Abstract
The Gag-Pol polyprotein of human immunodeficiency virus type 1 (HIV-1) is not required for efficient viral particle production. However, premature termination codons in pol, particularly in the integrase (IN)-coding region, can markedly impair HIV-1 particle formation, apparently due to the premature activation of the viral protease (PR). We now report that the IN domain of Gag-Pol is required for the incorporation of clathrin into HIV-1 virions. Significantly, PR-dependent effects of point mutations in IN on particle production correlated strictly with their effects on clathrin incorporation. A possible interpretation of these findings is that certain IN mutations impair particle production in a PR-dependent manner by promoting Gag-Pol dimerization, which also occludes a binding site for clathrin. Consistently with this model, the reverse transcriptase (RT) inhibitor efavirenz, which is thought to promote Gag-Pol dimerization, inhibited the incorporation of clathrin into HIV-1 virions. Clathrin-depleted cells produced normal amounts of HIV-1 virions; however, their infectivity was reduced. We also observed that HIV-2 and the simian immunodeficiency virus SIVmac interact with clathrin through one or two copies of a peptide motif in the p6 domain of Gag that resembles the clathrin box of cellular adaptor proteins. Furthermore, the substitution of the hydrophobic residues in the single clathrin box motif of SIVmac caused a replication defect in primary cells. Taken together, our results indicate that primate lentiviruses from two different subgroups functionally interact with clathrin during assembly.
INTRODUCTION
The formation of human immunodeficiency virus type 1 (HIV-1) virions is driven by the viral Gag protein, which assembles into a lipid envelope-covered protein shell underneath the plasma membrane. The association of Gag with the plasma membrane and with viral surface glycoproteins is mediated by its N-terminal matrix (MA) domain, the self association of Gag is predominantly driven by its capsid (CA) domain, and the recognition of viral nucleic acid occurs through its nucleocapsid (NC) domain (25). The Gag proteins of HIV-1 and other primate lentiviruses additionally contain a C-terminal p6 domain that mediates the incorporation of Vpr or of Vpr-like accessory proteins into assembling particles (2, 37). The p6 domain also binds to the cellular ESCRT pathway components Tsg101 and ALIX via so-called late domains to promote the detachment of the lipid envelope of assembled particles from the cell surface (5, 23).
Occasional ribosomal frameshifting near the end of the gag gene into the overlapping pol frame leads to the translation of Gag-Pol (26), in which the NC domain is followed by a transframe region and the essential viral enzymes protease (PR), reverse transcriptase (RT), and integrase (IN). Gag-Pol is incorporated into assembling particles via interactions with Gag, and the activation of its PR domain leads to Gag and Gag-Pol cleavage to yield the mature structural proteins and viral enzymes.
Surprisingly, mutations in the RT and IN domains of Gag-Pol can cause profound defects in HIV-1 particle production, even though Gag-Pol is dispensable for particle assembly and release (9, 12, 18, 19, 27). At least in some cases, particle production was restored to wild-type (WT) levels in the absence of PR, suggesting that the defect in particle production seen in the presence of PR was caused by its premature activation (9, 12). Since PR is an obligate dimer, in principle its premature activation could be caused by enhanced Gag-Pol dimerization. Consistently with this notion, premature Gag and Gag-Pol processing and, consequently, a defect in HIV-1 particle production, can be induced by efavirenz and other nonnucleoside RT inhibitors that promote RT homodimer formation (21, 43). Notably, the inhibitory effect of efavirenz on particle production depends on the presence of active PR, and it can be attenuated by mutations within a tryptophan repeat motif that is involved in RT dimerization (13, 21).
PR-dependent defects in HIV-1 particle production that are reminiscent of those induced by efavirenz also are caused by C-terminal truncations of Gag-Pol that remove most or all of the IN domain (9). To explain these observations, we had suggested that one function of the IN domain is to counteract RT domain-driven Gag-Pol dimerization and, thus, to prevent the premature activation of PR. Consistently with this model, particle production in the absence of IN could be rescued not only through the inactivation of PR but also through the shortening or removal of the RT domain (9). Interestingly, significant defects in HIV-1 particle production can be caused even by single-amino-acid substitutions in IN. Such mutants are unable to replicate irrespective of whether the catalytic activity of IN is affected (18).
In the present study, we report that the IN domain of HIV-1 Gag-Pol is required for the incorporation of a 180-kDa protein into HIV-1 particles. This protein, which is present in ample amounts in WT HIV-1 virions, was identified as the clathrin heavy chain. Significantly, the effects of point mutations in IN on clathrin incorporation, as determined in a PR-negative context to suppress particle production defects, correlated strictly with their effects on particle production in the WT context. The incorporation of clathrin into HIV-1 virions was inhibited by efavirenz, which is consistent with a model in which Gag-Pol dimerization reduces or prevents clathrin binding. The infectivity of HIV-1 virions produced in clathrin-depleted cells was reduced, indicating that the interaction with clathrin is functionally relevant. We also observed that HIV-2 and the related simian immunodeficiency virus SIVmac possess clathrin binding sites in p6 that closely resemble the clathrin box of adaptor proteins, and that SIVmac with a disrupted clathrin box was replication defective.
MATERIALS AND METHODS
Proviral constructs.
The parental, replication-competent proviral constructs used in this study were HXBH10 (24), which lacks vpr and nef, and HXBH10/R+ (16), a vpr-positive version of HXBH10. Point mutations in the IN coding region were generated by site-directed mutagenesis and inserted into HXBH10/R+ by standard cloning techniques. ΔMA/PR− is a version of HXBH10/R+ that harbors the gag gene of the previously described ΔMA HIV-1 proviral construct (40), which has all of the MA coding sequence replaced by a sequence encoding a heterologous myristylation signal. ΔMA/PR− also has the codon specifying Asp25 of PR replaced by a codon specifying Glu to prevent Gag and Gag-Pol processing, and it harbors a disrupted env gene. ΔMA/ΔIN/PR− was derived from ΔMA/PR− by replacing the first codon for IN with a premature termination codon.
The parental, replication-competent SIVmac proviral construct was pSIVmac239RQ (kindly provided by Riri Shibata), a derivative of the infectious full-length clone pMA239 (41). In contrast to pMA239, pSIVmac239RQ has an open nef reading frame. To introduce mutations into the p6 coding region, site-directed mutagenesis was performed on a subviral construct, and fragments harboring the desired mutations then were cloned into full-length pSIVmac239RQ. The mutations in p6 do not alter the overlapping pol reading frame. The LL→SQ mutant harbors the L52S and L53Q substitutions, and the LLHL→SQHH mutant additionally harbors the L55H substitution.
Viral particle analysis.
