Abstract
Helper-dependent adenovirus (hdAd) vectors have shown tremendous potential in animal models of human disease in numerous preclinical studies. Expression of a therapeutic transgene can be maintained for several years after a single administration of the hdAd vector. However, despite the long-term persistence of hdAd DNA in the transduced cell, little is known of the fate and structure of hdAd DNA within the host nucleus. In this study, we have characterized the assembly of hdAd DNA into chromatin in tissue culture. Eviction of the Ad DNA-packaging protein VII, histone deposition, and vector-associated gene expression all began within 2 to 6 h of host cell transduction. Inhibition of transcription elongation through the vector DNA template had no effect on the loss of VII, suggesting that transcription was not necessary for removal of the majority of protein VII. Vector DNA assembled into physiologically spaced nucleosomes within 6 h. hdAd vectors incorporated the histone H3 variant H3.3, which was dependent on the histone chaperone HIRA. Knockdown of HIRA reduced hdAd association with histones and reduced expression of the vector-carried transgene by 2- to 3-fold. Our study elucidates an essential role for hdAd DNA chromatinization for optimal vector gene expression.
INTRODUCTION
Fifty years of intense research has resulted in adenovirus (Ad) type 5 being among the best-characterized human viruses (4, 61). In part, these studies have contributed to its development as an effective gene delivery tool in many applications, including gene therapy (3). To increase the safety and efficacy of Ad vectors, we and others have developed Ad-based vectors devoid of all viral protein-coding sequences, termed helper-dependent adenovirus (hdAd) (47, 49). hdAd can mediate very-long-term expression of a therapeutic transgene in vivo in a number of animal models (up to several years in nonhuman primates) (47), while eliciting reduced deleterious acute and chronic toxicity and immune activation (40, 41). However, despite these very promising results, little is known about the structure of the hdAd within the infected cell and how this contributes to the persistence of the vector DNA and transgene expression. Indeed, despite our detailed knowledge of many aspects of the wild-type Ad life cycle, the fate and structure of Ad DNA within the nucleus of the infected cell are poorly characterized.
Within the capsid, Ad DNA is condensed within a subvirion structure called the Ad core by association with the following three highly basic protamine-like proteins that are encoded by the virus: V, VII, and mu (9). During wild-type Ad infection, the major DNA-binding protein, protein VII, enters the nucleus with the Ad DNA (9). However, conflicting data suggest that VII either stably associates with Ad DNA throughout the early phase of infection (9, 77) or is evicted within a few hours (70), and eviction of VII may require active transcription (10). Few studies have addressed whether Ad DNA in the nucleus directly interacts with histones or assembles into chromatin; indeed, conflicting data suggest that Ad DNA is (5, 13, 14, 66) or is not (76) assembled into chromatin. Histone H1 escorts the Ad core (Ad DNA bound to protein VII) through the nuclear pore, although this function for H1 appears independent of any role in condensing the viral DNA, and whether H1 continues to associate with the viral DNA within the nucleus is unknown (74). It is clear that infection of a cell by wild-type Ad causes a global change in the expression and epigenetic status of many cellular genes, primarily due to the ability of early region 1A (E1A) proteins to restrict p300/CBP histone acetyltransferase activity to a subset of E2F-responsive promoters (4, 18, 19, 25). Through this ability to modulate p300/CBP, E1A was also shown to enhance the acetylation of histone H4 at the Ad12 early region 2 (E2) promoter in plasmid-based reporter constructs and presumably also in the context of replicating virus (17). Recent studies have shown that, under nonreplication conditions (e.g., in nonpermissive cells in culture or mouse liver in vivo), early region 1 (E1)-deleted Ad and hdAd vectors remain primarily monomeric and episomal (16, 27), but whether these vectors are chromatinized is unknown.
In this study, we have analyzed the nucleoprotein structure of hdAd in infected cells and evaluated its impact on expression of vector-carried genes. Our results indicate that eviction of Ad DNA-packaging proteins, histone deposition, and transgene expression all begin within hours of host cell transduction. Vector DNA was assembled into physiologically spaced nucleosomes within 6 h postinfection (hpi). hdAd DNA associated with histone H3 variant H3.3 early after infection, and H3.3 deposition was dependent upon the histone chaperone HIRA. Finally, inhibition of H3.3 deposition reduced expression of vector-carried genes, indicating that assembly into chromatin is essential for efficient viral gene expression.
MATERIALS AND METHODS
Cell culture.
Propagation of 293 (a kind gift from Frank Graham, McMaster University) (22), 293-N3S (Microbix) (21), A549 (ATCC), and 116 (a kind gift from Philip Ng, Baylor College of Medicine) cells was performed as described previously (46, 59). HeLa cells (ATCC) were maintained in Dulbecco's modified Eagle's medium (DMEM) serum supplemented with 10% fetal bovine serum, 2 mM Glutamax, and 1× antibiotic-antimycotic (Invitrogen). pCMV-H3.3-YFP was obtained from Addgene. The H3.1 cDNA was amplified from human genomic DNA by PCR (using Platinum High-Fidelity Taq polymerase [Invitrogen]), subcloned into pCMV-H3.3-YFP (in place of H3.3), and confirmed by sequencing.
Pooled small interfering RNA (siRNA) targeting human HIRA and control nontargeting pooled siRNA were obtained from Dharmacon. Cells were transfected with 100 or 200 nM siRNA using Lipofectamine 2000. To analyze the effects of siRNA-mediated knockdown of HIRA on cell growth, HeLa cells in 35-mm dishes were transfected with 100 nM control or HIRA-directed siRNA and, 48 h later, replated at low density, and cell growth was monitored using a crystal violet staining protocol, as previously described (26). In experiments involving the transcription elongation inhibitor DRB (5,6-dichlorobenzimidazole 1-β-d-ribofuranoside; Sigma), cells were pretreated for 1 h with 500 μM DRB or dimethyl sulfoxide (DMSO) vehicle control in DMEM; all buffers and media were supplemented with 500 μM DRB throughout the duration of the experiments.
Virus culture.
hdAdΔ28lacZ was a kind gift from Philip Ng (Baylor College of Medicine), has been previously described (46), and is referred to as hdAd-lacZ in this study. hdAd-lacZ is devoid of all Ad protein-coding sequences but contains an ∼22-kb fragment of eukaryotic-derived, noncoding stuffer DNA and the Escherichia coli lacZ gene under regulation by the murine cytomegalovirus immediate-early promoter and enhancer and the simian virus 40 polyadenylation sequences. The lacZ expression cassette is located 7.8 kb from the left inverted terminal repeat and packaging element, and its transcription is directed rightward relative to the conventional Ad map. hdAdΔ28E4 is the parental vector from which hdAdΔ28lacZ was derived and lacks a transgene. hdAd-PGK-mSEAP contains the murine secreted alkaline phosphatase cDNA (12, 36) under regulation by the murine phosphoglycerate kinase promoter (41, 56). hdAd-PGK-mSEAP, hdAd-lacZ, and hdAdΔ28E4 were amplified with the helper virus AdNG163 (46) or AdJR46 using 116 cells and standard techniques (46, 49). AdJR46 is similar in structure to AdRP2050 (6) but is engineered to contain a FLAG epitope tag on the C terminus of core protein VII (67). The presence of the FLAG tag on core VII has no discernible deleterious effect on virus viability. The E1/E3-deleted Ad vector AdCA35 has been described previously (1) and carries a lacZ expression cassette identical to that of hdAd-lacZ. All E1-deleted Ads were propagated and their titers were determined in 293 cells, as described previously (59). All vectors were purified by cesium chloride buoyant density centrifugation, using standard procedures (46, 59). Infectious hdAd particles were scored through staining of infected 293 cells and visual inspection, using previously established protocols (36, 49). Particle counts were determined as previously described (59). Contamination of hdAd vector stocks with helper virus was determined by plaque assays and scored as the numbers of PFU (44, 59). Importantly, all hdAd vectors had similar ratios of hdAd to helper virus (approximately 0.05% helper virus contamination).
