vesicular stomatitis virus expressing green fluorescent protein (VSV-GFP) was provided by John Hiscott (56) and was propagated in Vero cells.
Mutations were introduced into the V gene by two-step PCR mutagenesis using the Advantage-HF PCR kit (Clontech Laboratories, Palo Alto, CA) and mutagenic primers. The mutated V gene was cloned into the AgeI and SacII sites of the rHPIV-2-P+V antigenomic sense cDNA (Fig. 1B). All regions of the mutated cDNAs that had been subjected to in vitro polymerase reactions were sequenced using a Perkin-Elmer ABI 3730 sequencer with the BigDye sequencing kit version 1.1 (Perkin-Elmer Applied Biosystems, Warrington, United Kingdom) as previously described (36).
Virus recovery and propagation.
rHPIV-2s were recovered from cDNA by reverse genetics in HEp-2 or BHK-T7 cells and propagated in LLC-MK2 cell monolayer cultures as described previously (43, 55). rHPIV-2 mutants were biologically cloned by serial terminal dilution on LLC-MK2 cells. To confirm that recovered recombinant viruses contained the introduced mutations, viral RNA was amplified by reverse transcriptase PCR (RT-PCR) using the SuperScript First-Strand synthesis system (Invitrogen) and Advantage-HF PCR kit (Clontech), as previously described (37, 55). Control reaction mixtures lacking RT confirmed that the PCR products were derived from RNA. Each virus preparation used in this study was sequenced in its entirety.
Purified virus stocks were obtained by infection of LLC-MK2 or Vero cells followed by centrifugation of culture supernatant containing virus in discontinuous 30%/60% (wt/vol) sucrose gradients. This was done to minimize contamination by cellular factors such as IFN. Virus titers were determined at 32°C by serial dilution on LLC-MK2 cells in 96-well plates using hemadsorption with guinea pig erythrocytes to detect infected cultures (19, 55). Titers are expressed as log10 TCID50/ml (50% tissue culture infectious dose/ml).
Kinetics of replication and temperature sensitivity of rHPIV-2 mutants.
Confluent monolayer cultures of LLC-MK2 cells in 6-well plates were infected in triplicate at a multiplicity of infection (MOI) of 0.01 or 5.0 TCID50/cell. After incubating for 1 h with virus, cultures were washed three times and incubated at 32°C. Aliquots of medium from each well were harvested and replaced with fresh medium at 24-hour intervals postinfection (p.i.). Virus present in each sample was quantified by serial dilution in 10-fold increments on 96-well LLC-MK2 cells, and infected monolayer cultures were detected by hemadsorption following a 7-day incubation.
Immunoblot analysis of protein expression.
Whole-cell lysates were prepared by lysing cells in lysis buffer containing 50 mM Tris-HCl (pH 8.0), 280 mM NaCl, 0.5% Igepal CA-360 (Sigma-Aldrich, St. Louis, MO), 10% glycerol, 0.2 mM EDTA, 2 mM EGTA, a Complete miniprotease inhibitor tablet (Roche, Indianapolis, IN), 50 mM NaF, and 1 mM Na3VO4 and clarifying by centrifugation. Lysates were heated to 70°C for 10 min in 1× NuPAGE LDS sample loading buffer (Invitrogen) and separated by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) in NuPAGE 4 to 12% Bis-Tris gels under denaturing and reducing conditions (Invitrogen). Proteins were transferred to Invitrolon polyvinylidene difluoride (PVDF) membranes (Invitrogen), blocked in phosphate-buffered saline (PBS) containing 0.1% Tween and 5% milk powder, and incubated with primary antibodies as indicated.
For native PAGE analysis of IRF3 monomers and dimers, LLC-MK2 cells were lysed in buffer containing 50 mM Tris-HCl (pH 8.0), 150 mM NaCl, 1% IGEPAL CA-360 (Sigma-Aldrich), 50 mM NaF, 5 mM Na3VO4, and a Complete miniprotease inhibitor tablet (Roche). Native PAGE was performed using 7% polyacrylamide gels and Tris-glycine running buffer (Bio-Rad, Hercules, CA) containing 0.2% sodium deoxycholate in the cathode chamber (60). Gels were first prerun for 30 min at 40 mA prior to electrophoresis of lysates at 25 mA.
Mouse monoclonal antibodies to the amino-terminal P/V common region (85A) (59) were kindly provided by Machiko Nishio of Mie University Graduate School of Medicine (Japan). Other primary antibodies used for immunoblotting were mouse monoclonal anti-glyceraldehyde-3-phosphate dehydrogenase (GAPDH)-peroxidase (GAPDH-71.1; Sigma-Aldrich), rabbit polyclonal anti-STAT2 (C-20; Santa Cruz Biotechnology), monoclonal anti-FLAG M2-peroxidase (Sigma), rabbit polyclonal anti-IRF3 (FL-425; Santa Cruz Biotechnology), and goat polyclonal anti-MDA5 (C-16; Santa Cruz Biotechnology). Proteins were detected by incubating membranes with horseradish peroxidase-conjugated donkey anti-goat IgG (Santa Cruz Biotechnology) and either goat anti-rabbit IgG (Kirkegaard & Perry Laboratories, Gaithersburg, MD) or goat anti-mouse IgG (Kirkegaard & Perry Laboratories) and were visualized using SuperSignal West Pico chemiluminescent substrate (Pierce, Rockford, IL).
Coimmunoprecipitation with myc–tagged V proteins.
The HPIV-2 V gene was codon optimized for efficient expression in mammalian cells, synthesized, and sequenced by GeneArt AG (Regensburg, Germany). The optimized V sequence, along with three sequential c-Myc epitope tags (MEQKLISEEDLHMH) at the N terminus, was cloned into the expression vector pcDNA3.1 (Invitrogen) to create pMyc-VWT. Mutations in the V gene were synthesized by GeneArt AG or were introduced by QuikChange XL site-directed mutagenesis (Stratagene, La Jolla, CA) using mutagenic primers and pMyc-VWT as a template. FLAG-MDA5 was kindly provided by Takashi Fujita (66).
Plasmids expressing Myc-tagged V proteins and FLAG-MDA5 were transfected into A549 cells using Lipofectamine 2000 (Invitrogen) according to the manufacturer's instructions. A549 cells were used because they transfect more efficiently than LLC-MK2 cells and are of human lung epithelial origin. A549 cells constitutively express MDA5, and endogenous MDA5 will coimmunoprecipitate with V-WT; MDA5 overexpression was used in these experiments to more easily visualize and compare immunoprecipitation results. Transfected cells were lysed at 48 h posttransfection in lysis buffer, and lysates were subjected to immunoprecipitation with anti-c-Myc agarose beads using the Pierce mammalian c-Myc tag IP/co-IP kit (Pierce Biotechnology) according to the manufacturer's instructions. c-Myc-glutathione S-transferase (GST) was included as a control (Pierce Biotechnology). Bound proteins were eluted by boiling beads in SDS gel loading buffer, treated with 100 mM dithiothreitol (DTT), and separated by SDS-PAGE. Immunoblot detection was performed as described above.
Real-time quantitative RT-PCR.
Cellular RNA was extracted using the RNeasy minikit (Qiagen), reverse transcribed using the high-capacity cDNA reverse transcription kit (Applied Biosystems, Foster City, CA), and amplified using TaqMan Universal PCR master mix (Applied Biosystems). The expression of rhesus monkey IFN-β mRNA (IFNB1, Rh03648734_s1) was analyzed relative to GAPDH mRNA (Rh02621745_g1) using TaqMan gene expression assays (Applied Biosystems). Samples were run on a 7900HT fast real-time PCR system (Applied Biosystems), and quantitative analysis was performed using SDS 2.3 and RQ Manager 1.2 (Applied Biosystems).
IFN-β ELISA.
Culture supernatant from LLC-MK2 cells was analyzed for IFN-β levels using the human IFN-β ELISA kit according to the manufacturer's protocol (Fujirebio, Inc., Tokyo, Japan). This enzyme-linked immunosorbent assay (ELISA) kit uses an antibody specific to the biologically active form of human IFN-β that cross-reacts with monkey IFN-β, which shares >95% amino acid identity with human IFN-β. We have previously determined in side-by-side comparisons that results using this ELISA correlate well with results from the standard type I IFN bioassay for rhesus LLC-MK2 IFN-β. Absorbance was measured using a microplate reader (VMax; Molecular Devices) and compared to a known concentration of IFN-β standard. IFN concentrations are expressed as means (pg/ml) ± the standard error (SE).