293T cells (3.5 × 106) were seeded into T80 flasks for incorporation experiments, and HeLa cells (3 × 105) were seeded into T25 flasks to examine effects on particle production. The next day, the cells were transfected with proviral DNA using a calcium phosphate precipitation technique. Where indicated, the transfected cells were metabolically labeled with [35S]methionine from 8 to 20 h posttransfection. Virions or virus-like particles (VLP) released into the culture medium were pelleted through sucrose and analyzed by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE), followed by autoradiography or Western blotting, as described previously (3). Proteins in the cell lysates were detected by Western blotting. The antibodies used for Western blotting were a rabbit anti-HIV CA serum (Advanced Biotechnologies, Columbia, MD), 183-H12-5C (11) against HIV-1 CA, TD.1 (Sigma-Aldrich, St. Louis, MO) and clone 23 (BD Transduction Laboratories, San Jose, CA) against the clathrin heavy chain, CON.1 (Santa Cruz Biotechnology, Santa Cruz, CA) against the clathrin light chains, AC-40 (Sigma-Aldrich) against actin, HA.11 (Covance, Berkeley, CA) against the hemagglutinin (HA) epitope, and Chessie 8 (1) against HIV-1 gp41. Western blots were quantified with ImageJ software.
Protein identification.
Bands were excised from Coomassie-stained gels and subjected to in-gel tryptic digestion. Extracted peptides were analyzed by microcapillary liquid chromatography coupled with tandem mass spectrometry (LC-MS/MS). Tandem mass spectra were searched against a human protein database with Sequest software.
Generation of HeLa cells stably expressing short hairpin RNAs (shRNAs).
293T cells were cotransfected with pHDMHgpm2 encoding codon-optimized HIV-1 Gag-Pol, LKO.1-based lentiviral vectors encoding shRNA targeting the clathrin heavy chain (clone TRCN0000007982; Open Biosystems, Huntsville, AL) or CD11A (clone TRCN0000029594; Open Biosystems), and vectors expressing HIV-1 Rev and the vesicular stomatitis virus G protein. Filtered supernatants then were used to transduce HeLa cells, followed by selection with 1 μg/ml puromycin.
GST pulldown assay.
A version of the bacterial expression vector pGEX-2TK (GE Healthcare, Piscataway, NJ) that encodes the full-length HIV-2ROD p6 domain fused to glutathione S-transferase (GST) has been described (42). The full-length p6 domain of SIVmac239 (residues 1 to 63) was amplified and inserted in frame into pGEX-2TK. Point mutations were introduced by site-directed mutagenesis. GST fusion proteins were expressed in strain BL21 and immobilized on glutathione-Sepharose beads (GE Healthcare). The beads then were incubated for 2 to 3 h at 4°C with hypotonic 293T cell lysates or with hypotonic lysates of 293T cells transiently expressing HA-ALIXΔPRD (42). The beads then were extensively washed in phosphate-buffered saline, and bound proteins were eluted by being boiled in SDS-PAGE sample buffer, resolved by SDS-PAGE, and visualized with colloidal Coomassie brilliant blue G-250. Captured clathrin and HA-ALIXΔPRD were detected by Western blotting.
Virus infectivity and replication assays.
Filtered supernatants from HeLa or HeLa-derived cells transfected with the nef-negative HIV-1 proviral clone HXBH10 were normalized for p24 antigen content and used to infect TZM-bl indicator cells (1 × 105) in 5 ml medium at 10 ng p24/ml. Two days postinfection, the indicator cells were lysed in 400 μl reporter lysis buffer supplied with the ß-galactosidase (ß-gal) enzyme assay system (Promega), and ß-gal activity induced by HIV-1 Tat as a consequence of infection was measured according to the manufacturer's instructions.
For SIV replication studies, peripheral blood mononuclear cells (PBMC) were isolated by Ficoll-Hypaque density gradient centrifugation from the blood of healthy rhesus macaques (obtained from the New England Regional Primate Research Center), stimulated for 2 days with 10 μg/ml concanavalin A (type IV; Sigma-Aldrich), washed, and resuspended in RPMI 1640 medium containing 15% fetal calf serum and 10% human lymphocyte-conditioned medium (interleukin-2; Hemagen Diagnostics, Columbia, MD) prior to infection. SIVmac virions were produced by transfecting 293T cells with wild-type or mutant versions of pSIVmac239RQ. Virus-containing supernatants were passed through 0.45-μm syringe filters, RT activity was determined as described previously (32), and 1 × 105 cpm of RT activity were used to infect 6 × 106 stimulated rhesus macaque PBMC. The cells were washed the next day to remove the virus inocula, and virus replication was monitored by measuring the RT activity in the culture supernatants at regular intervals.
RESULTS
HIV-1 incorporates clathrin in an IN-dependent manner.
We previously reported that the MA domain of HIV-1 Gag can be replaced by a foreign membrane-targeting sequence without compromising particle production (40). Because the ΔMA mutant used in that study had the entire MA domain replaced by a 15-amino-acid peptide, the Gag-Pol precursor produced by a PR-negative version was expected to migrate noticeably faster in SDS-PAGE than WT Gag-Pol. Surprisingly, SDS-PAGE analyses of [35S]methionine-labeled ΔMA/PR− virions reproducibly yielded two closely spaced bands of comparable intensity in the region where the Gag-Pol precursor was expected to migrate (Fig. 1A, lane 1, and data not shown). The position of the faster-migrating species was that predicted for the shortened Gag-Pol precursor produced by the ΔMA/PR− mutant, whereas the slower-migrating species (provisionally designated p180) exhibited an apparent molecular mass closer to that of the WT Gag-Pol precursor. These results raised the possibility that HIV-1 virions contain significant amounts of a protein that normally is obscured by the comigrating Gag-Pol precursor but becomes detectable by SDS-PAGE when the size of the Gag-Pol precursor is reduced.
Fig. 1.
IN domain-dependent incorporation of clathrin into HIV-1 particles. (A) 293T cells were transfected with PR-defective HIV-1 proviral DNAs expressing Gag and Gag-Pol proteins lacking MA. The ΔMA/ΔIN/PR− mutant (lane 2) also contains a premature termination codon in pol that prevents the translation of the IN domain of Gag-Pol. The transfected cells were labeled with [35S]methionine, and proteins incorporated into VLP were separated by SDS-PAGE, transferred to a nitrocellulose membrane, and visualized by autoradiography. The migration positions of molecular mass markers (in kilodaltons) are indicated on the left. (B) Western blots demonstrating the IN-dependent incorporation of the clathrin heavy chain (HC) and light chains (LCs). The same nitrocellulose membrane as that shown in panel A was examined to correlate [35S]methionine-labeled bands with those detected by Western blotting. To facilitate the use of different antibodies, the membrane was cut above the expected position of Gag and subsequently reassembled for autoradiography.