Expression assays and immunoblotting.
To assay reporter gene expression, crude cell lysates were harvested in 1× reporter lysis buffer (Promega) at the indicated time points and assayed for gene expression using a chemiluminescent β-galactosidase (β-Gal) assay kit (Roche). To prepare samples for immunoblotting, the cells were treated, and at the indicated time points, the medium was removed, and the cells were overlaid with 2× protein sample buffer. Immunoblotting was performed using standard techniques. For experiments involving protein extracts from isolated nuclei, cells were lysed and fractionated as described by Challberg and Kelly (8), with the exception that nuclei were further purified by centrifugation through a 0.88 M sucrose cushion. Nylon membranes were probed with anti-FLAG M2 (1:15,000 dilution; Sigma), anti-α-tubulin (1:10,000; Oncogene), anti-green fluorescent protein (GFP) (1:2,000; Invitrogen) (which cross-reacted with the yellow fluorescent protein [YFP] tag), or anti-Ad capsid protein (ab6982) (1:10,000; Abcam). Antibody to HIRA (clone WC119 [1:50]) was kindly provided by Peter Adams, Fox Chase Cancer Center, Philadelphia, PA (23). Antibody to core protein VII was a kind gift from S. J. Flint (Princeton) and was used at a dilution of 1/100. Of note, we have observed that several commercially available histone antibodies cross-react with Ad core protein V or VII; the anti-H3 antibody used in these experiments does not (catalog no. 2650; Cell Signaling).
ChIP.
Chromatin immunoprecipitation (ChIP) assays were performed using a modified version of the Upstate Biotechnology protocol. Lysis buffers and ChIP dilution buffer were supplemented with 1 mM phenylmethylsulfonyl fluoride (PMSF; OmniPur), 10 μg/ml leupeptin (Roche), and 10 μg/ml aprotinin (Roche). Confluent HeLa or A549 cells in 15-cm dishes (∼2.5 × 107 cells) were infected with virus at a multiplicity of infection (MOI) of 10, and at the indicated time points, formaldehyde was added to 1%, and the plates were incubated for 10 min. After 5 min of incubation in 0.125 M glycine, the monolayers were rinsed twice with cold phosphate-buffered saline (PBS) and scraped into PBS. The cells were pelleted (2,000 rpm, 10 min, 20°C), suspended in 0.5 ml cell lysis buffer {5 mM PIPES [piperazine-N,N′-bis(2-ethanesulfonic acid)] at pH 8, 85 mM KCl, 0.5% NP-40} and incubated for 45 min at 4°C with constant rotation. The nuclei were pelleted by centrifugation (5,000 rpm, 5 min, 4°C) and lysed in 0.25 ml nuclear lysis buffer (50 mM Tris-HCl at pH 8, 1% SDS, 10 mM EDTA). Chromatin was sheared to an average size of 500 bp by sonication, as follows: 5 to 10 pulses for 10 s with a Vibra-Cell VCX600 ultrasonic process (Sonics & Materials) equipped with a stepped microtip at a setting of 30% of maximal amplitude. Sheared chromatin was diluted by 10-fold with ChIP dilution buffer containing 22.2 μg/ml sheared herring sperm DNA and 1.11 mg/ml bovine serum albumin (BSA). Dilute chromatin was precleared with 50 μl of a 50% slurry of protein G beads for 2 h at 4°C with constant rotation. Precleared chromatin was incubated overnight with antibody and 50 μl of a 50% slurry of protein G beads at 4°C with constant rotation. Antibodies used for ChIP analysis were rabbit IgG anti-GFP (A-11122; Invitrogen), anti-FLAG M2 (Sigma), anti-H3 (catalog no. 2650; Cell Signaling), or control IgG. After immunoprecipitation (IP), beads were rinsed once with low-salt immune complex wash buffer (ICWB), high-salt ICWB, and lithium chloride ICWB and twice with Tris-EDTA (TE). The beads were incubated twice for 15 min with 0.25 ml immune complex elution buffer (100 mM sodium bicarbonate and 1% SDS). Eluates were combined, NaCl was added to 200 mM, and the samples were incubated for 4 to 6 h at 65°C. After cross-link reversal, 10 μl 0.5 mM EDTA, 20 μl 1 M Tris-HCl at pH 6.5, and 1 μl 20 mg/ml proteinase K were added to the samples and incubated for 2 h at 42°C. DNA was purified by phenol-chloroform extraction and precipitated with isopropanol and 20 μg glycogen. ChIP DNA was suspended in 20 μl H20, and input DNA was suspended in H20 (0.02% of input/μl). Alternatively, ChIP experiments were performed using the SimpleChIP enzymatic chromatin IP kit (with magnetic beads) obtained from Cell Signaling Technologies, according to the manufacturer's instructions. Quantitative PCR (qPCR) was performed using 1 μl of diluted DNA (1/50 for ChIP DNA and 1/200 for input) with SYBR green JumpStart Taq ReadyMix (Sigma), according to the manufacturer's instructions, with the exception that the reaction was performed in a total volume of 20 μl. The following primer sets for qPCR were used: lacZ, 5-TAC GCC AAT GTC GTT ATC CA and 5-CAC CAG CGA AAT GGA TTT TT; hexon, 5-CTT ACC CCC AAC GAG TTT GA and 5-GGA GTA CAT GCG GTC CTT GT; human β-globin, 5′-GAC AAC CTC AAG GGC ACC TTT and 5′-GAG GAT CCA CGT GCA GCT T. The PCR was performed on a MX3000P (Stratagene), with 40 amplification cycles and a 60°C annealing temperature. The threshold cycle (CT) values for each sample were compared to a dilution series of plasmids containing the target DNA and converted into an absolute DNA concentration.
Micrococcal nuclease accessibility assays.
A549 cells (∼2.5 × 107) in 15-cm dishes were infected with hdAd-PGK-mSEAP at an MOI of 500. At the indicated time points after infection, nuclei were isolated as described previously (42). Isolated nuclei (∼5 × 106) in aliquots of 0.2 ml were treated with 0, 1.5, 5, 10, or 25 units micrococcal nuclease (MNase; Roche) for 5 min at 22°C. Digests were terminated by addition of 0.2 ml stop solution (10 mM Tris-HCl at pH 7.6, 10 mM EDTA, 0.5% SDS, 0.4 mg/ml proteinase K), followed by overnight incubation at 42°C. DNA was purified by phenol-chloroform extraction and ethanol precipitated. Purified DNA (10 μg) from the 1.5-, 5-, and 10-unit treatments were separated on 20-cm 1.25% agarose gels, transferred to a nylon membrane, and subjected to Southern blotting (69) with a digoxigenin-labeled 2.15-kbp fragment from hdAd-PGK-mSEAP containing the PGK promoter and mSEAP open reading frame. DNA labeling and probe detection were performed using the digoxigenin (DIG) High Prime DNA labeling and detection starter kit II (Roche), according to the manufacturer's instructions.