Type I IFN signaling assay.
Vero cells were infected with rHPIV-2 at a multiplicity of infection (MOI) of 5.0 TCID50/cell for 24 h. Cells were then left untreated or treated with 100 or 1,000 IU/ml IFN-β (Avonex; Biogen, Inc.) for 24 h prior to infection with VSV-GFP at an MOI of 0.0005 PFU/cell. VSV-GFP-infected cells were cultured under 0.8% methylcellulose overlay prepared in MEM and supplemented with 50 μg/ml gentamicin sulfate and 4 mM l-glutamine. Plates were read for GFP expression 48 h after VSV-GFP infection using a Typhoon 8600 scanner (Molecular Dynamics), and VSV-GFP-positive foci were counted (60).
Evaluation of virus replication in AGMs.
This study was performed under an animal study protocol approved by the NIAID animal care and use committee. HPIV-2-seronegative African green monkeys (AGMs) were inoculated both intranasally (IN) and intratracheally (IT) with 1 ml of L-15 medium per site containing 106.0 TCID50 of rHPIV-2. Nasopharyngeal (NP) swabs were collected on days 0 to 10 p.i., and tracheal lavage (TL) samples were collected on days 2, 4, 6, 8, and 10 p.i. Virus titers were determined by serial dilution of the samples on LLC-MK2 cells at 32°C. The mean peak virus titer was calculated using the highest titer detected in each animal in the group, irrespective of sampling day. The lower limit of detection was 0.5 log10 TCID50/ml. Twenty-eight days after immunization, the monkeys were challenged IN and IT with 1 ml (106.0 TCID50) per site with rHPIV-2-WT. NP and TL samples were collected on days 2, 4, 6, and 8 postchallenge; the virus titer was determined as described above. Sera were collected from the monkeys on days 0 and 28, and the hemagglutination inhibition (HAI) antibody titer was determined as previously described (61).
RESULTS
Construction and recovery of mutant rHPIV-2 viruses.
Nine mutations were introduced individually into the C-terminal domain (CTD) of the HPIV-2 V protein at positions that are highly conserved among all paramyxovirus V proteins: six individual cysteine-to-serine substitutions (residues 193, 197, 209, 211, 214, and 218, corresponding to C1 to C6 in the conserved motif), one histidine-to-phenylalanine substitution (H174F), and two paired charge-to-alanine mutations (R175A/R176A and R205A/K206A), as indicated in Fig. 1C. Their positions in a proposed zinc-binding domain based on PIV5 (29) are shown in Fig. 1D. Codons specifying the intended substitutions were chosen so that each differed by as many nucleotides as possible from the codon encoding the respective WT assignment. This was done to reduce the frequency of same-site reversion and to increase the genetic stability of each mutation so that it could be used as an attenuating mutation in a live attenuated HPIV-2 vaccine virus (32). Sequences for the original and mutant codons are shown in Table 1. These mutations were introduced into a previously described version of the HPIV-2 antigenomic cDNA (HPIV-2-P+V) in which the P and V coding regions were engineered to be expressed as separate mRNAs (Fig. 1B); this was done to permit stabilization of the V mutations without altering the coding sequence of the phosphoprotein P. From the nine mutated cDNAs, six mutant viruses could be recovered (Table 1). Viruses containing any of the six cysteine substitutions were difficult to recover from cDNA, yielding only three viable viruses (the C193S, C214S, and C218S mutants), i.e., viruses with a substitution of the first, fifth, or sixth cysteine residue of the conserved motif (C1, C5, and C6, respectively). The other three mutants (the H174F, R175A/R176A, and R205A/K206A mutants) were successfully recovered. The genomes of the recovered viruses were sequenced in their entirety at passage level six or seven to confirm the presence of the engineered mutations and the absence of adventitious mutations. Each of the three recovered cysteine mutants had acquired one or more adventitious mutations during rescue and/or early passage (Table 1), including one substitution in V (T141P) in the C214S mutant. Both charge-to-alanine mutants (R175A/R176A and R205A/K206A) as well as the H174F mutant were free of adventitious mutations.
Table 1.
Recovery of rHPIV-2 V mutants
| V protein or V protein mutation (residue) | Codona |
No. of transfections yielding virus/no. of attempts (% frequency of virus recovery)b | Adventitious mutations in recovered virusc | In vitro growthd | |
|---|---|---|---|---|---|
| Wild type | Mutant | ||||
| WT | 34/34 (100) | None | ++++ | ||
| Zinc-binding motif | |||||
| H174F (H) | CAT | TTT | 1/14 (7) | None | ++++ |
| C214S (C5) | TGC | TCA | 1/22 (5) | V:T141P, M poly(A) | +++ |
| C193S (C1) | TGT | TCA | 1/8 (13) | M:V125E, L:G1888V | + |
| C218S (C6) | TGC | TCA | 1/17 (6) | P start signal | +/− |
| C197S (C2) | TGT | TCA | 0/16 (0) | − | − |
| C209S (C3) | TGC | TCA | 0/11 (0) | − | − |
| C211S (C4) | TGT | TCA | 0/12 (0) | − | − |
| Charge to alanine | |||||
| R175A/R176A (L1) | AGG-AGA | GCT–GCT | 1/2 (50) | None | +++ |
| R205A/K206A (L2) | CGC-AAG | GCT–GCT | 1/2 (50) | None | ++++ |
Nucleotides that differ from any codon specifying the wild-type assignment are underlined.
The number of transfections that resulted in recovery of mutant virus bearing the indicated mutation out of the total number of attempts in which the WT control virus was recovered in parallel.
Each recovered virus was sequenced in its entirety. Adventitious mutations are identified for the virus pool used in all subsequent experiments, sequenced at passage 6 or 7. Viruses that were not recovered are indicated by “−.”
Relative efficiency of growth in vitro for each mutant virus. + to ++++, relative growth rates ranging from very poor to very good growth. +/−, virus was recovered but could not be propagated. Viruses that were not recovered are indicated by “−.”
Replication of mutant rHPIV-2 viruses in vitro.
Multicycle replication of each mutant was assessed in LLC-MK2 cells, in which HPIV-2-WT replicates very efficiently. Upon infection at a low MOI (0.01 TCID50/cell), multicycle replication of the C218S mutant was below the limit of detection (data not shown), while replication of the C193S mutant was significantly reduced (>1,000-fold-lower peak titer compared to that of rHPIV-2-P+V) (Fig. 1E). The poor replication of the C218S and C193S mutants prevented further analysis of these viruses. The remaining mutants replicated to peak titers that were comparable to that of the rHPIV-2-P+V parent (Fig. 1E). These four mutants were also evaluated in single-step growth curves (see Fig. 3C), and their replication was equivalent to that of rHPIV-2-WT and rHPIV-2-P+V. None of the V mutants examined here induced cell death, in contrast to previous observations with the V knockout mutant rHPIV-2-Vko (data not shown) (53). Elevated temperatures to 39°C (the core body temperature of AGMs) did not affect the replication of the WT or mutant viruses in vitro (data not shown).
Fig. 3.
IFN-β induction during infection with rHPIV-2 in vitro. (A) Induction of IFN-β mRNA in HPIV-2-infected LLC-MK2 cells. Cells were infected at an MOI of 5.0 TCID50/cell and lysed on days 1 to 4 postinfection. IFN-β expression was measured by real-time RT-PCR. Expression values were first normalized against the GAPDH endogenous control and then normalized against values for mock-infected samples to obtain relative gene expression ratios. The mean values for 2 or 3 wells per virus ± SE are shown. Asterisks indicate samples in which IFN-β mRNA expression was significantly higher than those in WT- and rHPIV-2-P+V-infected samples at the same time point (** indicates a P value of <0.001; * indicates a P value of <0.01, two-way analysis of variance [ANOVA] with Bonferroni posttest). (B) Concentration of IFN-β in the supernatant of LLC-MK2 cells infected at an MOI of 5.0 TCID50/cell. Culture media were harvested and replaced with fresh media at days 1 to 5 postinfection, and aliquots were analyzed to determine ELISA IFN-β levels. Bar graphs show IFN-β concentrations, which were determined by comparison with an IFN-β standard curve and are expressed in pg/ml ± SE based on results for triplicate samples from two independent experiments. Asterisks indicate samples in which IFN-β levels were significantly higher than those in WT- and rHPIV-2-P+V-infected culture supernatants at the same time point (** indicates a P value of <0.001; * indicates a P value of < 0.05, two-way ANOVA with Bonferroni posttest). The lower limit of detection was 12.5 pg/ml. (C) Single-cycle replication in LLC-MK2 cells infected at an MOI of 5.0 TCID50/cell. Replication studies were performed in triplicate, and aliquots were taken at 24-h intervals. The mean titer for each time point is indicated.