To examine the relationship between the Gag-Pol precursor and p180, we truncated the Gag-Pol precursor produced by the ΔMA/PR− mutant from the C terminus to alter its electrophoretic mobility. In principle, truncating or removing the IN domain of Gag-Pol can markedly reduce particle production by otherwise intact HIV-1 proviruses, as we have shown previously (9). However, we also have observed that an active PR is required for these effects, and that particle production can be fully restored by inactivating PR (9). Therefore, particle production by the ΔMA/PR− mutant was not expected to be affected by a premature termination codon directly after the RT-coding sequence. Indeed, the resulting ΔMA/ΔIN/PR− mutant and the parental ΔMA/PR− construct produced comparable amounts of particles (Fig. 1A). Interestingly, the ΔIN mutation not only induced the expected shift in the migration of Gag-Pol but also led to the disappearance of p180 (Fig. 1A). Taken together, these data suggested that p180 was not derived from Gag-Pol but that its presence in viral particles nevertheless was dependent on the integrity of the IN domain of Gag-Pol.
To determine the identity of p180, the band was excised from a Coomassie-stained gel. Protein microsequencing indicated that p180 corresponds to the clathrin heavy chain, which has a calculated molecular mass of 191 kDa but is known to migrate at about 180 kDa (6). Western blotting with the monoclonal anti-clathrin heavy chain antibody TD.1 confirmed the presence of the clathrin heavy chain in particles produced by the ΔMA/PR− mutant (Fig. 1B). In contrast, the clathrin heavy chain was absent from ΔMA/ΔIN/PR− particles (Fig. 1B). Western blotting of the same particle preparations with anti-CA yielded Gag-Pol precursors of the expected size and confirmed that comparable amounts of Gag-Pol and Gag were present in each sample (Fig. 1B).
During clathrin-mediated endocytosis, the clathrin heavy chain polymerizes into lattices that coat the cytosolic side of endocytic vesicles. Clathrin assembly is controlled by clathrin light chains, which bind to the clathrin heavy chain and prevent its spontaneous polymerization (33). Most human cells express two light chains (LCa and LCb) that are encoded by different genes and exhibit about 60% homology (7). To determine whether HIV-1 particles contain clathrin light chains, we used the monoclonal antibody CON.1, which binds to both LCa and LCb (36). As shown in Fig. 1B, Western blotting of ΔMA/PR− particles with CON.1 yielded a prominent band, together with a weaker band of slightly faster electrophoretic mobility. The more prominent of the two bands had an apparent molecular mass of about 36 kDa (data not shown). Like the clathrin heavy chain, both bands were absent from ΔMA/ΔIN/PR− particles. Since the clathrin light chains migrate in the 32- to 36-kDa range in SDS-PAGE (29), we conclude that HIV-1 particles incorporate the clathrin heavy chain together with associated light chains in an IN-dependent manner.
Assembly-defective class II IN mutants do not incorporate clathrin.
Replication-defective IN mutants fall into two broad categories: class I mutants are specifically defective at the integration step and assemble virus particles normally, whereas class II mutants exhibit pleiotropic defects, such as those during reverse transcription or during virus morphogenesis (18). Because the morphogenesis defect of the ΔIN mutant is reminiscent of the phenotype of certain class II mutants, we examined the ability of a panel of class II mutants to incorporate clathrin.
The H16A mutation was selected for this analysis, because substitutions affecting His16 or any of the other zinc-coordinating residues in the N-terminal domain of HIV-1 IN yield class II mutants defective for viral DNA synthesis (30, 46). A class II mutant with a substitution in the catalytic domain of IN, HIV-1F185K, was included because of its previously documented strong defect at the level of assembly or release (20). Of note, the F185K mutation does not affect the in vitro enzymatic activity of HIV-1 IN, but full-length HIV-1 carrying the mutation is replication incompetent (27). Like the ΔIN mutant, the F185K mutant released particles at levels about 10-fold lower than those of the WT (9, 20). In contrast, HIV-1 carrying the F185A mutation in IN was released normally (20). Nevertheless, HIV-1F185A is considered a class II mutant, because F185A IN retains enzymatic activity even though the mutant is replication incompetent due to a defect in viral DNA synthesis (20, 46). Similarly, HIV-1K186Q is a class II IN mutant that shows no defect in virus release (34).
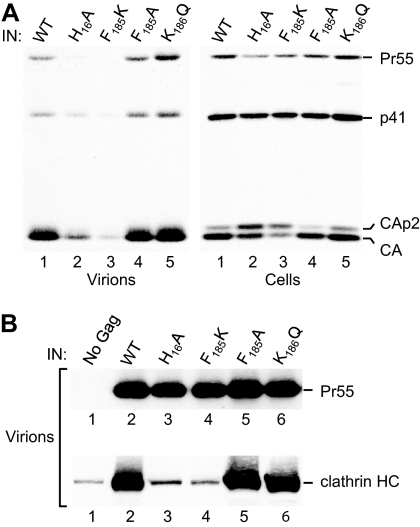
The ability of the point mutants to produce particles was examined in HeLa cells, because of the previously documented pronounced defect of certain IN mutants in that cell line (9, 20). As shown in Fig. 2A, the introduction of the H16A mutation into WT HIV-1 led to a reduction in the release of Gag proteins from transfected HeLa cells. An analysis of the cell lysates revealed that the H16A mutation had no significant effect on Gag expression levels. However, the mutation caused a Gag processing defect, because the cleavage intermediate CA-p2 accumulated in the transfected cells (Fig. 2A). We have shown previously that the ΔIN mutation has a similar effect on Gag processing (9). We conclude that the H16A mutation causes a defect at the level of virion morphogenesis in addition to its previously reported effect on reverse transcription (46). The F185K mutation caused an even more pronounced reduction in particle production, which was approximately 10-fold lower than the WT level, which is consistent with previous observations (20). Moreover, the F185K mutation had an effect on Gag processing similar to that of the H16A mutation. In contrast, the F185A and K186Q class II mutants processed Gag normally and produced close to WT levels of particles, as expected (Fig. 2A).
Fig. 2.
Particle production and clathrin incorporation by class II IN mutants. (A) Particle production and Gag processing were analyzed by transfecting HeLa cells with the parental HXBH10/R+ provirus (WT) or with the indicated class II IN mutants. Virion pellets and the cell lysates were analyzed by Western blotting with anti-CA antibody to detect the Gag precursor Pr55 and the Gag cleavage products p41, CA-p2, and CA. (B) The incorporation of clathrin into immature virions was examined by transfecting PR-negative versions of the proviral DNAs used for panel A into 293T cells. A full-length HIV-1 provirus with a premature termination codon and a frameshift in gag was used as a background control (lane 1). Virion pellets were analyzed by Western blotting to detect unprocessed Gag (Pr55) and the clathrin heavy chain as indicated.