RESULTS
Ad DNA-binding proteins dissociate from hdAd DNA soon after nuclear translocation.
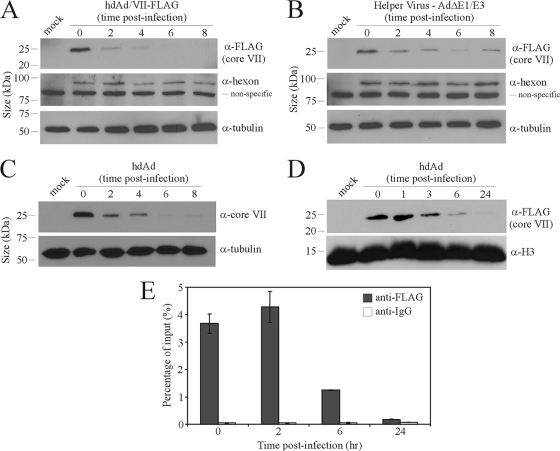
Ad DNA within the capsid is condensed by association with the virus-encoded protein VII. We generated an E1-deleted, first-generation Ad (fgAd) helper virus that encoded a FLAG-tagged VII protein and used it to generate hdAd with the FLAG-tagged core protein. We then examined the fate of VII within A549 cells at various times postinfection with the hdAd. Initially, we used a high MOI (5,000 infectious particles/cell) to allow for detection of protein VII from the infecting virions; however, subsequent studies revealed that a sufficient signal was achieved with a 10-fold-lower quantity of vector. Through immunoblot analysis of whole-cell protein extracts, we noted a dramatic loss of VII within the cells in the first 2 h following infection, with a more gradual but continual decline in the levels of VII up to our final time point of 8 hpi (Fig. 1A). We observed an identical decline in VII levels when the infected cells were lysed and boiled directly in SDS-protein loading buffer (data not shown), indicating that protein VII is degraded rather than becomes a part of the insoluble fraction. The levels of hexon protein remained relatively stable over this period, indicating that the decline in VII in the cells was not due to generalized degradation of infecting virions but rather a specific loss of VII. We observed a similar loss of VII from cells infected with the helper virus used to generate hdAd/VII-FLAG, although the level of VII appeared to rise at the 8 hpi time point, likely due to virus replication and the onset of late gene expression (Fig. 1B), as demonstrated previously for high MOIs of E1-deleted viruses in A549 cells (43). Consistent with this, we also observed an increase in hexon at 8 hpi in cells infected with the helper virus. We observed similar kinetics of loss of the wild-type VII (i.e., without a FLAG tag) from hdAd-infected cells, indicating that the presence of the FLAG tag had no deleterious effect on VII function or stability (Fig. 1C).
Fig. 1.
Ad core protein VII is rapidly removed from viral DNA after infection. (A) A549 cells were infected with hdAd-lacZ (5,000 infectious particles/cell), and whole-cell lysates were harvested at various times after infection and subjected to immunoblotting with anti-FLAG to detect core VII, an antibody recognizing all Ad capsid components (hexon is shown), or anti-α-tubulin. (B) A549 cells were infected with the E1-deleted helper virus AdJR46 (5,000 infectious particles/cell) and processed as described in the legend to panel A. (C) A549 cells were infected with hdAd-lacZ (5,000 infectious particles/cell) containing a wild-type VII protein, and whole-cell lysates were harvested at various times after infection and subjected to immunoblotting to detect core VII or α-tubulin. (D) A549 cells were infected with hdAd-lacZ (250 infectious particles/cell), nuclei were isolated, and protein extracts were prepared at various times after infection and subjected to immunoblotting with anti-FLAG to detect core VII or anti-H3 as a loading control. (E) A549 cells were infected with hdAd-lacZ (10 infectious particles/cell) and processed for ChIP with anti-FLAG or control IgG at the indicated time points. The resulting ChIP DNA was analyzed by quantitative PCR with primers for the β-galactosidase coding sequence (lacZ), and the fold of enrichment was calculated as described in Materials and Methods (error bars represent the ranges of values obtained from two replicate experiments).
The results presented in Fig. 1A to C are based on analysis of total protein within the cell (i.e., cytoplasm and nucleus) and may not accurately reflect the stability of core VII within the nucleus. We therefore isolated nuclei from cells infected with hdAd and examined the fate of protein VII. As shown in Fig. 1D, the majority of core protein VII was lost from the nucleus between 1 and 3 hpi; however, a fraction of core VII continued to persist in the nucleus until 24 hpi. We also directly examined the association of VII with the viral DNA by chromatin immunoprecipitation (ChIP). Association of vector DNA with core VII appeared to correlate with its persistence in the nucleus (Fig. 1E); we observed high association between VII and the viral DNA early after infection, which declined significantly by 6 hpi and further by 24 hpi. These data suggest that the majority of core VII is evicted from the viral DNA within the first few hours of infection but that a small proportion of VII remains in the nucleus and may continue to associate with the vector DNA until at least 24 hpi.
Transcription elongation is not required for removal of VII from hdAd DNA.
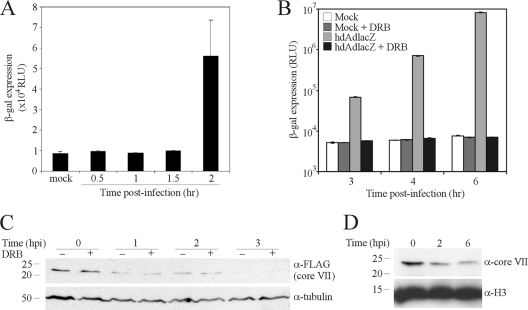
Data from immunofluorescence-based experiments suggested that core VII remains stably associated with wild-type Ad DNA throughout the early phase of infection, until the onset of late gene expression (∼10 to 12 hpi), and that transcription is necessary to evict core VII (10). Using hdAd which contained only noncoding DNA (i.e., lacked a transgene), VII was shown to remain stably associated with the vector DNA until at least 18 hpi by immunofluorescence analysis. The hdAd vector shown in Fig. 1 of our study contains the strong murine cytomegalovirus (CMV) promoter driving expression of lacZ (1, 46), suggesting that active transcription elongation could have been involved in the relatively early removal of VII from our vector, followed by its subsequent degradation. In support of this idea, initial detection of hdAd vector gene expression occurred at 2 hpi (Fig. 2A), a time frame similar to the onset of VII eviction and degradation (Fig. 1). To determine whether transcription was required for eviction of VII in our system, we examined the loss of VII from the cell in the absence or presence of a pharmacological inhibitor of transcription elongation, DRB (5,6-dichlorobenzimidazole 1-β-d-ribofuranoside). Pretreating cells with 500 μM DRB was sufficient to prevent transcription and expression from even the highly active murine CMV promoter for at least 6 hpi (Fig. 2B). A549 cells were pretreated for 1 h with DRB and infected with hdAd/VII-FLAG, and loss of VII was examined at various times postinfection by immunoblotting. As shown in Fig. 2C, inhibition of transcription elongation had no effect on the kinetics for loss of VII. We also examined loss of VII from cells infected with a vector lacking a transgene, hdAdΔ28E4. Once again, we observed that the majority of VII was lost from the cell within a few hours of infection (Fig. 2D). Taken together, these results indicate that the majority of core VII is removed from vector DNA within the first few hours of infection, which occurs independently of transcription elongation, but that a proportion of VII does remain associated with vector or viral DNA well into infection (∼10 to 24 h) (Fig. 1) (10).