Evaluation of IRF3 dimerization and IFN-β induction.
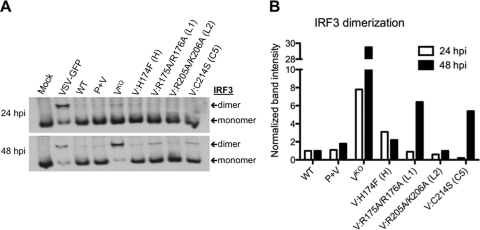
The V protein has been shown to prevent formation of IRF3 dimers, the active form of IRF3 that translocates into the nucleus to activate the IFN-β promoter (20, 48, 66). Therefore, we next examined whether the V protein CTD mutations altered the ability of HPIV-2 to inhibit dimerization of IRF3 and subsequent induction of IFN-β. Following virus infection of LLC-MK2 cells at an MOI of 5.0 TCID50/cell, mock-infected or virus-infected cells were lysed at 24 and 48 h p.i., and the lysates were subjected to native PAGE to separate dimeric and monomeric IRF3. Vesicular stomatitis virus expressing green fluorescent protein (VSV-GFP) was included as a positive control because it is a known inducer of IRF3 dimerization (56), and both monomeric IRF3 and dimeric IRF3 were detected in VSV-infected samples, as shown in Fig. 2A. In comparison, rHPIV-2-WT and rHPIV-2-P+V induced only low levels of IRF3 dimerization, demonstrating the ability of HPIV-2 to efficiently inhibit dimerization of IRF3. In contrast, the rHPIV-2-Vko mutant that does not express the V protein (53) induced levels of IRF3 dimerization that were almost 8-fold higher than that of the WT at 24 h p.i. and 30-fold higher than that of the WT at 48 h p.i. (Fig. 2B). All four V CTD mutants induced very little IRF3 dimerization at 24 h p.i., but at 48 h p.i., both the R175A/R175A and C214S mutants permitted approximately 3-fold more IRF3 dimerization than their P+V parent (Fig. 2B). Although IRF3 dimerization in response to the R175A/R176A and C214S mutants was approximately 5-fold less than that of rHPIV-2-Vko (Fig. 2B), both mutants were observed to be less effective than the WT at preventing IRF3 dimerization in two independent experiments. In contrast, there was little or no IRF3 dimerization in response to the H174F and R205A/K206A mutants, indicating that these mutants are as effective as the WT in preventing IRF3 dimer formation and suggesting that these residues may be less important in maintaining V protein control of IRF3 activation.
Fig. 2.
IRF3 dimerization during infection with rHPIV-2 in vitro. (A) IRF3 dimerization in HPIV-2-infected LLC-MK2 cells. Cells were infected at an MOI of 5.0 TCID50/cell and lysed at 24 and 48 h p.i. Cell lysates were resolved by native PAGE, and immunoblotting was used to identify both monomeric and dimeric forms of IRF3, as indicated by arrows. Immunoblots shown are representative of two independent experiments. (B) Relative levels of IRF3 dimerization from the immunoblot shown in panel A are indicated, with IRF3 dimer band intensity indicated relative to the IRF3 dimer band in the WT lane for each time point. Band quantification was performed on scanned immunoblot images using the ImageJ gel analysis tool (1).
To determine whether IRF3 dimerization was associated with IFN-β induction, we measured both IFN-β mRNA in LLC-MK2 cell lysates as well as IFN-β protein in the medium supernatants (Fig. 3). Cells were infected at an MOI of 5.0 TCID50/cell, and RNA was isolated on days 1 to 4 p.i. IFN-β (IFNB1) mRNA abundance was compared between lysates relative to the amount of GAPDH mRNA present in each sample. The level of IFNB1 mRNA is expressed as the fold increase over that detected in mock-infected samples from each time point (Fig. 3A). As expected, IFNB1 mRNA levels were significantly elevated in cells infected with the Vko mutant, with peak abundance at 48 h p.i. In the R175A/R176A- and C214S-mutant-infected cells, but not the H174F- and R205A/K206A-mutant-infected cells, IFNB1 expression was significantly higher (>2.5-fold at 48 h p.i., P < 0.001) than in cells infected with the P+V parent virus (Fig. 3A).
The pattern of IFN-β concentrations in the supernatant of infected LLC-MK2 monolayers was consistent with that observed for IRF3 dimerization and IFNB1 mRNA abundance (Fig. 3B). In comparison to IFN-β induction by rHPIV-2-Vko, which peaked on day 2 p.i., IFN-β induction by the H174F and R205A/K206A mutants was delayed and not significantly different from that of rHPIV-2-WT or rHPIV-2-P+V. In contrast, the R175A/R176A and C214S mutants induced levels of IFN-β that were significantly higher than those induced by rHPIV-2-WT or rHPIV-2-P+V (P < 0.01), with peak IFN-β expression at day 3 p.i. (165 and 325 pg/ml, respectively) (Fig. 3B). To confirm that the differences in IFN production were not the result of differences in replication, we also measured virus titers in supernatants. All of the V CTD mutants reached titers equivalent to those for rHPIV-2-WT and rHPIV-2-P+V, whereas the rHPIV-2-Vko titers were approximately 1,000-fold reduced (Fig. 3C).
Evaluation of type I IFN signaling.
To determine whether infection with the V CTD mutants affected type I IFN signaling and the establishment of an antiviral state, we performed an IFN signaling assay in which Vero cells were mock or rHPIV-2 infected for 24 h, treated with IFN-β for 24 h, and infected with VSV-GFP under methylcellulose overlay for 48 h. Vero cells were used for this assay because they readily respond to exogenous IFN-β but are unable to generate their own IFN due to a deletion in the IFN genes (9, 34, 63). If an rHPIV-2 mutant retains its ability to block IFN signaling, VSV-GFP will replicate efficiently. Conversely, if an HPIV-2 mutant lacks the ability to inhibit IFN signaling and prevent the induction of an antiviral state, the number of VSV-GFP foci will be reduced. We have previously shown that rHPIV-2-Vko and a substitution mutant, rHPIV-2-L101E/L102E, which bears an amino-terminal mutation in V that disrupts the V-DDB1 interaction and prevents STAT2 degradation, were both unable to block IFN signaling (52, 53).
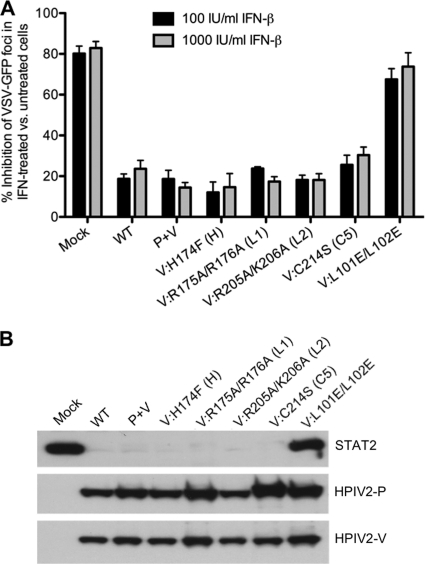
In mock-infected Vero cells, the addition of IFN-β at a concentration of 100 or 1,000 IU/ml led to an 80% decrease in the number of VSV-GFP foci (Fig. 4A), indicating that VSV-GFP replication was hindered by the establishment of an antiviral state. In addition to the reduced number of plaques, the average plaque size of the remaining plaques was reduced by approximately 85% in IFN-treated wells compared to untreated wells (not shown). In contrast, the addition of IFN-β to Vero cells infected with rHPIV-2-WT or rHPIV-2-P+V decreased the number of VSV-GFP foci by only 20% and did not reduce plaque size, demonstrating the ability of WT HPIV-2 to block IFN-β signaling. Similarly, the addition of IFN-β to cells infected with any of the V CTD mutants also decreased the number of VSV-GFP foci by no more than approximately 20%, indicating that these mutants do not interfere with HPIV-2's ability to control IFN-β signaling. In contrast, the previously described mutant that is unable to interact with DDB1 to degrade STAT2 (52), rHPIV-2-L101E/L102E, could not block IFN signaling in this assay (Fig. 4A).