To determine the effects of the mutations on clathrin incorporation, we generated PR-negative versions of the mutant viruses based on our observation that the inactivation of PR suppresses the particle production defect of IN deletion mutants (9). Furthermore, we produced virus particles in 293T cells, because we had previously found that the effect of deleting IN on particle production was less pronounced in these cells than in HeLa cells (5- versus 15-fold) (9). Importantly, under these conditions, the H16A and F185K mutants produced only about 2-fold smaller amounts of particles than the parental construct, and the F185A and K186Q mutants produced comparable amounts of particles (Fig. 2B). Interestingly, Western blotting with anti-clathrin heavy-chain antibody yielded prominent bands for the parental virus and for the F185A and K186Q mutants, whereas the H16A and particularly the F185K mutant yielded signals that were barely above the background level obtained with a proviral construct unable to express Gag (Fig. 2B). Thus, there was a strict correlation between the effects of the mutations on Gag processing and particle production in the PR-positive context and their effects on clathrin incorporation in the PR-negative background.
Efavirenz inhibits the incorporation of clathrin into HIV-1 virions.
The F185K mutation enhances the dimerization of IN (27, 28) and thus may impair viral particle production via the stimulation of Gag-Pol dimerization and the resulting premature activation of PR. Furthermore, an enhancement of Gag-Pol dimerization by the F185K mutation could account for its inhibitory effect on clathrin incorporation, provided that clathrin preferentially interacts with monomeric Gag-Pol.
To test the possibility that Gag-Pol dimerization interferes with clathrin incorporation, we used the nonnucleoside RT inhibitor efavirenz, which is known to induce both RT and Pol dimerization (21, 43, 44). Importantly, efavirenz is also thought to induce Gag-Pol dimerization in infected cells (21). Consistent with this notion, efavirenz inhibits HIV-1 particle production via the premature activation of PR (21). In contrast to efavirenz, the less potent nonnucleoside RT inhibitor nevirapine induces RT dimerization only weakly and has no significant effect on HIV-1 particle production (21, 44).
Because the inhibition of HIV-1 particle production by efavirenz is dependent on the presence of active PR, we used a PR-negative version of the full-length proviral clone HXBH10/R+ to examine the effect of efavirenz on clathrin incorporation. As shown in Fig. 3, efavirenz but not nevirapine inhibited the incorporation of clathrin into HIV-1 virions in a dose-dependent manner. We conclude that the ability of these drugs to induce Pol dimerization correlates with their ability to inhibit clathrin incorporation.
Fig. 3.
Effects of nonnucleoside RT inhibitors on the incorporation of clathrin into HIV-1 virions. 293T cells were transfected with a PR-negative version of the full-length HXBH10/R+ provirus, and virions were produced in the presence of DMSO (no drug), efavirenz (EFV), or nevirapine (NVP) at the indicated concentrations. Virion pellets were analyzed by Western blotting to detect unprocessed Gag (Pr55) and the clathrin heavy chain.
Clathrin knockdown reduces the infectivity of HIV-1 produced in HeLa cells.
Based on the results described above, we examined whether clathrin is critical for proper Gag processing and viral particle production. To this end, clathrin was knocked down in HeLa cells. To achieve a stable knockdown, we used a lentiviral vector (LKO.1) encoding an shRNA targeting the clathrin heavy chain. As shown in Fig. 4A, a more than 20-fold knockdown of clathrin heavy-chain expression at the protein level was observed in a polyclonal HeLa cell line stably transduced with this vector.
Fig. 4.
Clathrin knockdown in virus producer cells reduces the infectivity of progeny virions. (A) Western blot analysis of parental HeLa cells (lane 1) and of HeLa cells stably expressing an shRNA targeting the clathrin heavy chain (HeLa-sh-clathrin HC; lane 2). (B) Virus production by HeLa and HeLa-sh-clathrin HC cells transiently transfected with the full-length HIV-1HXBH10. Particle production, envelope glycoprotein incorporation, and Gag processing were compared by analyzing virion pellets and cell lysates by Western blotting with anti-CA or anti-gp41, as indicated. (C) Relative infectivities of HIV-1HXBH10 virions produced by HeLa or HeLa-sh-clathrin HC cells determined on TZM-bl indicator cells. (D) Relative infectivities of HIV-1HXBH10 virions produced by HeLa cells stably expressing a control shRNA targeting a protein not expressed in HeLa cells (CD11A) or by HeLa-sh-clathrin HC cells. Infectivities are the means plus standard deviations from triplicate experiments.
Importantly, the clathrin-depleted HeLa cell line remained transfectable under our conditions, even though clathrin-mediated endocytosis is thought to be a prerequisite for efficient transfection by some methods (49). To determine the impact of clathrin depletion on the assembly and release of HIV-1 particles, the parental HeLa cell line and the clathrin-depleted cells were transiently transfected with WT HXBH10 proviral DNA. As shown in Fig. 4B, both cell lines produced comparable amounts of virus, and Gag processing in the HeLa/clathrin knockdown cells appeared largely normal. Furthermore, virions produced in HeLa or HeLa/clathrin knockdown cells contained similar amounts of the transmembrane glycoprotein gp41 (Fig. 4B). These data suggest that clathrin is not required for virus assembly or release or for the incorporation of the viral envelope glycoproteins.
We also examined whether the depletion of clathrin in HeLa cells affected the infectivity of virus produced in these cells. To this end, virions released from HeLa or HeLa/clathrin knockdown cells transfected with nef-negative HIV-1HXBH10 were normalized for p24 content, and equal amounts were used to infect TZM-bl indicator cells. A nef-negative virus was used, because we have shown previously that the enhancement of HIV-1 infectivity by Nef depends on clathrin-mediated endocytosis (38). As shown in Fig. 4C, the stable depletion of the clathrin heavy chain led to an approximately 10-fold reduction in the infectivity of progeny virus. To control for off-target effects caused by the expression of shRNA, we stably transduced HeLa cells with LKO.1 encoding an shRNA targeting the leukocyte integrin CD11A, which is not expressed in HeLa cells (39). As shown in Fig. 4D, HIV-1HXBH10 produced in HeLa/clathrin knockdown cells was about 8-fold less infectious than the same virus produced in HeLa cells stably expressing the irrelevant shRNA. Taken together, these results indicate that clathrin can have an important role in HIV-1 infectivity even in the absence of Nef.
Clathrin interacts with the p6 domains of HIV-2 and SIVmac.
The N-terminal domain of clathrin is a ß-propeller that binds to five-residue clathrin box motifs within adaptor proteins that link clathrin to cargo (14, 45). Although HIV-1 Gag and Gag-Pol do not possess an obvious clathrin box, we noticed that the p6 domain of HIV-2 Gag contains a sequence (48LLHLE52) that fits the clathrin box consensus LLpL(−), where p is a polar residue and (−) is a negatively charged residue (29). Furthermore, HIV-2 p6 harbors a second sequence (66LLHLN70) that partially matches the clathrin box motif.
To examine whether HIV-2 p6 binds to clathrin, a fusion protein consisting of GST and HIV-2 p6 was used as bait in pulldown experiments. As shown in Fig. 5A, the fusion protein specifically pulled down a 180-kDa protein from 293T cell lysates in quantities that were easily sufficient for detection by Coomassie staining. The microsequencing of the 180-kDa band yielded 22 peptides that matched the human clathrin heavy chain (18% coverage). Furthermore, Western blotting confirmed that clathrin was pulled down in considerable quantities by GST-p6HIV-2 (Fig. 5B, lane 2) but was absent when GST was used instead (Fig. 5B, lane 1). Also, no clathrin was pulled down by GST-p6HIV-1 (data not shown).