Fig. 2.
Eviction of core protein VII is not dependent on active transcription. (A) A549 cells were infected with hdAd-lacZ (MOI = 100 infectious particles/cell), and β-Gal activity was determined at the indicated time points (expressed in relative light units [RLU]). Representative data from two experiments are displayed; error bars represent the ranges of values obtained from two replicate samples. (B) A549 cells were pretreated with 500 μM DRB (or DMSO vehicle) for 1 h and infected with hdAd-lacZ (100 infectious particles/cell), and β-Gal activity was determined at the indicated time points (expressed in relative light units [RLU]). (C) A549 cells were pretreated for 1 h with 500 μM DRB or vehicle (DMSO) and infected with hdAd-lacZ (250 infectious particles/cell), and at the indicated time points, whole-cell lysates were prepared and subjected to immunoblotting with anti-FLAG to detect core VII or anti-α-tubulin. (D) A549 cells were infected with a hdAd vector lacking a transgene (hdAdΔ28; 2,500 total particles/cell), and at the indicated time points, nuclei were isolated, and protein extracts were prepared and subjected to immunoblotting with anti-core VII or anti-H3 as a loading control.
hdAd DNA assembles into physiologically spaced nucleosomes.
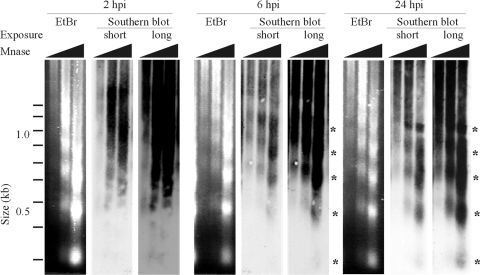
The dissociation of VII from vector DNA led us to examine whether VII is replaced by cellular histones, leading to assembly of hdAd DNA into chromatin. Southern analysis of MNase-digested nuclei isolated from hdAd-infected cells revealed that at the early time point of 2 hpi, vector DNA exhibited no evidence of a periodic oligomeric array (Fig. 3). However, by 6 hpi and continuing until 24 hpi, hdAd vector DNA exhibited MNase protection that was similar to that of bulk cellular chromatin. These data suggest that hdAd DNA-packaging proteins are replaced with cellular histones, which results in the assembly of vector DNA into physiologically spaced nucleosomes.
Fig. 3.
hdAd vector DNA assembles into physiologically spaced chromatin after host cell transduction. A549 cells were infected with hdAd-PGK-mSEAP (MOI = 500), and the samples were processed for micrococcal nuclease (MNase) assay as described in Materials and Methods. The ethidium bromide (EtBr)-stained agarose gel (showing bulk cellular chromatin) and short and long autoradiograph exposures (showing Ad DNA) are displayed. Bands representing DNA that was protected from MNase cleavage are indicated by asterisks.
hdAd chromatin incorporates histone H3.3.
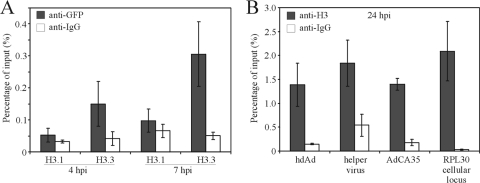
We have shown that the majority of the DNA-condensing core VII protein are removed from the viral DNA within the first few hours of infection (Fig. 1 and 2), which are then wrapped in structures resembling nucleosomes (Fig. 3). hdAd vectors are incapable of autonomous replication, and Jager and Ehrhardt (27) have shown that hdAd persists without undergoing replication by the host replicative machinery, suggesting that deposition of histones on hdAd DNA occurs via a replication-independent mechanism. Replication-independent assembly of chromatin in mammalian cells is generally viewed as a slow process (requiring ∼3 to 4 days [7]) that produces aberrantly spaced nucleosomes (reviewed in reference 68), which is in marked contrast to our observations (Fig. 3). To confirm that the hdAd vector underwent replication-independent chromatin assembly, we tested for the presence of markers of replication-coupled versus replication-independent chromatin assembly. Two major H3 variants are expressed in mammalian cells, as follows: H3.1, which is exclusively deposited during DNA replication, and H3.3, which is deposited in a replication-independent manner (2, 73). H3.3 deposition occurs primarily on actively transcribed genes or on incoming pronuclear DNA during fertilization (34, 65) and was shown recently to be deposited on telomeres (20, 75). To test whether hdAd DNA associated with H3.1 or H3.3, cells were transfected with plasmids encoding YFP-tagged H3.1 or H3.3, and we examined their association with hdAd vector DNA via ChIP at 4 and 7 hpi. Previous studies have shown that addition of an autofluorescent protein tag does not affect deposition patterns of H3 variants (2). hdAd vectors displayed a preference for association with H3.3 (Fig. 4A), suggesting that histone deposition on hdAd DNA occurred primarily via a replication-independent mechanism. Since association with H3.3 was detected as early as 4 hpi, these data suggest that removal of VII (Fig. 1), initiation of hdAd gene expression (Fig. 2), and deposition of cellular histones (Fig. 4) all occur with similar kinetics.
Fig. 4.
hdAd vector chromatin preferentially associates with histone variant H3.3. (A) HeLa cells were transfected with plasmids encoding H3.1-YFP or H3.3-YFP, and 24 h posttransfection, the cells were infected with hdAd-lacZ (MOI = 10). Four or seven hpi, the cells were processed for ChIP with the indicated antibodies, and the associated DNA was evaluated by quantitative PCR (n = 3). Of note, the GFP antibody cross-reacts with the YFP tag on H3.1 and H3.3. (B) HeLa cells were infected with hdAd-lacZ or AdCA35 (MOI = 10), and 24 hpi, the infected cells were processed for ChIP with anti-H3 or IgG antibody. The resulting ChIP DNA was analyzed by qPCR for the presence of hdAd DNA (lacZ amplicon), the E1-deleted helper virus (hexon amplicon), AdCA35 DNA (lacZ amplicon), or the cellular RPL30 locus and plotted as average values ± standard errors (n = 4).
Current methods of production for hdAd vectors result in stocks that contain a small amount of the E1-deleted helper virus (49). Therefore, we examined whether the E1-deleted helper virus also associated with cellular histones. As expected, we observed significant association of hdAd DNA with histone H3 at 24 hpi, and we also observed association of helper virus DNA with H3 (Fig. 4B). We also examined H3 association with a second E1-deleted Ad vector, AdCA35, which showed a level of association similar to those of the hdAd and helper, all of which showed a level of association similar to that of an endogenous locus, RPL30. Thus, under nonreplicating conditions, both hdAd and E1-deleted Ad associate with cellular histones to a degree similar to that of host DNA.
HIRA is necessary for deposition of H3.3 on hdAd vector DNA.