Fig. 4.
Type I IFN signaling and STAT2 degradation. (A) Vero cells in six-well plates were mock infected or infected with the indicated viruses at an MOI of 5.0 TCID50/cell for 24 h. Cells were then left untreated or treated with 100 or 1,000 IU/ml IFN-β. Twenty-four hours after IFN-β treatment, the cells were infected with VSV-GFP and incubated for 48 h under methylcellulose. A phosphorimager was used to visualize the VSV-GFP foci. The graph represents the percent reduction of VSV-GFP foci (± SE) in IFN-β-treated versus untreated cells based on results of three independent experiments. (B) LLC-MK2 cells infected at a high MOI (5.0 TCID50/cell) were lysed at day 5 p.i. and analyzed by SDS-PAGE with immunoblotting for endogenous cellular STAT2 and HPIV-2 P and V proteins.
Since the HPIV-2 V protein is known to target STAT2 for proteasomal degradation (2, 38, 41), we next confirmed that this function was unaltered in the V CTD mutants by immunoblotting infected Vero cell lysates to detect expression of STAT2 (Fig. 4B). Consistent with the results obtained in the IFN signaling assay, infection with rHPIV-2-WT, rHPIV-2-P+V, or any of the V CTD mutants caused efficient degradation of endogenous STAT2 (Fig. 4B). In contrast, cells infected with rHPIV-2-L101E/L102E, the amino-terminal mutant noted above that is unable to block IFN signaling, accumulated as much STAT2 as mock-infected cells (Fig. 4B). Taken together, these data indicate that the V CTD mutants inhibit IFN-β signaling and induction of an antiviral state with efficiency similar to that of HPIV-2-WT.
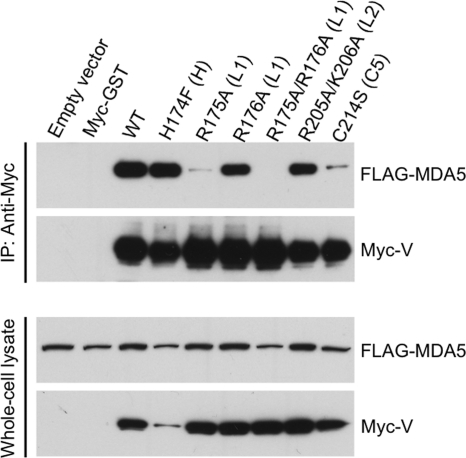
V protein mutations disrupt binding to MDA5.
Since the HPIV-2 V protein is known to interact with the cellular dsRNA sensor MDA5 (1a, 3, 5, 6, 27, 44, 66), we attempted to coimmunoprecipitate MDA5 with either WT or mutant V protein. A549 cells were transfected with plasmids expressing FLAG-tagged MDA5 and Myc-tagged WT or mutant V protein, immunoprecipitation was performed with anti-Myc antibody, and the immunoprecipitates were analyzed by Western blotting. As expected, the WT V protein efficiently coimmunoprecipitated MDA5 (Fig. 5). Two mutants, the H174F and R205A/K206A mutants, also efficiently coimmunoprecipitated MDA5. However, the R175A/R176A V protein was completely unable to coimmunoprecipitate MDA5, and the C214S V protein was much less able to coimmunoprecipitate MDA5 compared to WT V protein. The plasmid encoding the C214S V protein mutant did not contain the adventitious mutation in V (T141P) that was present in the recombinant C214S mutant virus, and therefore, only the effect of the C214S mutation is evaluated here. To determine whether the R175A and R176A substitutions each contributed to the loss of interaction with MDA5, plasmids expressing each individual substitution were tested. The R175A substitution eliminated most of the V-MDA5 coimmunoprecipitation, while the R176A substitution had little effect. Therefore, it appears that the R175A mutation is responsible for the phenotype of the R175A/R176A mutant, with little contribution from R176A.
Fig. 5.
Coimmunoprecipitation of MDA5 and the HPIV-2 V protein. A549 cells transiently expressing both FLAG-tagged MDA5 and Myc-tagged WT or mutant HPIV-2 V protein bearing the C-terminal mutations indicated were lysed at 48 h posttransfection and subjected to immunoprecipitation (IP) with antibodies specific to the Myc tag. Myc-immunoprecipitated proteins or total cell lysates were separated by SDS-PAGE and processed for immunoblotting with antiserum against MDA5 or HPIV-2 V protein, as indicated. Lysates from cells transfected with empty vector or with vector expressing Myc-tagged GST protein served as negative controls. Results are representative of several independent experiments.
Evaluation of replication of rHPIV-2 V mutants in vivo.
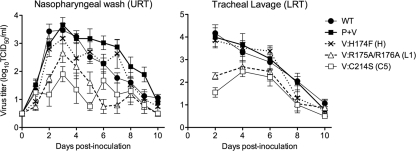
The H174F, R175A/R176A, and C214S mutant viruses were compared to the rHPIV-2-WT and rHPIV-2-P+V parent viruses for the ability to replicate in the respiratory tract of HPIV-2-seronegative AGMs. The R205A/K206A mutant was not evaluated with AGMs because its in vitro phenotype appeared unlikely to confer attenuation in vivo. AGMs were infected intranasally (IN) and intratracheally (IT) with 106.0 TCID50 per site of the indicated virus. Virus titers were determined in nasopharyngeal (NP) swab samples representing the upper respiratory tract (URT) and in tracheal lavage (TL) samples representing the lower respiratory tract (LRT). The H174F mutant replicated as efficiently at both sites as the rHPIV-2-WT and rHPIV-2-P+V parents (Fig. 6 and Table 2). In contrast, the mutants that were unable to physically interact with MDA5, namely, the R175A/R176A and C214S mutants, were restricted in replication in both the URT and the LRT of AGMs (Fig. 6). Compared to that of rHPIV-2-P+V, the mean peak titers in R175A/R176A- and C214S-mutant-infected monkeys were 10- to 40-fold lower in the URT and 40- to 60-fold lower in the LRT (Table 2). The possibility that mutations or reversions occurred during replication in vivo was excluded by sequencing virus isolated in respiratory specimens from each animal late in the course of infection (days 7 to 10). All virus isolates retained the introduced mutations in P and V. It can therefore be concluded that the mutations introduced into the CTD of V were able to attenuate HPIV-2 in the respiratory tract of nonhuman primates if and only if the mutation disturbed the ability of V to physically interact with MDA5 as assayed by coimmunoprecipitation.
Fig. 6.
HPIV-2 replication in the respiratory tract of AGMs. AGMs were inoculated IN and IT with 106.0 TCID50 of each virus. Mean daily virus titers ± SE in nasopharyngeal swabs (upper respiratory tract [URT]) and tracheal lavage fluid (lower respiratory tract [LRT]) were determined for each sampling day. The limit of detection was 0.5 log10 TCID50/ml.
Table 2.