Fig. 5.
Clathrin binds to clathrin box motifs in HIV-2 p6. (A) Glutathione-Sepharose beads decorated with bacterially expressed GST (lane 1), WT GST-p6HIV-2 (lane 2), or CB(1,2)− GST-p6HIV-2 (lane 3) were incubated with 293T cell lysates, and bound proteins were detected by SDS-PAGE and Coomassie staining. (B) GST pulldowns, performed as described for panel A, showing that both clathrin box motifs in HIV-2 p6 (CB1 and CB2) contribute to clathrin binding. The captured clathrin heavy chain was detected by Western blotting.
To determine the role of the 48LLHLE52 sequence within HIV-2 p6 in the interaction, we mutated each of the three hydrophobic residues in the motif. The resulting CB1− construct, which has 48LLHLE52 replaced by SQHHE, still pulled down clathrin, albeit with reduced efficiency (Fig. 5B, lane 3). We therefore considered the possibility that the 66LLHLN70 sequence accounted for the remaining affinity of the CB1− construct for clathrin. To examine this possibility, the 66LLHLN70 sequence was changed to SQHHN within the context of the CB1− construct. The resulting CB(1,2)− double mutant no longer bound clathrin, as determined by Coomassie staining (Fig. 5A, lane 3) and by Western blotting (Fig. 5B, lane 4).
The first of the two clathrin-binding sequences in HIV-2 p6 (LLHLE) is not present in SIVmac, even though both viruses belong to the same subgroup of primate lentiviruses. However, the distal LLHLH clathrin-binding sequence of HIV-2 p6 is conserved in SIVmac. We therefore asked whether SIVmac p6 interacts with clathrin. We found that SIVmac p6 fused to GST readily pulled down the endogenous clathrin heavy chain from 293T cell lysates (Fig. 6A, lane 1). Furthermore, the mutagenesis of the conserved LLHLH sequence (residues 52 through 56 of SIVmac p6) showed that this motif was required for clathrin binding in the GST pulldown assay. Indeed, even single-amino-acid substitutions that targeted the first two positions of the LLHLH sequence (L52S and L63Q) were sufficient to eliminate binding in this assay (Fig. 6A, lanes 2 and 3). Taken together, these data indicate that HIV-2 p6 harbors two functional clathrin-binding regions, whereas SIVmac p6 harbors only one.
Fig. 6.
Single clathrin binding motif in SIVmac p6 partially overlaps with the ALIX binding site. (A) GST pulldowns, performed as described for Fig. 5, showing that leucine residues in the 52LLHLH56 clathrin box of SIVmac p6 are individually critical for clathrin binding in vitro. (B) GST pulldowns showing that only the first of three leucine residues in the 52LLHLH56 clathrin binding motif is required for ALIX binding in vitro. Beads decorated with GST or GST-p6SIVmac fusion proteins were incubated with lysate from 293T cells expressing HA-tagged ALIXΔPRD, and captured ALIX was detected by Western blotting with anti-HA antibody.
The clathrin and ALIX binding sites in SIVmac p6 partially overlap.
HIV-1 p6 interacts with ALIX through an LYPxnL-type auxiliary late domain, which is not conserved in HIV-2 or SIVmac p6 (22, 23, 31, 35, 42, 47). Instead, the equivalent position of HIV-2 and SIVmac p6 is occupied by the clathrin-binding 52LLHLH56 sequence. Nevertheless, our previous observations indicated that the p6 domains of HIV-1, HIV-2, and SIVmac share the ability to bind to ALIX. Furthermore, residues 52 to 63 of SIVmac p6 appeared to be required for ALIX binding (42). Based on these considerations, we examined the possibility that the 52LLHLH56 sequence also is involved in ALIX binding. We focused on the three leucine residues of the motif, because leucine side chains of HIV-1 p6 are known to make critical contacts with ALIX (47). As shown in Fig. 6B, a point mutation that changed the first position of the motif (L52S) abolished the interaction of SIVmac p6 with ALIX in a GST pulldown assay. In contrast, the L53Q mutation at the second position, which also eliminated clathrin binding (Fig. 6A), had no effect on the interaction with ALIX (Fig. 6B). Similarly, ALIX binding was unaffected by the L55H mutation at the fourth position of the motif (Fig. 6B). We conclude that the clathrin binding site of SIVmac p6 partially overlaps with the ALIX binding site.
The clathrin binding region of SIVmac p6 is required for efficient virus replication in primary cells.
The presence of a clathrin binding site in the Gag protein of SIVmac raised the possibility that this virus incorporates significantly larger amounts of clathrin than HIV-1, which associates with clathrin via Gag-pol. We therefore compared the abilities of WT HIV-1 and SIVmac virions to incorporate clathrin. To this end, the full-length, replication-competent HXBH10 and pSIVmac293RQ proviral clones of HIV-1 and SIVmac were transfected into 293T cells. As a control, a version of HXBH10 with a disrupted gag gene also was transfected. Virions released by the transfected cells were partially purified through sucrose and analyzed by Western blotting with anti-HIV-1 CA antibody 183-H12-5C, which also recognizes SIVmac CA (Fig. 7A). In parallel, the virion samples were analyzed with an antibody against the clathrin heavy chain. As expected, HIV-1 and SIVmac virions both incorporated clathrin (Fig. 7A). However, assuming that 183-H12-5C does not recognize SIVmac CA more efficiently than HIV-1 CA, the data shown in Fig. 7A indicate that SIVmac virions do not incorporate larger amounts of clathrin than HIV-1 virions.
Fig. 7.
Clathrin incorporation into SIVmac virions. (A) Comparison of clathrin incorporation into WT HIV-1 and SIVmac virions. 293T cells were transfected with full-length, replication-competent proviral clones of HIV-1 or SIVmac. As a control, an HIV-1 mutant with a disrupted gag gene also was included. Virion pellets were analyzed by Western blotting with anti-CA and anti-clathrin heavy-chain antibodies. (B) Gag protein profile of virions produced by SIVmac p6 mutants. 293T cells transfected with the WT and the indicated mutant versions of pSIVmac239RQ were labeled with [35S]methionine, and virion-associated proteins were detected by autoradiography. (C) Clathrin incorporation by SIVmac p6 mutants. 293T cells transfected with the WT and the indicated mutant versions of pSIVmac239RQ were labeled with [35S]methionine. CA protein in virion pellets was detected by SDS-PAGE and autoradiography, and the clathrin heavy chain was detected by Western blotting.