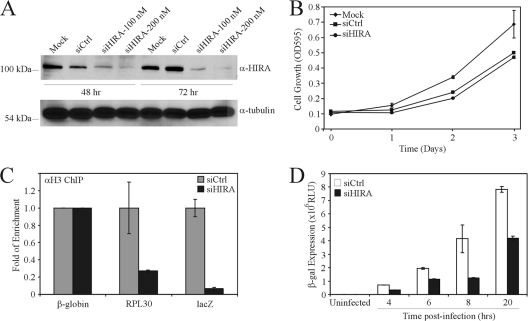
Biochemical analyses of human cells have shown that at least two H3.3 chaperones exist, HIRA (73) and DAXX (15). However, DAXX appears to be involved primarily in the deposition of H3.3 on pericentric DNA repeats and plays only a limited role in the deposition of H3.3 on active DNA templates (15). DAXX is actively degraded in wild-type Ad-infected cells (64). In contrast, HIRA is responsible for initial histone placement on incoming pronuclear DNA in Drosophila (34), and recent data have shown that knockdown of HIRA reduces H3.3 association with herpes simplex virus (HSV) DNA at early times of infection (i.e., before viral DNA replication [53]). Thus, we examined whether HIRA was involved in the deposition of H3.3 on hdAd DNA. We used small interfering RNA (siRNA) to deplete cells of HIRA to examine its role in the deposition of H3.3 on hdAd vector DNA. Treatment with siRNA targeting HIRA mRNA (siHIRA) resulted in a significant depletion of HIRA at both 48 and 72 h posttreatment (Fig. 5A). Since expression of virus-carried genes is intimately tied to the health of the host cell, we next tested whether siRNA-mediated knockdown of HIRA affected the ability of the cells to replicate. HeLa cells were treated with 100 nM control or HIRA-directed siRNA for 48 h and replated at low density, and the growth was followed over the next 3 days using a crystal violet staining assay, as we have described previously (26). Treatment of the cells with control or HIRA-directed siRNA did not result in any apparent reduction in cell viability but did cause a small decrease in the growth rate of the cells relative to that of untreated cells, but this was not specific to knockdown of HIRA (Fig. 5B).
Fig. 5.
Deposition of H3.3 promotes efficient expression of virus-carried genes. (A) HeLa cells were transfected with control siRNA (siCtrl) or siRNA targeting HIRA mRNA (siHIRA), and 48 or 72 h posttransfection, the cells were analyzed by immunoblotting for expression of HIRA or tubulin. (B) HeLa cells were transfected with 100 nM siCtrl or siHIRA and, 48 h later, replated to evaluate cellular growth kinetics. OD595, optical density at 595 nm. (C) HeLa cells were transfected with siCtrl or siHIRA, and 48 h later, the cells were infected with hdAd-lacZ (MOI = 10). Six hours later, the cells were processed for ChIP with anti-H3 or IgG antibody. The resulting ChIP DNA was analyzed by qPCR for the presence of hdAd DNA (lacZ amplicon), the cellular β-globin, or RPL30 loci. The level of enrichment with H3 is expressed relative to that of siCtrl (n = 4). (D) HeLa cells were transfected with siCtrl or siHIRA, infected 48 later with hdAd-lacZ (MOI = 10), and analyzed for β-Gal activity at various time postinfection (n = 3; means ± standard errors of the means).
To test whether HIRA depletion abrogates H3 deposition on hdAd DNA, we transfected cells with control siRNA (siCtrl) or siHIRA and used ChIP to detect the association of H3 with the incoming hdAd vector DNA at 6 hpi. As controls, we analyzed H3 association with the β-globin gene, which is not active in HeLa cells, and the ribosomal RPL30 locus, which is highly transcribed and thus predicted to demonstrate reduced association with H3.3 upon HIRA knockdown. Knockdown of HIRA resulted in a reduced association of hdAd DNA with H3 (Fig. 5C). Thus, HIRA is necessary for efficient deposition of H3 on hdAd vector DNA.
Assembly into chromatin promotes efficient expression of hdAd-carried genes.
Modified histones serve as docking sites for the recruitment of factors that promote or alter gene expression (reviewed in reference 28), suggesting that assembly into chromatin may be important for rapid and efficient initiation of expression of virus- or vector-carried genes. The H3/H4 tetramer forms the core of the nucleosome (35); therefore, disruption of H3 deposition should preclude nucleosome formation and chromatin assembly. We hypothesized that siRNA-mediated depletion of HIRA, and the resultant abrogation of H3.3 deposition, could be used to test whether assembly into chromatin affects hdAd-carried transgene expression in vivo. HeLa cells were pretreated for 48 h with 100 nM siRNA directed toward HIRA (or control siRNA) and infected with hdAd-lacZ, and transgene expression was analyzed at various times postinfection. Treatment of cells with siHIRA resulted in a 2- to 3-fold reduction in gene expression from the hdAd (P < 0.05) (Fig. 5D). This level of reduction in gene expression from the hdAd template is similar to the decline in viral gene expression that was observed upon HIRA knockdown in HSV-infected cells (53). Thus, these data indicate that de novo assembly of hdAd DNA into chromatin aids in the establishment of efficient gene expression.
DISCUSSION
In this study, we have examined the fate and structure of Ad vector DNA within the infected cell. Protein VII is a protamine-like protein and is responsible for wrapping and condensing the viral DNA in the infecting virion (39, 71), and it is this protein VII-wrapped DNA that enters the nucleus (9). This highly condensed nucleoprotein structure is refractory to viral gene expression, and for wild-type Ad, preventing decondensation of the Ad nucleoprotein core reduces expression of virus-carried genes and virus replication (24). Thus, Ad DNA must be remodeled from a core VII-wrapped DNA to a form more conducive to these processes.
We showed that the majority of core VII dissociates from the vector DNA and is degraded within the first few hours of reaching the nucleus (Fig. 1) and that transcription elongation on the viral DNA template is not required for this event (Fig. 2). However, even up to 24 hpi, a small proportion of VII does persist in the nucleus and may be associated with the vector DNA. Previous work by Chen and coworkers demonstrated that the level of VII within cells changed very little between 4 and 12 hpi and that foci of VII (presumably VII-associated viral genomes) could be detected in the nucleus by immunofluorescence for up to ∼12 hpi (10). However, that report did not examine the levels of VII at earlier time points (i.e., between 0 and 4 hpi) or whether the nuclear foci of VII contained Ad DNA completely or only partially wrapped in VII. siRNA-mediated knockdown of TAFI/SET1, a protein implicated in removing VII from viral DNA, delays viral gene expression, DNA replication, and yield (24), suggesting that timely removal of at least some of VII is required for optimal virus function. Komatsu et al. (31) showed that in a plasmid-based in vitro assay, addition of VII actually enhanced transcription and hypothesized that small quantities of VII may function in part to keep repressive histone/chromatin structures from forming on certain promoter regulatory elements. Indeed, based on ChIP/re-ChIP experiments, both VII and histones can be found to be associated with the same DNA molecule in the cell as early as 6 hpi (31). Thus, there may be a balance as to how much VII remains associated with viral DNA during the early stages of infection: sufficient VII must be removed to decondense the nucleoprotein structure to allow access to RNA polymerase and transcription factors, but some VII remains to stimulate transcription. Once initiated, transcription elongation may function to strip off the last of the remaining VII from the viral DNA.