Replication, immunogenicity, and protective efficacy in AGMs
| Immunizing virus (residue) | No. of animalsa | Mean peak virus titer (log10 TCID50/ml ± SE)b |
HAI antibody titer (reciprocal mean log2 ± SE)c | No. of animals challengedd | Mean peak challenge virus titer (log10 TCID50/ml ± SE)e |
||
|---|---|---|---|---|---|---|---|
| URT | LRT | URT | LRT | ||||
| rHPIV-2-WT | 21 | 3.9 ± 0.2 | 4.8 ± 0.3 | 5.6 ± 0.8 | 3 | ≤0.5 ± 0.0 | 0.8 ± 0.3 |
| rHPIV-2-P+V | 10 | 3.9 ± 0.2 | 4.4 ± 0.5 | 4.4 ± 0.5 | 2 | ≤0.5 ± 0.0 | 1.3 ± 0.8 |
| rHPIV-2 V H174F mutant (H) | 4 | 3.6 ± 0.3 | 3.9 ± 0.3 | 5.8 ± 0.3 | 0f | ND | ND |
| rHPIV-2 V R175A/R176A (L1) | 4 | 2.3 ± 0.2h | 2.8 ± 0.2 | 4.0 ± 0.0 | 4 | 0.6 ± 0.1 | 0.6 ± 0.1 |
| rHPIV-2 V C214S (C5) | 4 | 2.9 ± 0.2 | 2.6 ± 0.2 | 4.5 ± 0.3 | 0f | ND | ND |
| Control (L15)g | 8 | ≤0.5 ± 0.0 | ≤0.5 ± 0.0 | ≤1.0 ± 0.0 | 8 | 3.9 ± 0.4 | 4.6 ± 0.2 |
Total number of animals from several independent experiments.
AGMs in groups of 2 to 4 were immunized IN and IT on day 0 with 106.0 TCID50 of the indicated virus at each site. Nasopharyngeal swabs (upper respiratory tract [URT]) were collected on days 1 to 10 postinfection, and tracheal lavage samples (lower respiratory tract [LRT]) were collected on days 2, 4, 6, 8, and 10 postinfection. Virus titers were determined by limiting dilution on LLC-MK2 cells at 32°C. SE, standard error.
Mean serum HAI antibody titer from samples collected on day 28 postimmunization.
On day 28 after the initial immunization, the indicated number of monkeys were challenged IN and IT with 106.0 TCID50/ml of HPIV-2-WT per site.
URT (nasopharyngeal swabs) and LRT (tracheal lavage) samples were collected on days 2, 4, 6, and 8 postinfection, and virus titers were determined by serial dilution on LLC-MK2 cells at 32°C. The mean of the peak virus titers for the individual animals in each group irrespective of sampling day is shown. The lower limit of detection was 0.5 log10 TCID50/ml.
These groups did not receive challenge virus.
Control animals were challenged with HPIV-2-WT without receiving an initial immunization. ND, not done.
Replication of the viruses for which data are underlined was significantly reduced compared to that of rHPIV-2-WT by the use of the Neuman-Keuls multiple-comparison test, with a P value of <0.05.
In order to assess whether the attenuated HPIV-2 mutants were immunogenic and suitable for evaluation as vaccines, the serum antibody response following infection was assessed in a hemagglutination inhibition assay (HAI). Despite their restricted replication, R175A/R176A and C214S induced HAI titers similar to that of the P+V parent virus (Table 2). In addition, immunization with the R175A/R176A mutant restricted replication of the HPIV-2-WT challenge virus, administered 4 weeks postimmunization, as much as prior infection with rHPIV-2-WT or rHPIV-2-P+V (Table 2). The H174F and C214S mutants were not evaluated for protective efficacy because the lack of attenuation and the presence of adventitious mutations, respectively, ruled out their use as vaccines.
Identification of additional HPIV-2 V protein residues critical for physical interaction with MDA5.
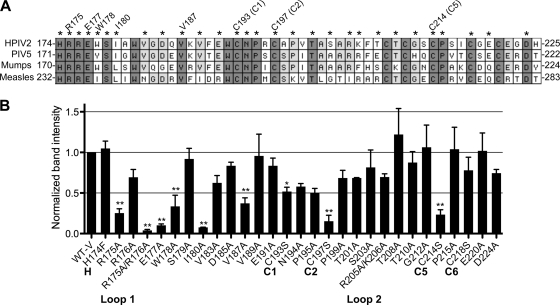
To identify additional V protein residues that may be involved in binding MDA5, we attempted to coimmunoprecipitate MDA5 with either WT V protein or with each of 29 different V protein mutants containing one or two different amino acid substitutions in its CTD. Mutated residues are indicated by asterisks in Fig. 7A; each was present as a single amino acid substitution, except for two paired substitutions: R175A/R176A and R205A/K206A. As in the experiment described for Fig. 5, A549 cells were transfected with plasmids expressing MDA5-FLAG and Myc-V and subjected to immunoprecipitation with anti-c-Myc antibody on agarose beads. Myc-immunoprecipitated proteins were separated by SDS-PAGE, and MDA5 binding was detected with anti-FLAG antibodies. Quantification of the MDA5 band intensity in each immunoprecipitation was used as a surrogate for the relative strength of binding of MDA5 to each mutant V protein, expressed as the fold difference over the intensity of the MDA5 band that coprecipitated with V-WT in each experiment. The mean data of two or more independent experiments for each mutant V protein are shown in Fig. 7B. Three Cys-to-Ser substitutions, at positions 193, 197, and 214 (C1, C2, and C5, respectively), significantly reduced MDA5 binding, i.e., reduced the intensity of the coimmunoprecipitated MDA5 band to 52%, 15%, and 23%, respectively. The substitution at position 218 (C6) did not have a significant effect on MDA5 binding (the MDA5 band intensity was an average of 78% of that seen with V-WT). The other Cys residues were not examined in this experiment. In addition to the conserved Cys residues, the region spanning residues 175 to 187 included several residues that are important for MDA5 binding, specifically the individual amino acids R175, E177, W178, I180, and V187 and the paired set of R175/R176. Each of these substitutions reduced the intensity of the coimmunoprecipitated MDA5 band to less than 40% of that seen with V-WT. The clustering of residues implicated in MDA5 binding suggests that this region (amino acids [aa] 175 to 180 or 187) may represent the binding site for MDA5 on the HPIV-2 V protein.
Fig. 7.
Association of HPIV-2 V protein CTD mutants with MDA5. (A) An alignment of the V-specific regions of four paramyxoviruses is shown, with amino acids numbered from the amino terminus of each V protein. Dark shading indicates amino acid residues that are 100% conserved; light shading indicates residues that are 75% conserved. Mutated residues are indicated by asterisks above the alignment. Residues that appear to be involved in MDA5 binding are indicated by annotations above the relevant amino acids. (B) Lysates of A549 cells transiently expressing both FLAG-tagged MDA5 and Myc-tagged WT or mutant HPIV-2 V protein bearing the indicated C-terminal mutations were subjected to immunoprecipitation with antibodies specific to the Myc tag as described in the legend to Fig. 5. Myc-immunoprecipitated proteins were separated by SDS-PAGE and processed for immunoblotting with antiserum for MDA5 and HPIV-2 V proteins. Relative values for binding of MDA5 to mutant V protein at each time point are normalized to the intensity of the MDA5 band coprecipitated with V-WT for each experiment. Band quantification was performed with scanned immunoblot images using the ImageJ gel analysis tool (1). Two or more independent experiments were performed for each mutant V protein, and the mean relative binding for each is shown ± SE. Asterisks indicate that binding of MDA5 to the indicated mutant V protein was significantly reduced compared to MDA5 binding to the V-WT using the Tukey-Kramer multiple-comparison test (** indicates a P value of <0.001; * indicates a P value of <0.01).
DISCUSSION
In this study, we sought to determine the effects of mutations in the CTD of the HPIV-2 V protein on its ability to bind MDA5, to function as an IFN antagonist, and to attenuate HPIV-2 to a degree suitable for use in a live attenuated vaccine. We hypothesized that mutation of the IFN antagonist V protein might increase expression of type I IFN in response to infection, which would be expected to induce a protective immune response and restrict virus replication. We were particularly interested in attenuating mutations that did not restrict replication in vitro, since that is essential for the manufacture of live vaccines. In addition to evaluating V mutations in live virus, we examined a larger panel of V protein mutants in a transient expression system to map residues involved in MDA5 binding.
We recovered four mutant viruses from cDNA that replicated well in vitro. Two of them, encoding a substitution of the fifth cysteine residue (C5, Cys-214) and the first pair of charged residues in the CTD (R175/R176), were attenuated enough in nonhuman primates to be considered candidate vaccines that deserve evaluation in human subject trials. Both of these attenuated mutants failed to bind MDA5 efficiently and failed to inhibit IFN-β induction, but both inhibited IFN signaling as efficiently as WT HPIV-2. However, the C214S mutant will likely not be considered due to the difficulty in recovering this virus and the presence of adventitious mutations, including one in V (T141P).