To determine the contribution of the 52LLHLH56 sequence within SIVmac p6 to the incorporation of clathrin, mutations that target the 52LLHLH56 sequence but do not alter the overlapping pol gene were introduced into pSIVmac239RQ. As shown in Fig. 7B, WT SIVmac and the 52LLHLH56 motif mutants produced comparable amounts of particles. However, the electrophoretic mobilities of the p6 protein bands varied, confirming the presence of mutations in p6. As is evident in Fig. 7C, the L52S and L53Q mutations, which abolished clathrin binding in vitro (Fig. 6A), did not eliminate the incorporation of clathrin into SIVmac virions. However, the clathrin content of the mutant virions was reduced by about 3- to 5-fold (Fig. 7C). A more significant, at least 10-fold, defect in clathrin incorporation was observed for the LLHL→SQHH mutant, which has all three leucine residues of the 52LLHLH56 sequence substituted without altering Gag-Pol (Fig. 7C, lane 4). Nevertheless, clathrin incorporation was not entirely abolished, since it exceeded the background level obtained with a mock-transfected control (Fig. 7C, lanes 5 and 6).
To determine the ability of these mutants to replicate, stimulated rhesus macaque PBMC were infected with viral stocks produced in 293T cells after normalization for RT activity. To monitor virus replication, we measured the release of RT activity by the infected cells over time. As shown in Fig. 8A, the L52S and L53Q mutants replicated with somewhat delayed kinetics compared to those of WT SIVmac. In contrast, the LLHL→SQHH mutant gave no consistent rise in RT activity in this experiment. We therefore examined the ability of the LLHL→SQHH mutant to replicate in PBMC from two additional monkeys, and we observed that the mutant replicated significantly slower than WT SIVmac in both cases (Fig. 8B and C).
Fig. 8.
Effects of mutations in the 52LLHLH56 motif of SIVmac p6 on virus replication in primary cells. (A to C) Macaque PBMC from three donors were exposed to equivalent amounts of WT or mutant SIVmac239 produced in 293T cells, and virus replication was monitored over time by measuring RT activity in the culture supernatants.
The LLHL→SQHH mutant harbors the L52S mutation, which abolished the SIVmac p6-ALIX interaction in vitro (Fig. 6B). However, a predicted defect in ALIX binding caused by the L52S mutation is unlikely to explain the pronounced replication defect of the LLHL→SQHH mutant, because the L52S mutation alone had only a comparatively modest effect on virus replication (Fig. 8A). We also note that the severity of the replication defect of the mutants analyzed correlated with their ability to incorporate clathrin. We thus infer that clathrin binding by SIVmac p6 is critical for virus replication in primary cells.
DISCUSSION
In addition to its essential enzymatic function during the early phase of the HIV-1 replication cycle, IN plays a poorly understood role in virus morphogenesis. In the present study, we found that the IN domain directs the incorporation of clathrin heavy and light chains into HIV-1 particles. The uptake of clathrin was specific, since none was detected in the absence of IN. Furthermore, our results indicate that HIV-1 virions incorporate clathrin in substantial amounts. Based on the intensities of the [35S]methionine-labeled clathrin heavy chain and Gag-Pol bands in Fig. 1A and on the number of methionine residues present in these molecules, we estimate that HIV-1 virions incorporate approximately one clathrin heavy-chain molecule for every two Gag-Pol molecules.
The best-characterized function of clathrin is in the formation of coated vesicles that retrieve membrane-anchored proteins from, for instance, the cell surface (29). Clathrin-coated vesicles also contain adaptor protein (AP) complexes that link clathrin to vesicular cargo (29). It has been reported that HIV-1 Gag interacts with the clathrin adaptor complexes AP1, AP-2, and AP-3, raising the possibility that the incorporation of clathrin observed in the present study occurs via AP complexes (4, 10, 15). However, the interaction with AP-1, AP-2, and AP-3 depends on the MA domain of HIV-1 Gag (4, 10, 15), which is, in its entirety, dispensable for clathrin incorporation (Fig. 1). Also, attempts to detect the γ subunit of AP-1 in MA-less virions by Western blotting were unsuccessful (data not shown), even though clathrin was readily detectable by the same method.
HIV-1 mutagenesis studies have revealed that certain point mutations in IN prevent virus replication at a step prior to integration, and that a subset of these so-called class II mutations also causes defects in particle production (18). In the present study, we observed a strict correlation between the effects of class II mutations on particle production and the effects of these mutations on clathrin incorporation, which was measured in a PR-negative context to restore particle production. Of the point mutants analyzed, the most dramatic defect in both particle production and clathrin incorporation was seen upon the replacement of phenylalanine 185 in the catalytic domain of IN by lysine. It has been shown that the activity of F185K IN is comparable to that of the WT protein in 3′ processing, strand transfer, and disintegration reactions (17, 20). Also, the structure of the catalytic domain of F185K IN is known, because the F185K change was found to improve solubility and thus enabled crystallization (17). The crystal structure of the catalytic domain of the F185K mutant is very similar to that of the F185H mutant (8), which is released normally and replicates with near-WT kinetics (20). Taken together, these observations argue against the possibility that the effects of the F185K mutation on virus morphogenesis and clathrin incorporation were due to the misfolding of the catalytic core of IN.
We note that the F185K mutation has been reported to stabilize the dimerization both of the isolated core domain and of full-length HIV-1 IN (27, 28). Thus, by promoting Gag-Pol dimerization in a proviral context, the F185K mutation could cause the premature activation of PR and, thus, assembly defects due to premature Gag processing. It also is conceivable that F185K-triggered Gag-Pol dimerization occludes an interface that is required for the association with clathrin. In support of the model that Gag-Pol dimerization occludes the clathrin binding site, we find that efavirenz, which is known to enhance RT and Pol dimerization (21), inhibits the incorporation of clathrin into HIV-1 virions. In contrast, nevirapine, a less potent enhancer of RT dimerization (21), had no significant effect on clathrin incorporation.
If class II IN mutations affect both the activation of PR and the incorporation of clathrin through effects on Gag-Pol dimerization, then reducing the association with clathrin by knocking down its cellular levels would not necessarily be expected to impair HIV-1 morphogenesis. Indeed, HIV-1 Gag processing and particle production appeared normal in cells depleted of the clathrin heavy chain, indicating that clathrin is not a release cofactor. However, we cannot exclude that even low levels of clathrin are sufficient to control Gag-Pol dimerization and thereby the activation of PR.
We have shown that the infectivity enhancement function of HIV-1 Nef depends on dynamin 2, which mediates the detachment of clathrin-coated vesicles (38). In that study, we also observed that the effect of Nef on HIV-1 infectivity was reduced in clathrin-depleted cells (38). The results of the present study indicate that clathrin additionally plays a Nef-independent role in the HIV-1 life cycle, because the infectivity of a nef-negative HIV-1 variant was significantly reduced when produced in HeLa cells stably depleted of clathrin. This contrasts with our previous observation that the low infectivity of nef-negative virus produced in Jurkat cells was not further reduced by a small interfering RNA targeting clathrin (38). These discrepancies may be attributable to differences between stable and transient depletion, the efficiency of the knockdown, or to cell type-specific differences.