We detected the association of hdAd DNA with cellular histones as early as 2 to 4 hpi (Fig. 4), and the DNA exhibited an MNase digestion pattern similar to that of cellular chromatin as early as 6 hpi (Fig. 3). That hdAd DNA preferentially associated with the H3 variant H3.3 suggests that chromatinization occurred by a replication-independent mechanism. H3.3 continued to be the predominant H3 variant associated with hdAd until at least 24 hpi (data not shown). Recent work has shown that histones are found to be associated with wild-type Ad DNA as early as 1 hpi and that all members of the nucleosome, H2A/H2B, H3 and H4, are present (31). These data, and previous work by others (5, 13, 14, 66), strongly suggest that hdAd and Ad DNA are wrapped in nucleosomes within the infected cell nucleus. The observation that both VII and histones can be found bound to the same DNA molecule (31) suggests that the processes of core VII eviction and histone placement may be coincident. The cellular chaperones implicated in removing VII from the incoming viral DNA, TAFI-III also known as B23/nucleophosmin (45), pp32 (77), SET (37), and NAP-1 (30), have also been implicated in the removal of protamines from incoming sperm DNA and in the assembly of sperm DNA into chromatin (38, 52, 60). HIRA, which we showed is necessary for placement of H3.3 on vector DNA (Fig. 5), is also involved in the placement of histones on pronuclear DNA (34). The similarity between the packaging and chromatinization of sperm and Ad DNA raises the intriguing possibility that Ad has coopted this highly efficient cellular process and now utilizes it to its own ends. Since HIRA was also shown to deposit H3.3 on HSV DNA at early times of infection (53), this may be a common mechanism for initial placement of histones on the genomes of nuclear DNA viruses.
Chromatinization of hdAd DNA is necessary for efficient expression of vector-carried genes (Fig. 5), and this event very likely contributes to their significant stability and efficacy in vitro and in vivo (27, 47). One of the advantages of hdAd is its large cloning capacity, which permits the use of large upstream regulator sequences or even whole genomic loci to permit tissue-specific gene expression (51, 62, 63). Chromatin plays an important role in gene regulation (28, 68), and indeed, proper placement of nucleosomes relative to the transcription start site of a gene is crucial for promoter fidelity (29, 33). The ability to incorporate large regulatory regions into hdAd coupled with its swift assembly into physiologically spaced nucleosomes likely contributes to the maintenance of a faithful expression profile from these control elements when contained in hdAd.
Chromatinization of infecting DNA also provides a template that the cell can modulate to epigenetically downregulate expression of invading DNA (58). Many hdAd vectors contain noncoding “stuffer” DNA to maintain the size of the vector DNA within appropriate virion packaging limits (50). hdAd containing 22 kb of prokaryotic stuffer DNA (hdAd-prok) expressed its transgene 10-fold less efficiently than a similar vector containing eukaryotic stuffer DNA (hdAd-euk) both in vitro and in vivo (48). Recently, we determined that, although both hdAd genomes associated with cellular histones to a similar degree, hdAd-prok chromatin was actively deacetylated (58). Insertion of an insulator element between the transgene and bacterial DNA derepressed hdAd-prok, suggesting that foreign/prokaryotic DNA nucleates repressive chromatin structures that spread to the transgene. Thus, the cell can epigenetically modulate expression from the vector through changes in histone modification, leading to inhibition of gene expression from certain invading DNAs. Epigenetic silencing is not unique to Ad and has been observed for both integrated (54, 55) and episomal (11, 57, 72) vectors. The fact that hdAd containing eukaryotic-derived stuffer DNA escapes epigenetic shutdown likely reflects its strong resemblance to cellular DNA (e.g., identical GC and CpG dinucleotide contents, correct nucleosome placement and phasing, etc.), thus contributing to the persistence of expression of their carried transgenes.
Both E1-deleted Ad (Fig. 4) and wild-type Ad (31) associate with cellular histones very early in infection. Thus, our results for hdAd likely also hold for replication-competent Ad (i.e., preferential association with H3.3 being dependent on HIRA), at least during the early phase of infection—before DNA replication and expression of late genes and de novo synthesis of core VII from the viral genome. HSV type 1 (HSV-1) DNA also associates with H3.3 at early times of infection, which is also dependent on HIRA, and inhibition of H3.3 deposition reduces mRNA expression (53). However, recent studies suggested that while HSV genomic DNA in latent infection is assembled into a regularly repeating nucleosome array, as we observed here with hdAd, during lytic infection, the nucleosomes on HSV DNA are “unstable” and form fragments of heterogeneous sizes upon MNase digestion (32). Whether replicating Ad assembles into stable or unstable chromatin remains to be determined. Electron microscopy analysis of viral genomes isolated late during infection (16 to 18 hpi) showed irregularly spaced nucleosome-like particles at about 1/10 the density of HeLa DNA (3 versus 26 per μm of DNA, respectively [5]). However, it is not clear whether this is due to “unstable” chromatin or the limited quantities of histones that are available late in Ad infection. Nonetheless, swift assembly of Ad DNA into chromatin may represent an important step in the early phase of infection that “kick-starts” the virus life cycle and is a common feature of other DNA viruses that replicate in the nucleus.
In summary, we have shown that hdAd DNA is wrapped in physiologically spaced nucleosomes soon after infection and that efficient chromatinization of the hdAd DNA template is crucial for efficient expression of vector-carried genes. Our study has elucidated an important event that contributes to the efficacy of Ad-based vectors.
ACKNOWLEDGMENTS
We thank Robert Meulenbroek, Karen Powell, Catherine Barrett, and Adam Smith for excellent technical assistance and Jeffrey Dilworth, David Picketts, Michael Rudnicki, and Rashmi Kothary for critical evaluation of the manuscript and helpful discussions. We thank Valerie Wallace, Jeffrey Dilworth, Marjorie Brand, Jane Flint, Philip Ng, and Peter Adams for supplying reagents.
This work was supported by grants from the Canadian Institutes of Health Research, Muscular Dystrophy Canada, the Amyotrophic Lateral Sclerosis Society of Canada, the Jesse Davidson Foundation for Gene and Cell Therapy, and the Cancer Research Society (Canada). P.J.R. was supported by a Postgraduate Scholarship and a Canada Graduate Scholarship from the Natural Science and Engineering Research Council of Canada.
Footnotes
Published ahead of print on 9 February 2011.