The V protein CTD contains several highly conserved residues that are thought to be essential for V protein IFN antagonist function. Among them are one histidine (H) and seven cysteine (C) residues that form a type of zinc finger resembling a RING domain. For PIV5, structural data indicated that the first histidine (residue H174 in HPIV-2) and C1, -6, and -7 coordinate one zinc atom (loop 1), while C2, -3, -4, and -5 are part of a smaller loop that coordinates a second zinc atom (loop 2) (Fig. 1D) (29). Nishio et al. had previously generated HPIV-2 mutants with Cys-to-Ala substitutions at C1+C2, C3+C4, and C6 (41). Although these mutants could be recovered from cDNA, they were highly restricted in replication in vitro (40, 41). In contrast, several of our mutants bearing individual Cys-to-Ser substitutions could not be recovered from cDNA at all (C2, C3, and C4), and none were recovered without adventitious mutations. Although we did not investigate whether these adventitious mutations were compensatory, the difficulty of recovering these mutants suggests that these substitutions conferred a more deleterious phenotype than the previously reported mutations. In our system, individual substitution of C2, -3, and -4 (loop 2) was lethal, and substitution of C1 and C6 (in loop 1 but adjacent to loop 2; Fig. 1D) restricted in vitro replication significantly. One possible explanation for the difference in the observed lethality of substitutions in loop 2 is that we introduced individual substitutions only, which may be more disruptive structurally than substitutions of Cys pairs, as reported by Nishio et al. (41). In this regard, we note that deletion of the entire V protein (rHPIV-2-Vko) (53) was tolerated more easily in our hands than substitutions of most of the individual cysteines, suggesting that the latter created a lethal gain-of-function phenotype, perhaps by enhancing binding of V mutants to a cellular or a viral protein, such as has been reported for the binding of WT HPIV-2 V to the viral N or L protein (39). Thus, for example, one could speculate that mutant V might bind more strongly to the L protein than WT V and thereby inhibit the polymerase function to a degree that is not compatible with virus rescue from cDNA, although this was not further examined. Future studies of V mutants that are under consideration as vaccine candidates will evaluate V protein's ability to interact with viral N and L proteins.
During preparation of the manuscript, Ramachandran and Horvath reported that mutation of C2 through C5 in the PIV5 V protein disrupted MDA5 binding, which is consistent with our observations for HPIV-2 (49). The rescue of PIV5 V protein mutant viruses from cDNA was not reported in that study, so the lethality of those mutations cannot be compared to ours. The substitution of the C1, C6, and C7 in PIV5 V had no effect on binding to MDA5, but in contrast, C1 and C2 were required for the MDA5 binding of measles and mumps virus V proteins. This indicated that some of the binding requirements are virus specific. While our data agree with the PIV5 V protein data with regard to the role of C2 and C5 in MDA5 binding, we observed that for HPIV-2, C1 was also important for MDA5 binding. Thus, in regard to MDA5 binding, HPIV-2 more closely resembled measles and mumps viruses, even though the HPIV-2 V protein is more closely related to the PIV5 V protein (44% amino acid identity) than to the measles and mumps V proteins (13% and 35% identity, respectively). Although we were unable to recover virus mutants expressing a C2, -3, or -4 substitution, we were able to recover a C5 mutant (i.e., a loop 2 mutant). It is noteworthy that transient expression assays are not necessarily predictive of the phenotype of a given mutation in infectious virus. For example, while a mutant of C1 did not bind MDA5 well and also was impaired for replication in vitro, a mutant of C6 that bound MDA5 efficiently also was, nonetheless, impaired for replication, indicating that neither loop assignment nor MDA5 binding capacity reliably predicted viability and growth in vitro.
Regarding other conserved noncysteine residues of the CTD, our HPIV-2 data agree with the recently reported PIV5 data, in that the first His and the second Arg residues (H174 and R176 in HPIV-2) are dispensable for MDA5 binding, whereas the first Arg (R175 in HPIV-2) is needed for binding. In addition, we showed that the first Glu and Ile residues in the CTD (E177 and I180 in HPIV-2) are important for MDA5 binding. In PIV5, mutation of the first Glu residue alleviated the inhibition of MDA5 signaling, yet the mutant protein appeared to retain the ability to bind to MDA5 (49).
Mutagenesis of the HPIV-2 V protein CTD showed that the amino-terminal part of this domain, residues 175 through 180 or 187, is heavily involved in MDA5 binding, suggesting a potential MDA5 binding domain that includes a number of highly conserved noncysteine residues. Despite its proximity to this putative MDA5 binding site and in spite of its participation in loop 1 of the RING domain, the H174F mutation did not disrupt V binding to MDA5. In addition, although the R175A/R176A mutation rendered the V protein wholly unable to bind MDA5, the second of the two charge-to-alanine substitutions, R205A/K206A, did not disrupt V-MDA5 binding. Both the H174F and the R205A/K206A mutants act as important controls in our study, containing mutations that not only permit V to bind MDA5, but also confer otherwise wild-type phenotypes, in terms of growth and IFN antagonist activity in vitro as well as wild-type growth in vivo in the case of H174F. The finding that some residues such as these in the CTD could be modified with little effect gives credence to the idea that the drastic effects observed upon mutation of nearby residues, such as residues 175, 177, and 180, suggest direct involvement of the latter residues. In sum, our data indicate that for the HPIV-2 V CTD, a number of highly conserved residues at the amino-terminal end of the domain (R175, E177, W178, and I180) as well as C1, C2, and C5 are needed for MDA5 binding but that other highly conserved residues (H174, R205, and R206) do not play an essential part in this interaction.
Although the R175A/R176A and C214S mutant viruses presented in this study disrupted binding to MDA5 and subsequently enhanced the induction of IFN in response to HPIV-2 infection, the IFN response was substantially weaker than following infection with rHPIV-2-Vko, and the attenuation in primates was also less marked than that previously observed with rHPIV-2-Vko (53). rHPIV-2-Vko could inhibit neither IFN production nor signaling, but it was also deficient with regard to V protein function in replication (39, 41) The IFN induction seen with the V CTD mutants that were impaired for binding to MDA5, which was less vigorous than that for rHPIV-2-Vko, may be attributable to the observation that the V CTD mutants retained a WT-like ability to inhibit IFN signaling, which is necessary for the full magnitude and breadth of the type I IFN response. It also is possible that the mutant V proteins were able to partially inhibit IFN induction via interactions with TBK1, IKKε, or IKKα (31, 47). Such interactions were not explored in detail, although the more limited IRF3 dimerization seen with these mutants, in comparison to that for rHPIV-2-Vko (Fig. 2), hints at an inhibition of the TBK/IKKε axis.
Our results indicate that the induction of IFN-β in response to infection with the R175A/R176A and C214S rHPIV-2 mutants correlates with the inability of their mutant V proteins to efficiently bind to MDA5. The resulting attenuation of these mutants in vivo points to a biological role of the V-MDA5 interaction in the pathogenesis of HPIV-2 infection. This is significant because several previous studies have suggested that RIG-I and MDA5 act in parallel, discriminating among different RNA ligands to initiate responses to unique sets of RNA viruses (12, 13, 50, 66), and RIG-I, rather than MDA5, is thought to be the primary sensor for paramyxoviruses (17, 25, 33, 51). However, it is MDA5, not RIG-I, that is targeted by paramyxovirus V proteins (5, 66). One study, using measles virus, suggested that the production of IFN-β was reduced in human epithelial cells in which either RIG-I or MDA5 had been knocked down, indicating that perhaps both proteins have a role in sensing paramyxoviruses (22). Although the mechanism of paramyxovirus V protein binding to MDA5 is well described (1a, 6), knowledge regarding the in vivo consequences of the V-MDA5 interaction has otherwise been very limited thus far. To our knowledge, this is the first report of a V protein-specific phenotype in vivo. In agreement with our finding that MDA5 binding is important for HPIV-2 virulence in vivo, MDA5 knockout mice were reported to exhibit increased morbidity and mortality following Sendai virus infection (14).