Interestingly, HIV-1 is not unique among primate lentiviruses in its ability to associate with clathrin during assembly. We find that HIV-2ROD possesses two clathrin binding motifs in a region of p6 that is duplicated in HIV-2 group A isolates. The duplication of the binding site likely explains why HIV-2ROD p6 interacts with clathrin considerably more avidly than SIVmac239 p6, which harbors only one clathrin binding motif (52LLHLH56). The clathrin binding motifs in both viruses resemble the clathrin box motifs of clathrin adaptors, which bind directly to the N-terminal domain of the clathrin heavy chain (45). It thus is tempting to speculate that HIV-2 and SIVmac Gag proteins mimic clathrin adaptors, perhaps to downregulate a factor that interferes with virus replication.
The clathrin box of SIVmac occupies a position equivalent to that of the ALIX binding site in HIV-1 p6. However, in full agreement with a very recent study (48), we find that only the first position of the SIVmac clathrin box 52LLHLH56 is involved in ALIX binding. Collectively, our mutagenesis data indicate that it is primarily the role of the 52LLHLH56 sequence in clathrin binding that is critical for SIVmac replication in primary cells. Importantly, the replication defect of the SIVmac clathrin box mutants cannot be attributed to defects in the uptake of Vpx or Vpr into virions, because the entire region occupied by the clathrin box is dispensable for the incorporation of these accessory viral proteins (2).
In summary, we find that primate lentiviruses from two different subgroups associate with clathrin via Gag or Gag-Pol, and we present evidence indicating that the engagement of clathrin is functionally relevant.
ACKNOWLEDGMENTS
We thank Steven Gygi for protein microsequencing, John Gray and Richard Mulligan for pHDMHgpm2, Riri Shibata for pSIVmac239RQ, and Ronald Desrosiers for rhesus macaque blood. The following reagents were obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: efavirenz, nevirapine, HIV gp41 Chessie 8 hybridoma from George Lewis, HIV-1 p24 hybridoma (183-H12-5C) from Bruce Chesebro, and TZM-bl from John C. Kappes, Xiaoyun Wu and Tranzyme Inc.
This work was supported by grant number R37AI029873 from the National Institute of Allergy and Infectious Diseases.
Footnotes
Published ahead of print on 2 February 2011.
REFERENCES
- 1. Abacioglu Y. H., et al. 1994. Epitope mapping and topology of baculovirus-expressed HIV-1 gp160 determined with a panel of murine monoclonal antibodies. AIDS Res. Hum. Retrovir. 10:371–381 [DOI] [PubMed] [Google Scholar]
- 2. Accola M. A., Bukovsky A. A., Jones M. S., Gottlinger H. G. 1999. A conserved dileucine-containing motif in p6(gag) governs the particle association of Vpx and Vpr of simian immunodeficiency viruses SIV(mac) and SIV(agm). J. Virol. 73:9992–9999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Accola M. A., Strack B., Gottlinger H. G. 2000. Efficient particle production by minimal gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J. Virol. 74:5395–5402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Batonick M., et al. 2005. Interaction of HIV-1 Gag with the clathrin-associated adaptor AP-2. Virology 342:190–200 [DOI] [PubMed] [Google Scholar]
- 5. Bieniasz P. D. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Brodsky F. M. 1988. Living with clathrin: its role in intracellular membrane traffic. Science 242:1396–1402 [DOI] [PubMed] [Google Scholar]
- 7. Brodsky F. M., et al. 1991. Clathrin light chains: arrays of protein motifs that regulate coated-vesicle dynamics. Trends Biochem. Sci. 16:208–213 [DOI] [PubMed] [Google Scholar]
- 8. Bujacz G., Alexandratos J., Qing Z. L., Clement-Mella C., Wlodawer A. 1996. The catalytic domain of human immunodeficiency virus integrase: ordered active site in the F185H mutant. FEBS Lett. 398:175–178 [DOI] [PubMed] [Google Scholar]
- 9. Bukovsky A., Gottlinger H. 1996. Lack of integrase can markedly affect human immunodeficiency virus type 1 particle production in the presence of an active viral protease. J. Virol. 70:6820–6825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Camus G., et al. 2007. The clathrin adaptor complex AP-1 binds HIV-1 and MLV Gag and facilitates their budding. Mol. Biol. Cell 18:3193–3203 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Chesebro B., Wehrly K., Nishio J., Perryman S. 1992. Macrophage-tropic human immunodeficiency virus isolates from different patients exhibit unusual V3 envelope sequence homogeneity in comparison with T-cell-tropic isolates: definition of critical amino acids involved in cell tropism. J. Virol. 66:6547–6554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chiang C. C., Wang S. M., Pan Y. Y., Huang K. J., Wang C. T. 2010. A single amino acid substitution in HIV-1 reverse transcriptase significantly reduces virion release. J. Virol. 84:976–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Chiang C. C., Wang S. M., Tseng Y. T., Huang K. J., Wang C. T. 2009. Mutations at human immunodeficiency virus type 1 reverse transcriptase tryptophan repeat motif attenuate the inhibitory effect of efavirenz on virus production. Virology 383:261–270 [DOI] [PubMed] [Google Scholar]
- 14. Dell'Angelica E. C., Klumperman J., Stoorvogel W., Bonifacino J. S. 1998. Association of the AP-3 adaptor complex with clathrin. Science 280:431–434 [DOI] [PubMed] [Google Scholar]
- 15. Dong X., et al. 2005. AP-3 directs the intracellular trafficking of HIV-1 Gag and plays a key role in particle assembly. Cell 120:663–674 [DOI] [PubMed] [Google Scholar]
- 16. Dorfman T., Gottlinger H. G. 1996. The human immunodeficiency virus type 1 capsid p2 domain confers sensitivity to the cyclophilin-binding drug SDZ NIM 811. J. Virol. 70:5751–5757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Dyda F., et al. 1994. Crystal structure of the catalytic domain of HIV-1 integrase: similarity to other polynucleotidyl transferases. Science 266:1981–1986 [DOI] [PubMed] [Google Scholar]
- 18. Engelman A. 1999. In vivo analysis of retroviral integrase structure and function. Adv. Virus Res. 52:411–426 [DOI] [PubMed] [Google Scholar]
- 19. Engelman A., Englund G., Orenstein J. M., Martin M. A., Craigie R. 1995. Multiple effects of mutations in human immunodeficiency virus type 1 integrase on viral replication. J. Virol. 69:2729–2736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Engelman A., Liu Y., Chen H., Farzan M., Dyda F. 1997. Structure-based mutagenesis of the catalytic domain of human immunodeficiency virus type 1 integrase. J. Virol. 71:3507–3514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Figueiredo A., et al. 2006. Potent nonnucleoside reverse transcriptase inhibitors target HIV-1 Gag-Pol. PLoS Pathog. 2:e119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Fisher R. D., et al. 2007. Structural and biochemical studies of ALIX/AIP1 and its role in retrovirus budding. Cell 128:841–852 [DOI] [PubMed] [Google Scholar]