REFERENCES
- 1. Addison C. L., Hitt M., Kunsken D., Graham F. L. 1997. Comparison of the human versus murine cytomegalovirus immediate early gene promoters for transgene expression by adenoviral vectors. J. Gen. Virol. 78:1653–1661 [DOI] [PubMed] [Google Scholar]
- 2. Ahmad K., Henikoff S. 2002. The histone variant H3.3 marks active chromatin by replication-independent nucleosome assembly. Mol. Cell 9:1191–1200 [DOI] [PubMed] [Google Scholar]
- 3. Amalfitano A., Parks R. J. 2002. Separating fact from fiction: assessing the potential of modified adenovirus vectors for use in human gene therapy. Curr. Gene Ther. 2:111–133 [DOI] [PubMed] [Google Scholar]
- 4. Berk A. J. 2007. Adenoviridae: the viruses and their replication, p. 2355–2394 In Knipe D. M., Howley P. M. (ed.), Fields virology, 5th ed. Lippincott Williams & Wilkins, Philadelphia, PA [Google Scholar]
- 5. Beyer A. L., Bouton A. H., Hodge L. D., Miller O. L., Jr 1981. Visualization of the major late R strand transcription unit of adenovirus serotype 2. J. Mol. Biol. 147:269–295 [DOI] [PubMed] [Google Scholar]
- 6. Bramson J. L., et al. 2004. Helper-dependent adenoviral vectors containing modified fibre for improved transduction of developing and mature muscle cells. Hum. Gene Ther. 15:179–188 [DOI] [PubMed] [Google Scholar]
- 7. Cereghini S., Yaniv M. 1984. Assembly of transfected DNA into chromatin: structural changes in the origin-promoter-enhancer region upon replication. EMBO J. 3:1243–1253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Challberg M. D., Kelly T. J., Jr 1979. Adenovirus DNA replication in vitro. Proc. Natl. Acad. Sci. U. S. A. 76:655–659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chatterjee P. K., Vayda M. E., Flint S. J. 1986. Identification of proteins and protein domains that contact DNA within adenovirus nucleoprotein cores by ultraviolet light crosslinking of oligonucleotides 32P-labelled in vivo. J. Mol. Biol. 188:23–37 [DOI] [PubMed] [Google Scholar]
- 10. Chen J., Morral N., Engel D. A. 2007. Transcription releases protein VII from adenovirus chromatin. Virology 369:411–422 [DOI] [PubMed] [Google Scholar]
- 11. Chen Z. Y., He C. Y., Meuse L., Kay M. A. 2004. Silencing of episomal transgene expression by plasmid bacterial DNA elements in vivo. Gene Ther. 11:856–864 [DOI] [PubMed] [Google Scholar]
- 12. Christou C., Parks R. J. 2011. Rational design of murine secreted alkaline phosphatase for enhanced performance as a reporter gene in mouse gene therapy preclinical trials. Hum. Gene Ther. [Epub ahead of print.] doi:10.1089/HUM-2010-171 [DOI] [PubMed] [Google Scholar]
- 13. Daniell E., Groff D. E., Fedor M. J. 1981. Adenovirus chromatin structure at different stages of infection. Mol. Cell. Biol. 1:1094–1105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Dery C. V., et al. 1985. The structure of adenovirus chromatin in infected cells. J. Gen. Virol. 66(Pt. 12):2671–2684 [DOI] [PubMed] [Google Scholar]
- 15. Drane P., Ouararhni K., Depaux A., Shuaib M., Hamiche A. 2010. The death-associated protein DAXX is a novel histone chaperone involved in the replication-independent deposition of H3.3. Genes Dev. 24:1253–1265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Ehrhardt A., Xu H., Kay M. A. 2003. Episomal persistence of recombinant adenoviral vector genomes during the cell cycle in vivo. J. Virol. 77:7689–7695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Fax P., Lehmkuhler O., Kuhn C., Esche H., Brockmann D. 2000. E1A12S-mediated activation of the adenovirus type 12 E2 promoter depends on the histone acetyltransferase activity of p300/CBP. J. Biol. Chem. 275:40554–40560 [DOI] [PubMed] [Google Scholar]
- 18. Ferrari R., et al. 2008. Epigenetic reprogramming by adenovirus e1a. Science 321:1086–1088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ghosh M. K., Harter M. L. 2003. A viral mechanism for remodeling chromatin structure in G0 cells. Mol. Cell 12:255–260 [DOI] [PubMed] [Google Scholar]
- 20. Goldberg A. D., et al. 2010. Distinct factors control histone variant H3.3 localization at specific genomic regions. Cell 140:678–691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Graham F. L. 1987. Growth of 293 cells in suspension culture. J. Gen. Virol. 68:937–940 [DOI] [PubMed] [Google Scholar]
- 22. Graham F. L., Smiley J., Russell W. C., Nairn R. 1977. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 36:59–74 [DOI] [PubMed] [Google Scholar]
- 23. Hall C., et al. 2001. HIRA, the human homologue of yeast Hir1p and Hir2p, is a novel cyclin-cdk2 substrate whose expression blocks S-phase progression. Mol. Cell. Biol. 21:1854–1865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Haruki H., Okuwaki M., Miyagishi M., Taira K., Nagata K. 2006. Involvement of template-activating factor I/SET in transcription of adenovirus early genes as a positive-acting factor. J. Virol. 80:794–801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Horwitz G. A., et al. 2008. Adenovirus small e1a alters global patterns of histone modification. Science 321:1084–1085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hubberstey A. V., Pavliv M., Parks R. J. 2002. Cancer therapy utilizing an adenoviral vector expressing only E1A. Cancer Gene Ther. 9:321–329 [DOI] [PubMed] [Google Scholar]
- 27. Jager L., Ehrhardt A. 2009. Persistence of high-capacity adenoviral vectors as replication-defective monomeric genomes in vitro and in murine liver. Hum. Gene Ther. 20:883–896 [DOI] [PubMed] [Google Scholar]
- 28. Jenuwein T., Allis C. D. 2001. Translating the histone code. Science 293:1074–1080 [DOI] [PubMed] [Google Scholar]
- 29. Jiang C., Pugh B. F. 2009. Nucleosome positioning and gene regulation: advances through genomics. Nat. Rev. Genet. 10:161–172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kawase H., et al. 1996. NAP-I is a functional homologue of TAF-I that is required for replication and transcription of the adenovirus genome in a chromatin-like structure. Genes Cells 1:1045–1056 [DOI] [PubMed] [Google Scholar]
- 31. Komatsu T., Haruki H., Nagata K. 2011. Cellular and viral chromatin proteins are positive factors in the regulation of adenovirus gene expression. Nucleic Acids Res. 39:889–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Lacasse J. J., Schang L. M. 2010. During lytic infections, herpes simplex virus type 1 DNA is in complexes with the properties of unstable nucleosomes. J. Virol. 84:1920–1933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lomvardas S., Thanos D. 2002. Modifying gene expression programs by altering core promoter chromatin architecture. Cell 110:261–271 [DOI] [PubMed] [Google Scholar]
- 34. Loppin B., et al. 2005. The histone H3.3 chaperone HIRA is essential for chromatin assembly in the male pronucleus. Nature 437:1386–1390 [DOI] [PubMed] [Google Scholar]
- 35. Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. 1997. Crystal structure of the nucleosome core particle at 2.8 A resolution. Nature 389:251–260 [DOI] [PubMed] [Google Scholar]
- 36. Maelandsmo G. M., et al. 2005. Use of a murine secreted alkaline phosphatase as a non-immunogenic reporter gene in mice. J. Gene Med. 7:307–315 [DOI] [PubMed] [Google Scholar]
- 37. Matsumoto K., Nagata K., Ui M., Hanaoka F. 1993. Template activating factor I, a novel host factor required to stimulate the adenovirus core DNA replication. J. Biol. Chem. 268:10582–10587 [PubMed] [Google Scholar]
- 38. McLay D. W., Clarke H. J. 2003. Remodelling the paternal chromatin at fertilization in mammals. Reproduction 125:625–633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mirza M. A., Weber J. 1982. Structure of adenovirus chromatin. Biochim. Biophys. Acta 696:76–86 [DOI] [PubMed] [Google Scholar]