The V protein CTD mutant viruses that disrupted MDA5 binding and permitted the induction of type I IFN had the effect of attenuating HPIV-2 for the respiratory tract of nonhuman primates. These viruses were strongly attenuated even though their ability to block IFN signaling appeared to be unaffected. This is in sharp contrast to a previous study in which we identified the converse situation: namely, HPIV-2 V mutant viruses whose ability to inhibit IFN signaling was lost, while IFN induction remained largely suppressed (52). Those mutant viruses were not attenuated in nonhuman primates. It is reasonable to suggest that the mutants that permit IFN signaling are likely not attenuated because their antiviral effects are active only in cells that are directly infected (52), while mutants that permit IFN induction are attenuated most likely because they permit the host to establish an antiviral state in a large number of (mostly uninfected) respiratory epithelial cells via the paracrine effect of type 1 IFNs. The targeted mutagenesis of the HPIV-2 V protein in a cDNA-derived virus that expresses the P and V proteins from separate gene units enabled an unequivocal assignment of this in vivo attenuation phenotype to the inability of the V protein to interact with MDA5 and represents the first evidence that V-MDA5 interactions are important for the virulence of HPIV-2 in primates.
ACKNOWLEDGMENTS
This research was supported by the Intramural Research Program of the National Institute of Allergy and Infectious Disease, National Institutes of Health, and was performed partly under a cooperative research and development agreement (CRADA) between NIAID and MedImmune, LLC (CRADA no. AI-5114), for the development of live attenuated virus vaccines for respiratory syncytial viruses, parainfluenza viruses, and human metapneumovirus.
We thank Ernest Williams and Fatemeh Davoodi (LID/NIAID) for performing the serology assays and Robin Kastenmayer and her team (CMB/NIAID) for performing the nonhuman primate studies. Antibodies to HPIV-2 P and V proteins were kindly provided by Machiko Nishio of Mie University Graduate School of Medicine in Japan. John Hiscott, McGill University, kindly provided the VSV-GFP virus. FLAG-MDA5 was kindly provided by Takashi Fujita, Kyoto University Institute for Virus Research.
Footnotes
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Abramoff M. D., Magelhaes P., Ram S. J. 2004. Image processing with ImageJ. Biophoton. Int. 11:36–42 [Google Scholar]
- 1a. Andrejeva J., et al. 2004. The V proteins of paramyxoviruses bind the IFN-inducible RNA helicase, mda-5, and inhibit its activation of the IFN-beta promoter. Proc. Natl. Acad. Sci. U. S. A. 101:17264–17269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Andrejeva J., Young D. F., Goodbourn S., Randall R. E. 2002. Degradation of STAT1 and STAT2 by the V proteins of simian virus 5 and human parainfluenza virus type 2, respectively: consequences for virus replication in the presence of alpha/beta and gamma interferons. J. Virol. 76:2159–2167 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Berghäll H., et al. 2006. The interferon-inducible RNA helicase, mda-5, is involved in measles virus-induced expression of antiviral cytokines. Microbes Infect. 8:2138–2144 [DOI] [PubMed] [Google Scholar]
- 4. Buchholz U. J., Finke S., Conzelmann K. K. 1999. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 73:251–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Childs K., et al. 2007. mda-5, but not RIG-I, is a common target for paramyxovirus V proteins. Virology 359:190–200 [DOI] [PubMed] [Google Scholar]
- 6. Childs K. S., Andrejeva J., Randall R. E., Goodbourn S. 2009. Mechanism of mda-5 inhibition by paramyxovirus V proteins. J. Virol. 83:1465–1473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Curran J., de Melo M., Moyer S., Kolakofsky D. 1991. Characterization of the Sendai virus V protein with an anti-peptide antiserum. Virology 184:108–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Didcock L., Young D. F., Goodbourn S., Randall R. E. 1999. The V protein of simian virus 5 inhibits interferon signalling by targeting STAT1 for proteasome-mediated degradation. J. Virol. 73:9928–9933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Emeny J. M., Morgan M. J. 1979. Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J. Gen. Virol. 43:247–252 [DOI] [PubMed] [Google Scholar]
- 10. Fensterl V., Sen G. C. 2009. Interferons and viral infections. Biofactors 35:14–20 [DOI] [PubMed] [Google Scholar]
- 11. Ferko B., et al. 2004. Immunogenicity and protection efficacy of replication-deficient influenza A viruses with altered NS1 genes. J. Virol. 78:13037–13045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Foy E., et al. 2005. Control of antiviral defenses through hepatitis C virus disruption of retinoic acid-inducible gene-I signaling. Proc. Natl. Acad. Sci. U. S. A. 102:2986–2991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Gitlin L., et al. 2006. Essential role of mda-5 in type I IFN responses to polyriboinosinic:polyribocytidylic acid and encephalomyocarditis picornavirus. Proc. Natl. Acad. Sci. U. S. A. 103:8459–8464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Gitlin L., et al. 2010. Melanoma differentiation-associated gene 5 (MDA5) is involved in the innate immune response to Paramyxoviridae infection in vivo. PLoS Pathog. 6:e1000734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Goodbourn S., Didcock L., Randall R. E. 2000. Interferons: cell signalling, immune modulation, antiviral response and virus countermeasures. J. Gen. Virol. 81:2341–2364 [DOI] [PubMed] [Google Scholar]
- 16. Gottlieb J., et al. 2009. Community-acquired respiratory viral infections in lung transplant recipients: a single season cohort study. Transplantation 87:1530–1537 [DOI] [PubMed] [Google Scholar]
- 17. Guix S., et al. 2007. Norwalk virus RNA is infectious in mammalian cells. J. Virol. 81:12238–12248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hai R., et al. 2008. Influenza B virus NS1-truncated mutants: live-attenuated vaccine approach. J. Virol. 82:10580–10590 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hall S. L., et al. 1992. Cold-passaged human parainfluenza type 3 viruses contain ts and non-ts mutations leading to attenuation in rhesus monkeys. Virus Res. 22:173–184 [DOI] [PubMed] [Google Scholar]
- 20. He B., et al. 2002. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology 303:15–32 [DOI] [PubMed] [Google Scholar]
- 21. Horvath C. M. 2004. Weapons of STAT destruction. Interferon evasion by paramyxovirus V protein. Eur. J. Biochem. 271:4621–4628 [DOI] [PubMed] [Google Scholar]
- 22. Ikegame S., et al. 2010. Both RIG-I and MDA5 RNA helicases contribute to the induction of alpha/beta interferon in measles virus-infected human cells. J. Virol. 84:372–379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kapikian A., et al. 1963. An outbreak of parainfluenza 2 (croup-associated) virus infection. JAMA 183:324–330 [DOI] [PubMed] [Google Scholar]
- 24. Karron R. A., Collins P. L. 2007. Parainfluenza viruses, p. 1497–1526 In Knipe D. M., Howley P. M., Roizman B., Straus S. E. (ed.), Fields virology, 5th ed., vol. 1 Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 25. Kato H., et al. 2006. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature 441:101–105 [DOI] [PubMed] [Google Scholar]
- 26. Kawano M., et al. 2001. Recovery of infectious human parainfluenza type 2 virus from cDNA clones and properties of the defective virus without V-specific cysteine-rich domain. Virology 284:99–112 [DOI] [PubMed] [Google Scholar]
- 27. Komatsu T., Takeuchi K., Gotoh B. 2007. Bovine parainfluenza virus type 3 accessory proteins that suppress beta interferon production. Microbes Infect. 9:954–962 [DOI] [PubMed] [Google Scholar]
- 28. Komatsu T., Takeuchi K., Yokoo J., Gotoh B. 2004. C and V proteins of Sendai virus target signaling pathways leading to IRF-3 activation for the negative regulation of interferon-beta production. Virology 325:137–148 [DOI] [PubMed] [Google Scholar]
- 29. Li T., Chen X., Garbutt K. C., Zhou P., Zheng N. 2006. Structure of DDB1 in complex with a paramyxovirus V protein: viral hijack of a propeller cluster in ubiquitin ligase. Cell 124:105–117 [DOI] [PubMed] [Google Scholar]