- 23. Fujii K., Hurley J. H., Freed E. O. 2007. Beyond Tsg101: the role of Alix in “ESCRTing” HIV-1. Nat. Rev. Microbiol. 5:912–916 [DOI] [PubMed] [Google Scholar]
- 24. Göttlinger H. G., Dorfman T., Cohen E. A., Haseltine W. A. 1993. Vpu protein of human immunodeficiency virus type 1 enhances the release of capsids produced by gag gene constructs of widely divergent retroviruses. Proc. Natl. Acad. Sci. U. S. A. 90:7381–7385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Gottlinger H. G., Weissenhorn W. 2010. Assembly and release, p. 187–215In Kurth R., Bannert N. (ed.), Retroviruses: molecular microbiology and genomics. Caister Academic Press, Norfolk, United Kingdom [Google Scholar]
- 26. Jacks T., et al. 1988. Characterization of ribosomal frameshifting in HIV-1 gag-pol expression. Nature 331:280–283 [DOI] [PubMed] [Google Scholar]
- 27. Jenkins T. M., Engelman A., Ghirlando R., Craigie R. 1996. A soluble active mutant of HIV-1 integrase: involvement of both the core and carboxyl-terminal domains in multimerization. J. Biol. Chem. 271:7712–7718 [DOI] [PubMed] [Google Scholar]
- 28. Jenkins T. M., et al. 1995. Catalytic domain of human immunodeficiency virus type 1 integrase: identification of a soluble mutant by systematic replacement of hydrophobic residues. Proc. Natl. Acad. Sci. U. S. A. 92:6057–6061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kirchhausen T. 2000. Clathrin. Annu. Rev. Biochem. 69:699–727 [DOI] [PubMed] [Google Scholar]
- 30. Leavitt A. D., Robles G., Alesandro N., Varmus H. E. 1996. Human immunodeficiency virus type 1 integrase mutants retain in vitro integrase activity yet fail to integrate viral DNA efficiently during infection. J. Virol. 70:721–728 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Lee S., Joshi A., Nagashima K., Freed E. O., Hurley J. H. 2007. Structural basis for viral late-domain binding to Alix. Nat. Struct. Mol. Biol. 14:194–199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Li J., Lord C. I., Haseltine W., Letvin N. L., Sodroski J. 1992. Infection of cynomolgus monkeys with a chimeric HIV-1/SIVmac virus that expresses the HIV-1 envelope glycoproteins. J. Acquir. Immune Defic. Syndr. 5:639–646 [PubMed] [Google Scholar]
- 33. Liu S. H., Wong M. L., Craik C. S., Brodsky F. M. 1995. Regulation of clathrin assembly and trimerization defined using recombinant triskelion hubs. Cell 83:257–267 [DOI] [PubMed] [Google Scholar]
- 34. Lu R., et al. 2004. Class II integrase mutants with changes in putative nuclear localization signals are primarily blocked at a postnuclear entry step of human immunodeficiency virus type 1 replication. J. Virol. 78:12735–12746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Munshi U. M., Kim J., Nagashima K., Hurley J. H., Freed E. O. 2007. An Alix fragment potently inhibits HIV-1 budding: characterization of binding to retroviral YPXL late domains. J. Biol. Chem. 282:3847–3855 [DOI] [PubMed] [Google Scholar]
- 36. Näthke I. S., et al. 1992. Folding and trimerization of clathrin subunits at the triskelion hub. Cell 68:899–910 [DOI] [PubMed] [Google Scholar]
- 37. Paxton W., Connor R. I., Landau N. R. 1993. Incorporation of Vpr into human immunodeficiency virus type 1 virions: requirement for the p6 region of gag and mutational analysis. J. Virol. 67:7229–7237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pizzato M., et al. 2007. Dynamin 2 is required for the enhancement of HIV-1 infectivity by Nef. Proc. Natl. Acad. Sci. U. S. A. 104:6812–6817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Puig-Kröger A., et al. 2000. Polyomavirus enhancer-binding protein 2/core binding factor/acute myeloid leukemia factors contribute to the cell type-specific activity of the CD11a integrin gene promoter. J. Biol. Chem. 275:28507–28512 [DOI] [PubMed] [Google Scholar]
- 40. Reil H., Bukovsky A. A., Gelderblom H. R., Gottlinger H. G. 1998. Efficient HIV-1 replication can occur in the absence of the viral matrix protein. EMBO J. 17:2699–2708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shibata R., et al. 1991. Generation of a chimeric human and simian immunodeficiency virus infectious to monkey peripheral blood mononuclear cells. J. Virol. 65:3514–3520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Strack B., Calistri A., Craig S., Popova E., Gottlinger H. G. 2003. AIP1/ALIX is a binding partner for HIV-1 p6 and EIAV p9 functioning in virus budding. Cell 114:689–699 [DOI] [PubMed] [Google Scholar]
- 43. Tachedjian G., Moore K. L., Goff S. P., Sluis-Cremer N. 2005. Efavirenz enhances the proteolytic processing of an HIV-1 pol polyprotein precursor and reverse transcriptase homodimer formation. FEBS Lett. 579:379–384 [DOI] [PubMed] [Google Scholar]
- 44. Tachedjian G., Orlova M., Sarafianos S. G., Arnold E., Goff S. P. 2001. Nonnucleoside reverse transcriptase inhibitors are chemical enhancers of dimerization of the HIV type 1 reverse transcriptase. Proc. Natl. Acad. Sci. U. S. A. 98:7188–7193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. ter Haar E., Harrison S. C., Kirchhausen T. 2000. Peptide-in-groove interactions link target proteins to the beta-propeller of clathrin. Proc. Natl. Acad. Sci. U. S. A. 97:1096–1100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wu X., et al. 1999. Human immunodeficiency virus type 1 integrase protein promotes reverse transcription through specific interactions with the nucleoprotein reverse transcription complex. J. Virol. 73:2126–2135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhai Q., et al. 2008. Structural and functional studies of ALIX interactions with YPX (n) L late domains of HIV-1 and EIAV. Nat. Struct. Mol. Biol. 15:43–49 [DOI] [PubMed] [Google Scholar]
- 48. Zhai Q., Landesman M. B., Robinson H., Sundquist W. I., Hill C. P. 2011. Identification and structural characterization of the ALIX-binding late domains of simian immunodeficiency virus SIVmac239 and SIVagmTan-1. J. Virol. 85:632–637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Zuhorn I. S., Kalicharan R., Hoekstra D. 2002. Lipoplex-mediated transfection of mammalian cells occurs through the cholesterol-dependent clathrin-mediated pathway of endocytosis. J. Biol. Chem. 277:18021–18028 [DOI] [PubMed] [Google Scholar]