- 40. Morral N., et al. 1998. High doses of a helper-dependent adenoviral vector yield supraphysiological levels of alpha1-antitrypsin with negligible toxicity. Hum. Gene Ther. 9:2709–2716 [DOI] [PubMed] [Google Scholar]
- 41. Muruve D. A., et al. 2004. Helper-dependent adenovirus vectors elicit intact innate but attenuated adaptive host immune responses in vivo. J. Virol. 78:5966–5972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mymryk J. S., Fryer C. J., Jung L. A., Archer T. K. 1997. Analysis of chromatin structure in vivo. Methods 12:105–114 [DOI] [PubMed] [Google Scholar]
- 43. Nelson J. E., Kay M. A. 1997. Persistence of recombinant adenovirus in vivo is not dependent on vector DNA replication. J. Virol. 71:8902–8907 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ng P., Parks R. J., Graham F. L. 2002. Preparation of helper-dependent adenoviral vectors. Methods Mol. Med. 69:371–388 [DOI] [PubMed] [Google Scholar]
- 45. Okuwaki M., Iwamatsu A., Tsujimoto M., Nagata K. 2001. Identification of nucleophosmin/B23, an acidic nucleolar protein, as a stimulatory factor for in vitro replication of adenovirus DNA complexed with viral basic core proteins. J. Mol. Biol. 311:41–55 [DOI] [PubMed] [Google Scholar]
- 46. Palmer D., Ng P. 2003. Improved system for helper-dependent adenoviral vector production. Mol. Ther. 8:846–852 [DOI] [PubMed] [Google Scholar]
- 47. Palmer D. J., Ng P. 2005. Helper-dependent adenoviral vectors for gene therapy. Hum. Gene Ther. 16:1–16 [DOI] [PubMed] [Google Scholar]
- 48. Parks R. J., Bramson J. L., Wan Y., Addison C. L., Graham F. L. 1999. Effects of stuffer DNA on transgene expression from helper-dependent adenovirus vectors. J. Virol. 73:8027–8034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Parks R. J., et al. 1996. A helper-dependent adenovirus vector system: removal of helper virus by Cre-mediated excision of the viral packaging signal. Proc. Natl. Acad. Sci. U. S. A. 93:13565–13570 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Parks R. J., Graham F. L. 1997. A helper-dependent system for adenovirus vector production helps define a lower limit for efficient DNA packaging. J. Virol. 71:3293–3298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Pastore L., et al. 1999. Use of a liver-specific promoter reduces immune response to the transgene in adenoviral vectors. Hum. Gene Ther. 10:1773–1781 [DOI] [PubMed] [Google Scholar]
- 52. Philpott A., Leno G. H. 1992. Nucleoplasmin remodels sperm chromatin in Xenopus egg extracts. Cell 69:759–767 [DOI] [PubMed] [Google Scholar]
- 53. Placek B. J., et al. 2009. The histone variant H3.3 regulates gene expression during lytic infection with herpes simplex virus type 1. J. Virol. 83:1416–1421 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Poleshko A., et al. 2010. Identification of a functional network of human epigenetic silencing factors. J. Biol. Chem. 285:422–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Poleshko A., et al. 2008. Identification of cellular proteins that maintain retroviral epigenetic silencing: evidence for an antiviral response. J. Virol. 82:2313–2323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Poulin K. L., et al. 2010. Retargeting of adenovirus vectors through genetic fusion of a single-chain or single-domain antibody to capsid protein IX. J. Virol. 84:10074–10086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Riu E., Grimm D., Huang Z., Kay M. A. 2005. Increased maintenance and persistence of transgenes by excision of expression cassettes from plasmid sequences in vivo. Hum. Gene Ther. 16:558–570 [DOI] [PubMed] [Google Scholar]
- 58. Ross P. J., Kennedy M. A., Parks R. J. 2009. Host cell detection of noncoding stuffer DNA contained in helper-dependent adenovirus vectors leads to epigenetic repression of transgene expression. J. Virol. 83:8409–8417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Ross P. J., Parks R. J. 2009. Construction and characterization of adenovirus vectors. Cold Spring Harb. Protoc. 2009:pdb.prot5011 [DOI] [PubMed] [Google Scholar]
- 60. Ruiz-Lara S. A., Cornudella L., Rodriguez-Campos A. 1996. Dissociation of protamine-DNA complexes by Xenopus nucleoplasmin and minichromosome assembly in vitro. Eur. J. Biochem. 240:186–194 [DOI] [PubMed] [Google Scholar]
- 61. Russell W. C. 2009. Adenoviruses: update on structure and function. J. Gen. Virol. 90:1–20 [DOI] [PubMed] [Google Scholar]
- 62. Schiedner G., et al. 2002. Variables affecting in vivo performance of high-capacity adenovirus vectors. J. Virol. 76:1600–1609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Schiedner G., et al. 1998. Genomic DNA transfer with a high-capacity adenovirus vector results in improved in vivo gene expression and decreased toxicity. Nat. Genet. 18:180–183 [DOI] [PubMed] [Google Scholar]
- 64. Schreiner S., et al. 2010. Proteasome-dependent degradation of Daxx by the viral E1B-55K protein in human adenovirus-infected cells. J. Virol. 84:7029–7038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Schwartz B. E., Ahmad K. 2005. Transcriptional activation triggers deposition and removal of the histone variant H3.3. Genes Dev. 19:804–814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Sergeant A., Tigges M. A., Raskas H. J. 1979. Nucleosome-like structural subunits of intranuclear parental adenovirus type 2 DNA. J. Virol. 29:888–898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Smith A. C., Poulin K. L., Parks R. J. 2009. DNA genome size affects the stability of the adenovirus virion. J. Virol. 83:2025–2028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Smith C. L., Hager G. L. 1997. Transcriptional regulation of mammalian genes in vivo. A tale of two templates. J. Biol. Chem. 272:27493–27496 [DOI] [PubMed] [Google Scholar]
- 69. Southern E. M. 1975. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J. Mol. Biol. 98:503–517 [DOI] [PubMed] [Google Scholar]
- 70. Spector D. J., Johnson J. S., Baird N. L., Engel D. A. 2003. Adenovirus type 5 DNA-protein complexes from formaldehyde cross-linked cells early after infection. Virology 312:204–212 [DOI] [PubMed] [Google Scholar]
- 71. Sung M. T., Cao T. M., Coleman R. T., Budelier K. A. 1983. Gene and protein sequences of adenovirus protein VII, a hybrid basic chromosomal protein. Proc. Natl. Acad. Sci. U. S. A. 80:2902–2906 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Suzuki M., Kasai K., Saeki Y. 2006. Plasmid DNA sequences present in conventional herpes simplex virus amplicon vectors cause rapid transgene silencing by forming inactive chromatin. J. Virol. 80:3293–3300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Tagami H., Ray-Gallet D., Almouzni G., Nakatani Y. 2004. Histone H3.1 and H3.3 complexes mediate nucleosome assembly pathways dependent or independent of DNA synthesis. Cell 116:51–61 [DOI] [PubMed] [Google Scholar]
- 74. Trotman L. C., Mosberger N., Fornerod M., Stidwill R. P., Greber U. F. 2001. Import of adenovirus DNA involves the nuclear pore complex receptor CAN/Nup214 and histone H1. Nat. Cell Biol. 3:1092–1100 [DOI] [PubMed] [Google Scholar]
- 75. Wong L. H., et al. 2010. ATRX interacts with H3.3 in maintaining telomere structural integrity in pluripotent embryonic stem cells. Genome Res. 20:351–360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Wong M. L., Hsu M. T. 1988. Psoralen-cross-linking study of the organization of intracellular adenovirus nucleoprotein complexes. J. Virol. 62:1227–1234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Xue Y., Johnson J. S., Ornelles D. A., Lieberman J., Engel D. A. 2005. Adenovirus protein VII functions throughout early phase and interacts with cellular proteins SET and pp32. J. Virol. 79:2474–2483 [DOI] [PMC free article] [PubMed] [Google Scholar]