- 30. Lin G. Y., Lamb R. A. 2000. The paramyxovirus simian virus 5 V protein slows progression of the cell cycle. J. Virol. 74:9152–9166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lu L. L., Puri M., Horvath C. M., Sen G. C. 2008. Select paramyxoviral V proteins inhibit IRF3 activation by acting as alternative substrates for inhibitor of kappaB kinase epsilon (IKKe)/TBK1. J. Biol. Chem. 283:14269–14276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McAuliffe J. M., et al. 2004. Codon substitution mutations at two positions in the L polymerase protein of human parainfluenza virus type 1 yield viruses with a spectrum of attenuation in vivo and increased phenotypic stability in vitro. J. Virol. 78:2029–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. McCartney S. A., et al. 2008. MDA-5 recognition of a murine norovirus. PLoS Pathog. 4:e1000108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Mosca J. D., Pitha P. M. 1986. Transcriptional and posttranscriptional regulation of exogenous human beta interferon gene in simian cells defective in interferon synthesis. Mol. Cell. Biol. 6:2279–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Murphy B. R. 1988. Current approaches to the development of vaccines effective against parainfluenza viruses. Bull. World Health Organ. 66:391–397 [PMC free article] [PubMed] [Google Scholar]
- 36. Newman J. T., et al. 2004. Generation of recombinant human parainfluenza virus type 1 vaccine candidates by importation of temperature-sensitive and attenuating mutations from heterologous paramyxoviruses. J. Virol. 78:2017–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Newman J. T., et al. 2002. Sequence analysis of the Washington/1964 strain of human parainfluenza virus type 1 (HPIV1) and recovery and characterization of wild-type recombinant HPIV1 produced by reverse genetics. Virus Genes 24:77–92 [DOI] [PubMed] [Google Scholar]
- 38. Nishio M., Garcin D., Simonet V., Kolakofsky D. 2002. The carboxyl segment of the mumps virus V protein associates with Stat proteins in vitro via a tryptophan-rich motif. Virology 300:92–99 [DOI] [PubMed] [Google Scholar]
- 39. Nishio M., Ohtsuka J., Tsurudome M., Nosaka T., Kolakofsky D. 2008. Human parainfluenza virus type 2 V protein inhibits genome replication by binding to the L protein: possible role in promoting viral fitness. J. Virol. 82:6130–6138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Nishio M., Tsurudome M., Ishihara H., Ito M., Ito Y. 2007. The conserved carboxyl terminus of human parainfluenza virus type 2 V protein plays an important role in virus growth. Virology 362:85–98 [DOI] [PubMed] [Google Scholar]
- 41. Nishio M., et al. 2005. Identification of paramyxovirus V protein residues essential for STAT protein degradation and promotion of virus replication. J. Virol. 79:8591–8601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Nolan S. M., et al. 2007. Recombinant human parainfluenza virus type 2 vaccine candidates containing a 3′ genomic promoter mutation and L polymerase mutations are attenuated and protective in non-human primates. Vaccine 25:6409–6422 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Nolan S. M., Amaro-Carambot S. S. E., Collins P. L., Murphy B. R., Skiadopoulos M. H. 2005. Live-attenuated intranasal parainfluenza virus type 2 vaccine candidates developed by reverse genetics containing L polymerase protein mutations imported from heterologous paramyxoviruses. Vaccine 39:4765–4774 [DOI] [PubMed] [Google Scholar]
- 44. Parisien J. P., et al. 2009. A shared interface mediates paramyxovirus interference with antiviral RNA helicases MDA5 and LGP2. J. Virol. 83:7252–7260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Parrott R., Vargosko A. K. H. W., Bell J., Chanock R. 1962. Myxoviruses. III. Parainfluenza. Am. J. Public Health 52:907–917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Paterson R., Leser G., Shaughnessy M., Lamb R. 1995. The paramyxovirus SV5 V protein binds two atoms of zinc and is a structural component of virions. Virology 208:121–131 [DOI] [PubMed] [Google Scholar]
- 47. Pfaller C. K., Conzelmann K. K. 2008. Measles virus V protein is a decoy substrate for IkappaB kinase alpha and prevents Toll-like receptor 7/9-mediated interferon induction. J. Virol. 82:12365–12373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Poole E., He B., Lamb R. A., Randall R. E., Goodbourn S. 2002. The V proteins of Simian virus 5 and other paramyxoviruses inhibit induction of interferon-beta. Virology 303:33–46 [DOI] [PubMed] [Google Scholar]
- 49. Ramachandran A., Horvath C. M. 2010. Dissociation of paramyxovirus interferon evasion activities: universal and virus-specific requirements for conserved V protein amino acids in MDA5 interference. J. Virol. 84:11152–11163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Saito T., Gale M., Jr 2008. Differential recognition of double-stranded RNA by RIG-I-like receptors in antiviral immunity. J. Exp. Med. 205:1523–1527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Saito T., Owen D. M., Jiang F., Marcotrigiano J., Gale M., Jr 2008. Innate immunity induced by composition-dependent RIG-I recognition of hepatitis C virus RNA. Nature 454:523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Schaap-Nutt A., et al. 2010. Recombinant human parainfluenza virus type 2 with mutations in V that permit cellular interferon signaling are not attenuated in non-human primates. Virology 406:65–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Schaap-Nutt A., et al. 2010. Human parainfluenza virus type 2 V protein inhibits interferon production and signaling and is required for replication in non-human primates. Virology 397:285–298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Reference deleted.
- 55. Skiadopoulos M. H., et al. 2003. The genome length of human parainfluenza virus type 2 follows the rule of six, and recombinant viruses recovered from non-polyhexameric-length antigenomic cDNAs contain a biased distribution of correcting mutations. J. Virol. 77:270–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Stojdl D. F., Lichty B. D., R. tenOever B., et al. 2003. VSV strains with defects in their ability to shutdown innate immunity are potent systemic anti-cancer agents. Cancer Cell 4:263–275 [DOI] [PubMed] [Google Scholar]
- 57. Talon J., et al. 2000. Influenza A and B viruses expressing altered NS1 proteins: a vaccine approach. Proc. Natl. Acad. Sci. U. S. A. 97:4309–4314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Teng M. N., et al. 2000. Recombinant respiratory syncytial virus that does not express the NS1 or M2-2 protein is highly attenuated and immunogenic in chimpanzees. J. Virol. 74:9317–9321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Tsurudome M., Nishio M., Komada H., Bando H., Ito Y. 1989. Extensive antigenic diversity among human parainfluenza type 2 virus isolates and immunological relationships among paramyxoviruses revealed by monoclonal antibodies. Virology 171:38–48 [DOI] [PubMed] [Google Scholar]
- 60. Van Cleve W., et al. 2006. Attenuating mutations in the P/C gene of human parainfluenza virus type 1 (HPIV1) vaccine candidates abrogate the inhibition of both induction and signaling of type I interferon (IFN) by wild-type HPIV1. Virology 352:61–73 [DOI] [PubMed] [Google Scholar]
- 61. van Wyke Coelingh K. L., et al. 1990. Antibody responses of humans and nonhuman primates to individual antigenic sites of the hemagglutinin-neuraminidase and fusion glycoproteins after primary infection or reinfection with parainfluenza type 3 virus. J. Virol. 64:3833–3843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wansley E. K., Parks G. D. 2002. Naturally occurring substitutions in the p/v gene convert the noncytopathic paramyxovirus simian virus 5 into a virus that induces alpha/beta interferon synthesis and cell death. J. Virol. 76:10109–10121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Wathelet M. G., Berr P. M., Huez G. A. 1992. Regulation of gene expression by cytokines and virus in human cells lacking the type-I interferon locus. Eur. J. Biochem. 206:901–910 [DOI] [PubMed] [Google Scholar]
- 64. Whitehead S. S., et al. 1999. Recombinant respiratory syncytial virus bearing a deletion of either the NS2 or SH gene is attenuated in chimpanzees. J. Virol. 73:3438–3442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Wright P. F., et al. 2006. The interferon antagonist NS2 protein of respiratory syncytial virus is an important virulence determinant for humans. J. Infect. Dis. 193:573–581 [DOI] [PubMed] [Google Scholar]
- 66. Yoneyama M., et al. 2005. Shared and unique functions of the DExD/H-box helicases RIG-I, MDA5, and LGP2 in antiviral innate immunity. J. Immunol. 175:2851–2858 [DOI] [PubMed] [Google Scholar]
- 67. Young D. F., Didcock L., Goodbourn S., Randall R. E. 2000. Paramyxoviridae use distinct virus-specific mechanisms to circumvent the interferon response. Virology 269:383–390 [DOI] [PubMed] [Google Scholar]